Abstract
There are more than 2,500 known Salmonella serovars, and some of these can be further subclassified into groups of strains that differ profoundly in their gene content. We refer to these groups of strains as “genovars.” A compilation of comparative genomic hybridization data on 291 Salmonella isolates, including 250 S. enterica subspecies I strains from 32 serovars (52 genovars), was used to select a panel of 384 genes whose presence and absence among serovars and genovars was of potential taxonomic value. A subset of 146 genes was used for real-time PCR to successfully identify 12 serovars (16 genovars) in 24 S. enterica strains. A further subset of 64 genes was used to identify 8 serovars (9 genovars) in 12 multiplex PCR mixes on 11 S. enterica strains. These gene panels distinguish all tested S. enterica subspecies I serovars and their known genovars, almost all by two or more informative markers. Thus, a typing methodology based on these predictive genes would generally alert users if there is an error, an unexpected polymorphism, or a potential new genovar.
The classification of infectious bacteria below the subspecies level is often important for clinical or epidemiological investigations. In general, classification involves one or more measurements, each with its own associated error rate due to technical or biological variation. These errors occasionally lead to incorrect or inadequate classification of clinical strains. Here we present steps toward a PCR-based typing system for Salmonella enterica in which possible errors in typing can be recognized and do not result in incorrect strain classification.
Different members of the bacterial species Salmonella enterica cause a surprising variety of diseases in both human and animal hosts. These range from asymptomatic persistence through gastric infections to potentially fatal systemic disease (including typhoid fever). Different salmonellae have diverse host ranges—some isolates are able to infect a broad range of animal and plant hosts, while others are very host specific. In the United States, there are an estimated 1.4 million annual cases of human nontyphoid salmonellosis (39), with 400 deaths. The death toll is much higher in third-world countries, where typhoid fever is a major killer due to poor sanitary conditions.
S. enterica is subdivided into six subspecies: enterica (I), salamae (II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV), and indica (VI). Of these, subspecies I is responsible for the overwhelming majority of human and domestic animal infections. Based on the bacterium's O (surface polysaccharide) and H (flagellar) antigens, S. enterica is classified into more than 2,500 serovars, approximately 60% of which have been identified from subspecies I isolates (23, 24). Typing of S. enterica based on these antigens is the current gold standard of Salmonella identification in U.S. state health department laboratories and worldwide and employs more than 150 O and H antigens.
Comparative genomic hybridization (CGH) assays on whole-genome microarrays have shown that genomic differences generally correlated well with serotype assignments, with a few notable exceptions (25). Some serovars can be further subclassified into genovars based on substantial differences in genetic content. Conversely, two or more distinct serovars can have almost identical genovars. Some genovars group into clades with entirely different serogroups, indicating that genes determining serotypes may be laterally transferred into different genovars (25).
A number of methods have been developed to complement serology. These include pulsed-field gel electrophoresis (14, 33, 35), multilocus enzyme electrophoresis (34), variable-number-of-tandem-repeat analysis (2, 37), and multilocus sequence typing (6, 36). In addition, several PCR-based methods were investigated, targeting genes specific to some or all salmonellae (7, 9, 10, 17, 19, 38). Recent approaches using multiplex PCR are based on variable numbers of probes, ranging from 7 to 12 (13, 17).
Salmonella oligonucleotide microarrays have been designed to assay variable genes and antibiotic resistance markers within Salmonella (20, 21). Some of these focused entirely on sequence variations within the O and H antigens (40). A liquid-microsphere suspension array-based protocol was also developed, where seven specific probes distinguished the six most common O serogroups in the United States (8). However, the genes encoding serologically relevant antigens have been shown to be extremely variable and highly prone to recombination (18, 32). It would therefore be valuable to develop a molecular identification scheme that included both serotype and genetic background to complement serology.
PCR, oligonucleotide array, and microsphere array protocols usually rely on exact matches of oligonucleotide sequences. Thus, point mutations can result in misassignments of strains. The variation in sequence among strains in the same serovar will remain largely unknown, but it is certain that some strains will deviate from the known sequence at some of the sites inspected. In order to circumvent this issue, a scheme that involves error detection (11) is highly desirable. This scheme would not rely on a single character state to distinguish any two different serovars and would avoid misinterpretation due to a single aberrant data point. We have explored this possibility using the currently available Salmonella genome sequences and CGH data from 291 Salmonella strains, generated in our lab. We have compiled gene selections capable of distinguishing all 32 S. enterica subspecies I serovars (52 genovars) investigated, in both uniplex real-time PCR and multiplex PCR assays. These selections included sufficient information to detect technical errors, unexpected polymorphisms, and novel strains. They encompassed two or more gene differences in 99.5% of pairwise comparisons between these genovars, and point mutations would therefore not lead to misclassification of a strain.
MATERIALS AND METHODS
Gene selection.
In order to maximize predictive values of marker genes, these genes needed to differ substantially from each other in their distribution among the different Salmonella taxa. Therefore, the “genetic distance” between genes was determined using the CGH data of 291 Salmonella strains. Each of the genes was treated as if it was a taxon, and the gene presence/absence prediction in each strain was treated as a character; in essence, the data matrix was rotated. “Uncertain” gene predictions were treated as missing data. Next, a genetic distance tree was constructed using the PAUP (phylogenetic analysis using parsimony) software program (Sinauer Associates, Sunderland, MA). This “tree” effectively clustered genes that have similar taxonomic distributions. Subsequently, 384 genes were manually selected based on coverage of widely distributed and deep branches of the “tree,” avoiding genes that clustered together. This selection captured most of the variation in taxonomic distribution observed in the CGH data but was just 1 of billions of possible combinations of 384 poorly clustering genes.
Strains and primers.
A list of all S. enterica subspecies I strains employed for CGH experiments is presented in Table 1. Using subsets of the selected genes, 24 S. enterica isolates were analyzed by real-time PCR and 11 were analyzed by multiplex PCR. All strains were grown to stationary phase under standard conditions, in Luria-Bertani broth at 37°C with shaking at 180 rpm. Bacterial genomic DNA was prepared using the Sigma GenElute kit according to the manufacturer's recommendations. Table S1 in the supplemental material shows the primers used successfully for real-time PCR and in the multiplex PCR assays.
TABLE 1.
Salmonella enterica subspecies I strains used in comparative genomic hybridizations, real-time PCR, and multiplex assays
| Genovar no.a | Genovar | Serovar | No. of strains used
|
||
|---|---|---|---|---|---|
| CGH | real-time PCR | Multiplex PCR | |||
| 1 | Abortusovis 1 | Abortusovis | 6 | ||
| 2 | Abortusovis 2 | Abortusovis | 1 | ||
| 3 | Agona 1 | Agona | 3 | ||
| 4 | Bovismorbificans 1 | Bovismorbificans | 2 | ||
| 5 | California 1 | California | 2 | ||
| 6 | Choleraesuis 1 | Choleraesuis | 5 | ||
| 7 | Cubana 1 | Cubana | 1 | ||
| 8 | Derby 1 | Derby | 1 | ||
| 9 | Dublin 1 | Dublin | 10 | 1 | 1 |
| 10 | Dublin 2 | Dublin | 1 | 1 | |
| 11 | Enteritidis 1 | Enteritidis | 31 | 2 | 2 |
| 12 | Enteritidis 2 | Enteritidis | 1 | 1 | |
| 13 | Gallinarum 1 | Gallinarum (including biovar Pullorum) | 8 | 1 | |
| 14 | Gallinarum 2 | Gallinarum (including biovar Pullorum) | 1 | ||
| 15 | Gallinarum 3 | Gallinarum (including biovar Pullorum) | 1 | ||
| 16 | Hadar 1 | Hadar | 3 | ||
| 17 | Hadar 2 | Hadar | 1 | ||
| 18 | Heidelberg 1 | Heidelberg | 5 | ||
| 19 | Heidelberg 2 | Heidelberg | 1 | ||
| 20 | Infantis 1 | Infantis | 3 | 1 | |
| 21 | Infantis 2 | Infantis | 1 | 1 | |
| 22 | Javiana 1 | Javiana | 1 | ||
| 23 | Kentucky 1 | Kentucky | 1 | ||
| 24 | Miami 1 | Miami | 2 | 1 | 1 |
| 25 | Miami 2 | Miami | 1 | ||
| 26 | Montevideo 1 | Montevideo | 4 | ||
| 27 | Montevideo 2 | Montevideo | 1 | ||
| 28 | Muenchen 1 | Muenchen | 3 | 1 | |
| 29 | Muenchen 2 | Muenchen | 1 | 1 | |
| 30 | Muenster 1 | Muenster | 1 | ||
| 31 | Muenster 2 | Muenster | 1 | ||
| 32 | Newport 1 | Newport | 9 | ||
| 33 | Newport 2 | Newport | 1 | ||
| 34 | Newport 3 | Newport | 1 | ||
| 35 | Oranienburg 1 | Oranienburg | 2 | ||
| 36 | Paratyphi A 1 | Paratyphi A | 13 | 2 | 1 |
| 37 | Paratyphi B 1 | Paratyphi B (including biovar tartrate + [Java]) | 7 | 2 | 1 |
| 38 | Paratyphi B 2 | Paratyphi B (including biovar tartrate + [Java]) | 1 | 1 | |
| 39 | Paratyphi B 3 | Paratyphi B (including biovar tartrate + [Java]) | 3 | ||
| 40 | Paratyphi C 1 | Paratyphi C | 2 | 1 | 1 |
| 41 | Paratyphi C 2 | Paratyphi C | 1 | ||
| 42 | Reading 1 | Reading | 1 | ||
| 43 | Saint Paul 1 | Saint Paul | 7 | ||
| 44 | Saint Paul 2 | Saint Paul | 1 | ||
| 45 | Sendai 1 | Sendai | 2 | ||
| 46 | Sendai 2 | Sendai | 1 | ||
| 47 | Sendai 3 | Sendai | 1 | ||
| 48 | Senftenberg 1 | Senftenberg | 2 | ||
| 49 | Thompson 1 | Thompson | 2 | ||
| 50 | Typhi 1 | Typhi | 21 | 5 | 2 |
| 51 | Typhimurium 1 | Typhimurium | 67 | 2 | 2 |
| 52 | Typhisuis 1 | Typhisuis | 1 | ||
Numbers correspond to those in Fig. 1.
Definition of genovars.
The term genovar (as opposed to serovar) was coined (25) to characterize isolates that belong to a certain serovar but have a markedly different genetic repertoire from other isolates in that same serovar. A “genovar” is therefore merely a practical division indicating substantial genetic difference. Here we use the term “genovar” for isolates of the same serovars that differ in genetic content by more than 90 nonphage serovar Typhimurium LT2 chromosomal genes. This definition led to the classification of 52 genovars in the 32 S. enterica subspecies I serovars investigated here.
Real-time PCR.
Real-time PCR was performed using the ABI Prism 7900 sequence detection system (Applied Biosystems). PCRs contained 10-μl total volumes dispensed into each of the 384 wells of a thin-walled microAmp optical plate (Applied Biosystems). Reactions contained 5 ng of genomic DNA, 0.2 mM deoxynucleoside triphosphates, 0.25 U of Promega Taq polymerase, 4 mM MgCl2, 0.35 μM 6-carboxyl-X-rhodamine (ROX), and 0.4× (final concentration) Sybr green (Invitrogen, Carlsbad, CA). PCR was performed at 95°C for 3 min and 40 cycles of 20 s at 95°C, 20 s at 57°C, and 2 min at 72°C. A dissociation step of 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C finished the procedure. All reactions were carried out in triplicate on different days. The real-time PCR data were analyzed using the ABI Prism 7000 SDS v2.1 data analysis software program (Applied Biosystems). Median threshold cycle (CT) values were used for absence/presence predictions (see below).
Multiplex PCR.
A modification of the Primer3 software program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi/%20primer3_www.cgi) was used to select groups of compatible primer pairs to amplify up to 25 genes in the same PCR. PCR products were required to be 50 to 600 bases in length. The 3′ ends of the primers were not allowed to represent the third position of a codon to reduce the chances of polymorphisms in different strains, possibly causing PCR failure. A complete match of the five bases on the 3′ end of any primer was not allowed in any other PCR product in the multiplex. PCR products in one multiplex differed by at least 3% (and at least 4 bp) from each other in length to ensure resolution on electrophoresis platforms. Using these criteria, all 146 candidate genes were covered in fewer than 30 multiplexes of up to 25 genes. Twelve subsets of eight genes were selected from these lists to be tested.
Each 10-μl reaction mixture consisted of 10 ng of genomic DNA, 0.25 μM primers, appropriate concentrations of 16S control primers (varying from 0.001 μM to 0.1 μM), 0.05 U Takara Ex Taq polymerase, and 0.25 mM deoxynucleoside triphosphates in 1× Ex Taq buffer (Takara). PCRs were carried out using 95°C for 3 min, followed by 35 cycles of 15 s at 95°C, 15 s at 55°C, and 30 s at 72°C, and finished with a 7-min incubation at 72°C. Products were subsequently resolved on a 3.3% agarose gel. Sixty-four of the 96 amplified PCR products were successfully resolved under these standard electrophoresis conditions and subsequently used for scoring as outlined below.
Absence/presence scoring.
CGH assays on custom-made whole-genome PCR product arrays were performed as previously described (26). Predictions of gene presence/absence were calculated using generic cutoff values: if a gene that was present in the control strain exhibited a normalized signal ratio R(query strain/control strain) of >0.67, the gene was called present (“2”). If the signal ratio was an R(query strain/control strain) value of <0.33, the gene's status was defined as absent (“0”); and if the ratio was between these two thresholds, the status was classified as uncertain (“1”). Note that these predictions are not perfectly accurate. Hybridizations of sequenced Salmonella genomes to the array platforms confirmed an error rate of approximately 1% (data not shown) (26).
In real-time PCR predictions, four ubiquitously present genes were used as positive controls: STM2072 (hisD), STM0736 (sucA), STM2384 (aroC), and STM3837 (dnaN). PCR products of these controls were generally detected to be logarithmically amplified at control CT (CTC) cycle numbers of ≤18. After careful examination of final PCR products on agarose gels, thresholds for successful real-time PCRs were set as follows: a product was considered present (“2”) when for at least three of four positive controls the following conditions were fulfilled: CTP ≤ CTC + 8. An intermediate status (“1”) was defined for products detected under the following conditions: CTC + 8 < CT ≤ CTC + 12. Products were considered absent (“0”) when their CT values for detection fulfilled the following conditions: CT > CTC + 12. Agarose electrophoresis of the real-time PCR products for strain SARB64 (R09) confirmed that these predictions were incorrect for only 6 of the 146 absence/presence calls (i.e., less than 5%) (data not shown).
In multiplex PCR experiments, products were visually scored after ethidium bromide staining and scores of “2,” “1,” and “0” were assigned to strong, weak, and no-product bands of the appropriate size, respectively.
Genetic distances between the strains investigated by CGH, multiplex PCR, and real-time PCR were then estimated based on the encoded data. Genetic distances between data sets were computed using an R script (http://www.r-project.org/), giving true differences (absence versus presence predictions) double weight and all other distinctions (absent versus uncertain and uncertain versus present) single weight. The weighting was implemented to reflect the lower degree of confidence in the status predictions of “uncertain” genes.
RESULTS
CGH.
Data on the diversity of gene content among serovars of Salmonella come from two sources; first, the sequenced genomes, which are rapidly increasing in number; and second, CGH data. Table S2 in the supplemental material contains CGH data obtained in our laboratory from 291 Salmonella strains, including 250 S. enterica subspecies I isolates, representing 32 serovars. These data were obtained using an array of PCR products representing almost all serovar Typhimurium LT2 and serovar Typhi CT18 genes. Data for 178 strains of this matrix have previously been published (1, 3, 16, 22, 25-31). The isolates include all major serovars that cause infection in humans and domestic animals in the United States and worldwide.
Resolution of model genovar isolates by gene selections.
After parsimony analysis (using the PAUP software program) as described in Material and Methods, 384 open reading frames [ORFs] were picked to represent each “pseudoclade” of genes with similar taxonomic distribution, starting with the deepest branches and working toward the smaller branches of the phylogenetic tree, where genes had increasingly similar distributions. In addition, four control ORFs were selected based on representation in all sequenced and hybridized isolates. In this demonstration project, 384 genes were selected because of easy applicability to 384-well real-time PCR assays. These genes distinguish between all model isolates for each of the 52 genovars, and the number of differences between each pair of genovar model strains generally exceeds three. The model strains were those isolates with the best quality score of the hybridization within each genovar. The only such pairwise comparison that resulted in only two differences (genovars Hadar 1 and Heidelberg 1) may represent a recent transfer of gene clusters into the respective genetic background of the other serovar, a recent point mutation in these gene clusters, or a serotyping error.
The number of ORF status differences was calculated for CGH data using the different subsets of genes to be used later in this report (the 384 genes of the phylogenetic tree set, the 146 successful reporters in the real-time PCR set, the 64 resolved products of the multiplex set, and the 12-probe set used in reference 17). The elements of the four different gene panels discussed in this report are shown in Table 2.
TABLE 2.
Gene panels used for DNA-based typing of Salmonella enterica subspecies I isolates
| Panel in Fig. 1 | Description | ORFsa |
|---|---|---|
| D | Probe set used in reference 19 | STM0716, STY0346, STY2349, STM0839, STM1350, STM2150, STM3845, STM4525, STM4538, STY0311, STY0312, STY0347, STY2299, SEN1381+SEN1381-SEN1382 intergenic region |
| C | 64-gene multiplex PCR set | STM0059, STM0068, STM0177, STM0198, STM0287, STM0288, STM0327, STM0516, STM0610, STM0927, STM0947, STM1025, STM1029, STM1035, STM1045, STM1521, STM1534, STM1549, STM1579, STM1673, STM1869, STM1869A, STM2030, STM2033, STM2055, STM2094, STM2177, STM2438, STM2591, STM2601, STM2700, STM2904, STM3033, STM3034, STM3125, STM3358, STM3546, STM3652, STM3653, STM3671, STM3771, STM3859, STM4066, STM4206, STM4215, STM4218, STM4447, STM4483, STM4498, STM4528, PSLT040, PSLT060, PSLT107, STY1049, STY2077, STY2364, STY3088, STY3688, STY4412, STY4414, STY4706, STY4849, STM0716, STY0346; control 16SrRNA gene |
| B | 146-gene real-time PCR set | All ORFs of the 64-gene set plus the following ORFs: STM0056, STM0269, STM0279, STM0284, STM0295, STM0330, STM0335, STM0521, STM0895, STM0897, STM0904, STM0908, STM0917, STM0921, STM0929, STM1007, STM1024, STM1416, STM1547, STM1550, STM1551, STM1619, STM1867, STM1897, STM1998, STM2005, STM2018, STM2027, STM2134, STM2176, STM2225, STM2237, STM2452, STM2455, STM2456, STM2508, STM2610, STM2618, STM2630, STM2698, STM2709, STM2728, STM2733, STM2773, STM2901, STM2902, STM3170, STM3258, STM3666, STM3756, STM3767, STM3784, STM3906, STM3941, STM4031, STM4191, STM4197, STM4209, STM4317, STM4375, STM4381, STM4523, STM4592, STM4594, PSLT016, PSLT018, PSLT025, PSLT085, STY1605, STY2296, STY2350, STY2880, STY3087, STY3666, STY3670, STY4521, STY4650, STY4669; control genes STM0736, STM2072, STM2384 and STM3837 |
| A | 384-gene phylogenetic tree set | All ORFs of the 146-gene set except the 4 control genes plus the following ORFs: STM0024, STM0030, STM0032, STM0036, STM0042, STM0051, STM0055, STM0061, STM0063, STM0073, STM0077, STM0162, STM0163, STM0175, STM0197, STM0256, STM0257, STM0268, STM0271, STM0286, STM0292, STM0301, STM0303, STM0325, STM0333, STM0337, STM0341, STM0343, STM0345, STM0351, STM0353, STM0426, STM0427, STM0428, STM0429, STM0431, STM0511, STM0519, STM0526, STM0530, STM0531, STM0550, STM0557, STM0558, STM0572, STM0574, STM0611, STM0652, STM0656, STM0659, STM0722, STM0726, STM0762, STM0764, STM0770, STM0809, STM0857, STM0911, STM1002, STM1031, STM1055, STM1088, STM1096, STM1130, STM1188, STM1240, STM1284, STM1285, STM1352, STM1360, STM1380, STM1381, STM1525, STM1533, STM1537, STM1540, STM1543, STM1554, STM1557, STM1559, STM1561, STM1577, STM1582, STM1614, STM1615, STM1625, STM1630, STM1634, STM1637, STM1670, STM1675, STM1677, STM1741, STM1864, STM1868, STM2007, STM2008, STM2026, STM2050, STM2051, STM2052, STM2084, STM2085, STM2092, STM2099, STM2133, STM2135, STM2151, STM2152, STM2232, STM2240, STM2243, STM2274, STM2289, STM2290, STM2315, STM2377, STM2397, STM2421, STM2423, STM2453, STM2460, STM2514, STM2574, STM2575, STM2594, STM2644, STM2695, STM2705, STM2722, STM2740, STM2747, STM2753, STM2767, STM2816, STM2914, STM2917, STM2922, STM2941, STM2960, STM3024, STM3026, STM3028, STM3036, STM3082, STM3120, STM3253, STM3254, STM3256, STM3257, STM3260, STM3355, STM3356, STM3357, STM3518, STM3529, STM3548, STM3549, STM3599, STM3600, STM3620, STM3637, STM3638, STM3639, STM3658, STM3673, STM3678, STM3691, STM3697, STM3753, STM3770, STM3775, STM3780, STM3782, STM3822, STM3823, STM3826, STM3828, STM3831, STM3833, STM3834, STM3860, STM4012, STM4032, STM4046, STM4052, STM4067, STM4068, STM4072, STM4074, STM4075, STM4078, STM4102, STM4113, STM4115, STM4205, STM4261, STM4279, STM4306, STM4319, STM4373, STM4413, STM4420, STM4426, STM4445, STM4484, STM4486, STM4493, STM4499, STM4524, STM4527, STM4537, STM4573, STM4574, STM4589, STM4590, PSLT006, PSLT031, PSLT104, STY0314, STY0324, STY1016, STY1601, STY1630, STY2004, STY2026, STY2044, STY2069, STY2349, STY2419, STY2420, STY2629, STY2884, STY2887, STY3344, STY3674, STY3682, STY3684, STY3690, STY4756, STY4834, STY4853 |
STM, ORFs from S. enterica serovar Typhimurium LT2; PSLT, ORFs from LT2 virulence plasmid pSLT; STY, ORFs from S. enterica serovar Typhi CT18; SEN, ORFs from S. enterica serovar Enteritidis PT4.
For this calculation, representative isolates of all 52 subspecies I genovars present in our CGH data collection were compared with each other, and the number of differences is illustrated in Fig. 1. A difference was defined as a gene that was predicted to be present in one strain but predicted to be absent in the other. Uncertain gene status predictions did not contribute to the number of differences. All numbers from this analysis are depicted in Fig. S1 in the supplemental material. Table S3 in the supplemental material shows the same comparison for every S. enterica subspecies I strain that had been subjected to CGH (250 strains) for the 384 genes of the phylogenetic tree set.
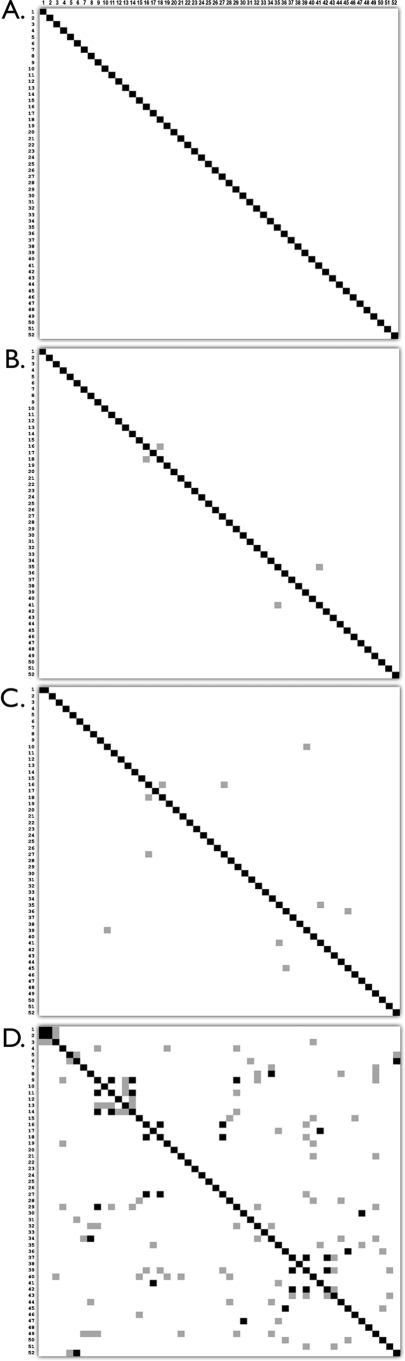
FIG. 1.
Analysis of genetic differences (whole ORFs) between 32 Salmonella serovars (52 genovars). Black, no difference; gray, 1 difference; white, ≥2 differences. (A) Three hundred eight-four genes selected initially for real-time PCR. (B) One hundred forty-six genes reporting consistently in real-time PCR assays. (C) Sixty-four multiplexed genes scored on agarose gels. (D) Thirteen S. enterica serovar Typhimurium and serovar Typhi genes used in reference 17. Genovar numbers are shown on the top and the left side of each panel and correspond to column 1 of Table 1. Absence/presence data were obtained from microarray-based CGH analyses, as described in Materials and Methods. The number of differences in gene predictions (absence versus presence) was calculated in pairwise comparisons between model strains for each genovar. These model strains were those isolates with the best quality score of the hybridization within each genovar. The raw numbers of differences are shown in Fig. S1 in the supplemental material. Note: reference 17 used an additional region specific to S. enterica serovar Enteritidis that was not sampled in our CGH experiments and therefore could not be included in this analysis.
Unlike the few probes used in reference 17, our gene sets distinguish almost all model isolates for the different genovars of our database by more than one character state (Fig. 1). In our lowest gene selection pool, the 64 genes used to evaluate the multiplex PCR assays, all 52 subspecies I genovars were distinguished, and of these, all but 5 of the 1,326 pairwise genovar comparisons could be resolved by more than 1 gene prediction. The five similar genovars are as follows: a, Hadar 1 and Heidelberg 1; b, Paratyphi C 2 and Oranienburg 1; c, Hadar 1 and Montevideo 2; d, Dublin 2 and Paratyphi B 3; and e, Sendai 1 and Paratyphi A 1. In comparison, the genes used by Kim et al. (17), excluding the one serovar Enteritidis PT4 region that was not interrogated in our CGH data, resulted in 16 unresolved pairwise genovar comparisons (no difference) and 49 such comparisons that were distinguished by only one character state.
Real-time PCR.
The PCR product for each gene on the array used for CGH is generally derived from a primer positioned at the beginning of the gene and another primer positioned at the end of the gene. These primer pairs were screened in proof-of-principle real-time assays against LT2 and CT18 DNA to select primer pairs with the best PCR amplification characteristics. One hundred forty-six of these primer pairs resulted in correct products when tested against CT18 and LT2 and were selected to interrogate 24 Salmonella genomes (R01 to R24 [Table 3]), representing 12 serovars. Based on CT values (the number of amplification cycles needed to detect the gene), genes were binned into “present”, “absent,” and “intermediate” (see Materials and Methods). Table S2 in the supplemental material contains these data. Subsequently, relationships (genetic distances) of these test strains to strains in the CGH database were calculated.
TABLE 3.
Top matches from the CGH database in correlation analysis of real-time and multiplex PCR assays of Salmonella strainsa
| Strain name | CGH strain no. | Genovar | Real-time PCR result
|
Multiplex PCR result
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-time-PCR strain no. | Best hit in CGH database | Corr coeff | Detection of correct genovar | Multi-plex strain no. | Best hit in CGH database | Corr coeff | Detection of correct genovar | |||
| SARB14 | C024 | Dublin 2 | R01 | C024/Dublin 2 | 0.81 | Yes | ||||
| SARB26 | C085 | Infantis 1 | R02 | C085/Infantis 1 | 0.84 | Yes | ||||
| SARB27 | C086 | Infantis 2 | R03 | C086/Infantis 2 | 0.78 | Yes | ||||
| SARB47 | C138 | Paratyphi B 2 | R04 | C138/Paratyphi B 2 | 0.85 | Yes | ||||
| SARB32 | C101 | Muenchen 1 | R05 | C101/Muenchen 1 | 0.81 | Yes | ||||
| SARB34 | C103 | Muenchen 2 | R06 | C103/Muenchen 2 | 0.85 | Yes | ||||
| SL1344 | C225 | Typhimurium 1 | R07 | C204/Typhimurium 1 | 0.92 | Yes | S01 | C162/Typhimurium 1 | 0.94 | Yes |
| CT18 | Typhi 1 | R08 | C243/Typhi 1 | 0.84 | Yes | S07 | C243/Typhi 1 | 0.88 | Yes | |
| Ty2 | Typhi 1 | R09 | C243/Typhi 1 | 0.83 | Yes | S08 | C245/Typhi 1 | 0.88 | Yes | |
| SARB64 | C243 | Typhi 1 | R10 | C243/Typhi 1 | 0.83 | Yes | ||||
| CDC1707 | C233 | Typhi 1 | R11 | C243/Typhi 1 | 0.83 | Yes | ||||
| TYT1669 | C250 | Typhi 1 | R12 | C244/Typhi 1 | 0.79 | Yes | ||||
| PT4 | Enteritidis 1 | R13 | C040/Enteritidis 1 | 0.89 | Yes | S03 | C034/Enteritidis 1 | 0.91 | Yes | |
| 49-2778 | C052 | Enteritidis 1 | R14 | C064/Enteritidis 1 | 0.86 | Yes | S04 | C034/Enteritidis 1 | 0.91 | Yes |
| SARB17 | C061 | Enteritidis 2 | R15 | C061/Enteritidis 2 | 0.83 | Yes | ||||
| 02-0341 | C093 | Miami 1 | R16 | C093/Miami 1 | 0.87 | Yes | S05 | C093/Miami 1 | 0.80 | Yes |
| SARB12 | C022 | Dublin 1 | R17 | C022/Dublin 1 | 0.87 | Yes | S06 | C027/Dublin 1 | 0.85 | Yes |
| SARA52 | Paratyphi B 1 | R18 | C134/Paratyphi B 1 | 0.86 | Yes | |||||
| SARB42 | C118 | Paratyphi A 1 | R19 | C119/Paratyphi A 1 | 0.88 | Yes | S09 | C119/Paratyphi A 1 | 0.90 | Yes |
| SGSC2276 | C119 | Paratyphi A1 | R20 | C119/Paratyphi A 1 | 0.88 | Yes | ||||
| SARB43 | C136 | Paratyphi B 1 | R21 | C134/Paratyphi B 1 | 0.89 | Yes | S10 | C134/Paratyphi B 1 | 0.77 | Yes |
| SARB48 | C142 | Paratyphi C 1 | R22 | C143/Paratyphi C 1 | 0.85 | Yes | S11 | C229/Typhisuis 1 | 0.70 | Yesb |
| CDC2606 | C220 | Typhimurium 1 | R23 | C167/Typhimurium 1 | 0.92 | Yes | C142/Paratyphi C 1 | 0.70 | ||
| 281/91 | C065 | Gallinarum 1 | R24 | C065/Gallinarum 1 | 0.90 | Yes | ||||
| LT2 | Typhimurium 1 | S02 | C162/Typhimurium 1 | 0.94 | Yes | |||||
Corr coeff, coefficient of correlation.
The best hit also included an incorrect genovar.
Overall, among 20 strains that were used in both CGH and real-time PCR analysis, less than 6% of total gene absence/presence predictions were incongruent and about two-thirds of these discrepancies were called absent in real-time PCR but present in CGH. For all strains, genetic distance calculations identified isolates from the correct serovar as the closest match in the CGH data matrix (Table 3). One sample, strain R18, was supposedly an S. enterica serovar Gallinarum isolate but correlated instead with serovar Paratyphi B isolates in the CGH data. To determine if this misassignment was due to limitations in the real-time PCR assay, we performed a subsequent comparative genomic hybridization experiment using the same DNA preparation. The CGH data revealed the DNA to indeed be of serovar Paratyphi B origin. Therefore, real-time PCR was able to correctly identify the closest genovar and the correct serovar for all 24 subspecies I isolates. However, the almost 5% divergence of Salmonella bongori orthologues from their subspecies I gene counterparts (unpublished data) precluded successful amplification of several products from the S. bongori genome (not shown), emphasizing the effect of point mutations in the use of PCR product-based assays for genome classification.
Isolates of different genovars within the same serovar were also tested by real-time PCR. Strains representing two different genovars within five serovars were investigated (Table 3). In all cases, the closest matches in the CGH data compilation were with isolates of not only the correct serovar but also the correct genovar within that serovar.
Multiplex PCR.
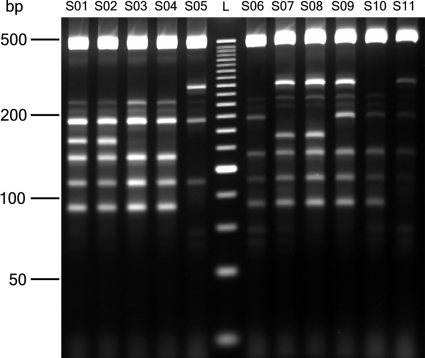
To test the feasibility of an assay that includes the presence of multiple reporters in one tube, the performance of a subset of genes was investigated in a multiplex PCR format. A series of 12 multiplexes was developed, each containing 8 pairs of PCR primers amplifying genes that were least likely to interfere with each other (minimized 3′ matches with other primers and PCR products present in each reaction mix). In addition, primer pairs amplifying the 16S rRNA gene were included as a positive control in each multiplex reaction. When tested against serovar Typhimurium LT2 and serovar Typhi CT18, 64 of these 96 primer pairs produced expected amplification products that could be resolved on a 3.3% standard agarose gel. A higher-resolution platform, such as polyacrylamide or capillary gel electrophoresis, may resolve more products. With cost effectiveness in mind (a factor that is most relevant, especially in developing countries), we continued to determine the performance of the multiplex mixtures on agarose gels. These 64 genes were used to identify sero- and genovars of 11 S. enterica strains. An image of 1 reaction mix on all 11 test isolates is depicted in Fig. 2. Absence/presence calls generated from these electrophoresis images are included in Table S2 in the supplemental material. All but one of the S. enterica strains investigated correlated best with isolates of the correct genovar in the CGH data collection (see Table S3 in the supplemental material). In one example, strain S11, the genetic distance of the correct genovar (Paratyphi C 1) was the same as that of a serovar (Typhisuis 1) known to be very similar (25). However, based on our CGH data, 4 differences exist among these 64 genes that would distinguish these serovars, and differentiation would easily be achieved when a multiplex database is established. As in the real-time assays, an S. bongori isolate was distinguished by the failure of many primer pairs due to sequence divergence (not shown).
FIG. 2.
Multiplex PCRs with 11 Salmonella enterica strains. Strain designations are shown in Table 3. Primer mix 10 is characterized in Table S1 in the supplemental material. L, DNA size standard.
DISCUSSION
While useful, serotyping does not necessarily provide insights into evolutionary relationships of isolates and is difficult to automate. Here we have concentrated on one of the alternatives, PCR. Similar DNA-based strategies to detect and identify Salmonella isolates have been explored in the past (5, 7, 9, 10, 12, 13, 19, 38). These protocols usually rely on exact matches of oligonucleotide sequences. The most successful of these strategies is a multiplex PCR approach based on 12 probes that represent 7 serovar Typhimurium LT2 genes, 6 serovar Typhi CT18 genes, and 1 serovar Enteritidis-specific region in PT4 (17). This simple assay was able to classify 19 of the top 20 serotypes in the United States, representing more than 75% of all reported Salmonella infections in the country. A blind test using 111 strains (17 serotypes) resulted in a 97% correct serotype assignment based on the PCR pattern generated. However, when all but one of the probes of this study were used in in silico tests on the 52 genovars we investigated here, they were unable to resolve 16 comparisons between genovars (Fig. 1) (see Fig. S1 in the supplemental material). Inclusion of the remaining serovar Enteritidis-specific probe would not have improved the outcome dramatically.
Our approach is different from all these previous studies because it sought to address the problem of point mutations leading to incorrect classification of isolates. Any individual PCR primer may fail due to a previously unknown single base polymorphism in the primer binding sequences. Thus, point mutations can result in misassignments of strains when only a few markers are used. This affects both real-time PCR and multiplex PCR. The amount of a particular product in a multiplex assay can also be influenced by the presence or absence of other PCR products in the multiplex which are competing for resources. In addition, all technologies include technical error. Therefore, it is necessary to have the number of markers used for classification exceed the theoretical minimum number. The additional information can then be used to recognize these “misreports” immediately and initiate a follow-up analysis for the isolate in question. In short, we wanted to be able to distinguish between serovars and genovars by more than one genetic marker.
We used information gleaned from our comparative genome hybridization database on 291 Salmonella isolates to classify serovars and all the major genome differences (down to the subserovar level, at genovar resolution). Three hundred eighty-four genes were manually picked from among those with potentially informative distribution patterns for taxonomy among the salmonellae. This panel of genes distinguished all investigated S. enterica subspecies I serovars and genovars by more than one genetic marker. Real-time PCR was tested using primers designed for PCR amplification of entire ORFs. One hundred forty-six primer pairs that had the best technical performance in real-time PCR were successfully tested on a variety of Salmonella genomes. A subset of these 146 genes was also used for multiplex PCR assays, and 64 reporters that could be scored consistently after separation of products on an agarose gel were used to classify Salmonella genomes. Based on CGH data, we showed that these 64 genes (which perform well on both real-time and multiplex PCR platforms) are able to unambiguously identify all genovars and moreover still contain 2 or more gene differences that distinguish between 99.5% of all 52 genovars investigated. Only five cross-genovar comparisons could not be resolved by more than one marker. In the future, these and other imperfectly resolved genovars and serovars can be addressed by adding additional PCR products as reporters.
The few discrepancies between multiplex PCR, real-time PCR, and CGH data were reliable within each platform in repeat experiments. Reliable reporting differences can be caused by a number of factors, particularly sequence polymorphisms in primer binding sites. For CGH, sequence homology requirements for a positive signal permit at least 5% divergence, whereas both PCR assays require almost total sequence homology in the short priming regions. In addition, the multiplex format requires the amplification product to be of a certain expected size and will therefore not tolerate insertions or deletions within the genetic segment interrogated. When real-time PCR or multiplex assays are performed on a large number of strains in many serovars, the resulting database for each platform will provide the best benchmark for subsequent strain analysis. The comparison with CGH data is a temporary expedient used here for proof of principle. This will be unnecessary when a database is established containing real-time or multiplex PCR profiles—correlations would then reach near-perfection for isolates that belonged to the same genovar as an isolate already present in the database. No interplatform comparisons would then be required.
Recently liquid-microsphere suspension array-based molecular protocols (8, 38) have been explored for typing purposes. This technology may in the future replace serology when targeted at the O- and H-antigen gene clusters. An O-group-specific Bio-Plex assay using seven specific probes distinguished the six most common serogroups in the United States (8) and correctly reported for 98.7% of the 384 tested isolates. Protocols utilizing that technology could have wide applicability for public health laboratories and are much cheaper than standard printed arrays. The Luminex-based technology allows for maximally 100 different analytes, which would likely be sufficient to distinguish all S. enterica subspecies I serovars based on gene presence. Such a liquid bead-based assay should be easier to design than an assay based on single nucleotide polymorphisms (such as those targeting the serological antigens). Ultimately, it will be desirable to integrate both the serological antigen markers and genovar markers to classify Salmonella. A microsphere-based hybridization platform using probes based on our error-detecting selection of genes, perhaps in combination with serological markers, may have great potential for a quick, reliable, and cost-effective sequence-based determination of Salmonella genovars and of many other pathogens.
As costs fall and automation improves, the balance of cost effectiveness and speed may soon tip in favor of DNA-based assays compared to serology. Among improvements that could increase speed and potentially decrease costs is the use of larger multiplexes. Multiplexes of 35 genes or more are possible, and fluorescent universal oligonucleotides can be used, reducing the time from colony picking to data analysis (4, 15). It is also conceivable that appropriately processed clinical samples might contain enough Salmonella DNA for direct analysis without culturing.
The classification schemes and gene panels we present here not only differentiate every S. enterica subspecies I serovar and respective genovar that we have tested but also do so with more than one distinguishing character state in 99.5% of all intergenovar comparisons. These panels can be incrementally improved as new data are acquired. If new high-quality CGH or sequencing data suggest that a strain in a previously unstudied serovar or genovar cannot be distinguished from other serovars or genovars by more than one marker already in the panel, then one would add an additional marker(s) to facilitate this distinction. The genes added to the panel would ideally also add additional discrimination power for other serovars.
Raising the number of genetic marker regions from 12 (covering 13 genes and one serovar Enteritidis-specific region, as used in reference 17) to 64 ensured resolution of all S. enterica serovars and genovars tested here and introduced the capacity to detect technical error and unexpected polymorphisms when classifying the vast majority of them. Further refinements in gene selection from the 384-gene panel and incorporation of more sequencing data as they become available may facilitate the design of a probe set that results in error-tolerant classification for all known Salmonella serovars and most genovars with fewer than 100 probes.
Supplementary Material
Acknowledgments
This work was funded in part by NIH grants R01AI034829, R01AI52237, and R21AI057733 and by the generous support of Sidney Kimmel.
We thank Carlos “Cliff” Santiviago for many helpful discussions, Monica Ponder for CGH data generation of non-subspecies I isolates, and YiPeng Wang for the correlation analysis using R.
Footnotes
Published ahead of print on 4 June 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alvarez, J., S. Porwollik, I. Laconcha, V. Gisakis, A. B. Vivanco, I. Gonzalez, S. Echenagusia, N. Zabala, F. Blackmer, M. McClelland, A. Rementeria, and J. Garaizar. 2003. Detection of a Salmonella enterica serovar California strain spreading in Spanish feed mills and genetic characterization with DNA microarrays. Appl. Environ. Microbiol. 697531-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boxrud, D., K. Pederson-Gulrud, J. Wotton, C. Medus, E. Lyszkowicz, J. Besser, and J. M. Bartkus. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 45536-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, E. F., S. Porwollik, F. Blackmer, and M. McClelland. 2003. Differences in gene content among Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 413823-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Q. R., G. Vansant, K. Oades, M. Pickering, J. S. Wei, Y. K. Song, J. Monforte, and J. Khan. 2007. Diagnosis of the small round blue cell tumors using multiplex polymerase chain reaction. J. Mol. Diagn. 980-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echeita, M. A., S. Herrera, J. Garaizar, and M. A. Usera. 2002. Multiplex PCR-based detection and identification of the most common Salmonella second-phase flagellar antigens. Res. Microbiol. 153107-113. [DOI] [PubMed] [Google Scholar]
- 6.Falush, D., M. Torpdahl, X. Didelot, D. F. Conrad, D. J. Wilson, and M. Achtman. 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3612045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell, J. J., L. J. Doyle, R. M. Addison, L. B. Reller, G. S. Hall, and G. W. Procop. 2005. Broad-range (pan) Salmonella and Salmonella serotype typhi-specific real-time PCR assays: potential tools for the clinical microbiologist. Am. J. Clin. Pathol. 123339-345. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, C., M. Collins, S. van Duyne, M. Mikoleit, T. Brown, and P. Fields. 2007. Multiplex, bead-based suspension array for molecular determination of common salmonella serogroups. J. Clin. Microbiol. 453323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, C., L. Gheesling, M. Collins, and P. I. Fields. 2006. Sequence analysis of the rfb loci, encoding proteins involved in the biosynthesis of the Salmonella enterica O17 and O18 antigens: serogroup-specific identification by PCR. Appl. Environ. Microbiol. 727949-7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, C., R. Sherwood, L. L. Gheesling, F. W. Brenner, and P. I. Fields. 2003. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl. Environ. Microbiol. 696099-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamming, R. W. 1986. Coding and information theory, 2nd ed. Prentice-Hall, Upper Saddle River, NJ.
- 12.Herrera-Leon, S., J. R. McQuiston, M. A. Usera, P. I. Fields, J. Garaizar, and M. A. Echeita. 2004. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 422581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera-Leon, S., R. Ramiro, M. Arroyo, R. Diez, M. A. Usera, and M. A. Echeita. 2007. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res. Microbiol. 158122-127. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, C. R., P. J. Fedorka-Cray, N. Wineland, J. D. Tankson, J. B. Barrett, A. Douris, C. P. Gresham, C. Jackson-Hall, B. M. McGlinchey, and M. V. Price. 2007. Introduction to United States Department of Agriculture VetNet: status of Salmonella and Campylobacter databases from 2004 through 2005. Foodborne Pathog. Dis. 4241-248. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, P. H., R. P. Walker, S. W. Jones, K. Stephens, J. Meurer, D. A. Zajchowski, M. M. Luke, F. Eeckman, Y. Tan, L. Wong, G. Parry, T. K. Morgan, Jr., M. A. McCarrick, and J. Monforte. 2002. Multiplex gene expression analysis for high-throughput drug discovery: screening and analysis of compounds affecting genes overexpressed in cancer cells. Mol. Cancer Ther. 11293-1304. [PubMed] [Google Scholar]
- 16.Kang, M. S., T. E. Besser, D. D. Hancock, S. Porwollik, M. McClelland, and D. R. Call. 2006. Identification of specific gene sequences conserved in contemporary epidemic strains of Salmonella enterica. Appl. Environ. Microbiol. 726938-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S., J. G. Frye, J. Hu, P. J. Fedorka-Cray, R. Gautom, and D. S. Boyle. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 443608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, J., K. Nelson, A. C. McWhorter, T. S. Whittam, and R. K. Selander. 1994. Recombinational basis of serovar diversity in Salmonella enterica. Proc. Natl. Acad. Sci. USA 912552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk, J. M., U. Kongmuang, P. R. Reeves, and A. A. Lindberg. 1993. Selective amplification of abequose and paratose synthase genes (rfb) by polymerase chain reaction for identification of Salmonella major serogroups (A, B, C2, and D). J. Clin. Microbiol. 312118-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majtan, T., L. Majtanova, J. Timko, and V. Majtan. 2007. Oligonucleotide microarray for molecular characterization and genotyping of Salmonella spp. strains. J. Antimicrob. Chemother. 60937-946. [DOI] [PubMed] [Google Scholar]
- 21.Malorny, B., C. Bunge, B. Guerra, S. Prietz, and R. Helmuth. 2007. Molecular characterisation of Salmonella strains by an oligonucleotide multiprobe microarray. Mol. Cell Probes 2156-65. [DOI] [PubMed] [Google Scholar]
- 22.Nair, S., S. Alokam, S. Kothapalli, S. Porwollik, E. Proctor, C. Choy, M. McClelland, S. L. Liu, and K. E. Sanderson. 2004. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J. Bacteriol. 1863214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popoff, M. Y., J. Bockemuhl, and L. L. Gheesling. 2004. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155568-570. [DOI] [PubMed] [Google Scholar]
- 24.Popoff, M. Y., and L. Le Minor. 2001. Antigenic formulas of the Salmonella serovars, 8th revision. WHO Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France.
- 25.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 1865883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 311869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porwollik, S., C. A. Santiviago, P. Cheng, L. Florea, and M. McClelland. 2005. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 1876545-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porwollik, S., R. M. Wong, R. A. Helm, K. K. Edwards, M. Calcutt, A. Eisenstark, and M. McClelland. 2004. DNA amplification and rearrangements in archival Salmonella enterica serovar Typhimurium LT2 cultures. J. Bacteriol. 1861678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 998956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porwollik, S., R. M. Wong, S. H. Sims, R. M. Schaaper, D. M. DeMarini, and M. McClelland. 2001. The DeltauvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat. Res. 4831-11. [DOI] [PubMed] [Google Scholar]
- 31.Reen, F. J., E. F. Boyd, S. Porwollik, B. P. Murphy, D. Gilroy, S. Fanning, and M. McClelland. 2005. Genomic comparisons of Salmonella enterica serovar Dublin, Agona, and Typhimurium strains recently isolated from milk filters and bovine samples from Ireland, using a Salmonella microarray. Appl. Environ. Microbiol. 711616-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves, P. 1993. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 917-22. [DOI] [PubMed] [Google Scholar]
- 33.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 359-67. [DOI] [PubMed] [Google Scholar]
- 34.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tankouo-Sandjong, B., A. Sessitsch, E. Liebana, C. Kornschober, F. Allerberger, H. Hachler, and L. Bodrossy. 2007. MLST-v, multilocus sequence typing based on virulence genes, for molecular typing of Salmonella enterica subsp. enterica serovars. J. Microbiol. Methods 6923-36. [DOI] [PubMed] [Google Scholar]
- 37.Torpdahl, M., G. Sorensen, B. A. Lindstedt, and E. M. Nielsen. 2007. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg. Infect. Dis. 13388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tracz, D. M., H. Tabor, M. Jerome, L. K. Ng, and M. W. Gilmour. 2006. Genetic determinants and polymorphisms specific for human-adapted serovars of Salmonella enterica that cause enteric fever. J. Clin. Microbiol. 442007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3)S127-S134. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida, C., K. Franklin, P. Konczy, J. R. McQuiston, P. I. Fields, J. H. Nash, E. N. Taboada, and K. Rahn. 2007. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J. Microbiol. Methods 70261-271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.