Abstract
The accurate and rapid identification of bacteria isolated from the respiratory tract of patients with cystic fibrosis (CF) is critical in epidemiological studies, during intrahospital outbreaks, for patient treatment, and for determination of therapeutic options. While the most common organisms isolated from sputum samples are Pseudomonas aeruginosa, Staphylococcus aureus, and Haemophilus influenzae, in recent decades an increasing fraction of CF patients has been colonized by other nonfermenting (NF) gram-negative rods, such as Burkholderia cepacia complex (BCC) bacteria, Stenotrophomonas maltophilia, Ralstonia pickettii, Acinetobacter spp., and Achromobacter spp. In the present study, we developed a novel strategy for the rapid identification of NF rods based on Fourier transform infrared spectroscopy (FTIR) in combination with artificial neural networks (ANNs). A total of 15 reference strains and 169 clinical isolates of NF gram-negative bacteria recovered from sputum samples from 150 CF patients were used in this study. The clinical isolates were identified according to the guidelines for clinical microbiology practices for respiratory tract specimens from CF patients; and particularly, BCC bacteria were further identified by recA-based PCR followed by restriction fragment length polymorphism analysis with HaeIII, and their identities were confirmed by recA species-specific PCR. In addition, some strains belonging to genera different from BCC were identified by 16S rRNA gene sequencing. A standardized experimental protocol was established, and an FTIR spectral database containing more than 2,000 infrared spectra was created. The ANN identification system consisted of two hierarchical levels. The top-level network allowed the identification of P. aeruginosa, S. maltophilia, Achromobacter xylosoxidans, Acinetobacter spp., R. pickettii, and BCC bacteria with an identification success rate of 98.1%. The second-level network was developed to differentiate the four most clinically relevant species of BCC, B. cepacia, B. multivorans, B. cenocepacia, and B. stabilis (genomovars I to IV, respectively), with a correct identification rate of 93.8%. Our results demonstrate the high degree of reliability and strong potential of ANN-based FTIR spectrum analysis for the rapid identification of NF rods suitable for use in routine clinical microbiology laboratories.
The major causes of morbidity and mortality in patients with cystic fibrosis (CF) are chronic lung infections caused by Pseudomonas aeruginosa, Staphylococcus aureus, and Haemophilus influenzae (20, 46). Nevertheless, in recent decades an increasing fraction of CF patients has been colonized by other emerging nonfermenting gram-negative bacilli, such as members of the Burkholderia cepacia complex (BCC), Stenotrophomonas maltophilia, Achromobacter xylosoxidans, Ralstonia pickettii, Pandoraea spp., and Acinetobacter spp. (9, 10, 17, 47). Advances in the taxonomy of BCC revealed that it comprises at least nine distinct species (previously designated genomovars) which are very closely related (11, 34, 58). Furthermore, some Burkholderia species that do not belong to BCC, like B. gladioli, and species from other genera, such as Pandoraea and Ralstonia, are commonly grouped with and referred to as B. cepacia-like organisms and share genotypic and phenotypic characteristics with BCC bacteria (7, 8). Even though the frequency of infection with these emerging species is relatively low (20), they represent a significant challenge to clinical laboratories due to the misidentifications that often occur (11, 47). Therefore, it has been reported that the microbiological detection of microorganisms in sputum samples from CF patients is one of the most labor-intensive, expensive, and sometimes unreliable procedure performed in microbiological laboratories (37). All nine species belonging to BCC are capable of colonizing CF patients, and some of them produce devastating consequences when they are the cause of lung infections in these patients (11, 57). The accurate and rapid identification of the respiratory tract pathogens is required for the proper clinical management of these patients, for the study of intrahospital outbreaks, and to improve the understanding of the changing epidemiology of the microbiology in the lungs of CF patients (37, 46, 58). In most clinical laboratories, the identification of microorganisms isolated from sputum samples from CF patients is generally performed by using a combination of selective media and commercially available phenotypic identification systems, such as the API 20NE system (bioMérieux, Marcy l'Etoile, France) or Vitek GNI Plus or Vitek 2 ID-GNB cards (bioMérieux) (3, 47). For final microbiological detection, several additional phenotypic tests are included for the differentiation among B. gladioli, Pandoraea spp., R. pickettii, A. xylosoxidans, and some members of BCC (11, 26). However, misidentification may still occur at the species level, particularly among bacteria belonging to BCC (11, 60). Consequently, isolates that are considered putative members of BCC should be further examined by genotypic methods.
Various genotypic approaches are currently applied for the identification of bacteria isolated from sputum specimens and are used as alternatives or complements to phenotypic identification procedures. In this regard, 16S rRNA gene sequence analysis has been a widely accepted tool for molecular identification (17). Other techniques, such as recA-based PCR (recA PCR) followed by restriction fragment length polymorphism (RFLP) analysis and sequence analysis, have also been used to identify BCC clinical isolates to the species level (33, 43, 56, 57). Although these techniques are simple and initially useful, they have become confusing with the increasing species diversity in BCC. Therefore, a new strategy involving multilocus sequence typing (MLST) has been applied and validated for the identification of BCC. This powerful technique allows both species and strain differentiation; and the sequence data are highly transferable, unambiguous, and useful for phylogenetic analysis (1). Although genotypic approaches to the identification of clinically relevant microorganisms are finding their way into the field of clinical diagnostic microbiology, they are still expensive and laborious and are considered unattractive for routine application in busy clinical laboratories.
Modern Fourier transform infrared (IR) spectroscopy (FTIR) techniques constitute radically different approaches for the identification of microorganisms. The IR spectra of intact bacteria provide highly specific patterns that allow microbial cells to be distinguished at different taxonomic levels and have frequently been employed for the rapid and accurate identification of microorganisms even to the strain level (24, 29, 35, 45). FTIR is easy to implement, allows the analysis of small quantities of biomass, and requires no consumables or reagents (38, 52). Standardization of the cultivation conditions and the sampling and measurement parameters enabled the creation of reference libraries containing spectra for well-identified microbes. These databases, which can be analyzed by using different algorithms, such as hierarchical cluster analysis (HCA) (2, 29), linear discriminant analysis (36), or analysis with artificial neural networks (ANNs) (45, 49, 55), make the identification of unknown microorganisms possible. In particular, the computer-based ANN pattern recognition method was reported to reliably solve problems with the identification of closely related microorganisms (45).
In this report we describe for the first time a new approach based on FTIR combined with ANNs for the discrimination and identification of nonfermentative gram-negative bacilli recovered from cultures of sputum from CF patients hospitalized in different CF centers and hospitals in Argentina between 2004 and 2006. This approach constitutes a reliable, rapid, and specific means of identification of P. aeruginosa, R. pickettii, A. xylosoxidans, S. maltophilia, Acinetobacter spp., and the clinically relevant species of BCC isolated from CF patients.
MATERIALS AND METHODS
Bacterial strains.
A total of 15 references strain (kindly provided by Laura Galanternik, Laboratory for Microbiology, Gutierrez Hospital, Argentina) and 169 clinical isolates of aerobic nonfermenting gram-negative rods recovered from sputum samples from 150 CF patients attending three different CF centers and hospitals in Argentina between 2004 and 2006 were used in this study (Table 1). The CF centers and hospitals are located in Buenos Aires Province (La Plata Children's Hospital, Sor María Ludovica), Córdoba Province in central Argentina (Santísima Trinidad Hospital, HST), and Buenos Aires City (Buenos Aires Clinical Hospital). Additional information on the origins of the strains and the identification methods applied to each strain is listed in Table 1.
TABLE 1.
Nonfermentative gram-negative bacterial strains used in this study
| Species | No. of isolates | Origin/culture collection no.a | Identification methodb |
|---|---|---|---|
| P. aeruginosac | 2 | ATCC 9027, ATCC 27853 | |
| 15 | HNLP | ||
| BCCd | 1 | HST 8684 | RFLP with HaeIII/species-specific recA PCR |
| 1 | HC BC05 | RFLP with HaeIII/species-specific recA PCR | |
| B. cepacia | 1 | ATCC 25416 | RFLP with HaeIII |
| 3 | HST | RFLP with HaeIII/species-specific recA PCR | |
| 6 | HC | RFLP with HaeIII/species-specific recA PCR | |
| B. multivorans | 2 | LMG 13010, ATCC 17616 | RFLP with HaeIII |
| 1 | HST | RFLP with HaeIII/species-specific recA PCR | |
| B. cenocepacia lineage IIIA | 1 | LMG 18863 | RFLP with HaeIII |
| 37e | HNLP | RFLP with HaeIII/species-specific recA PCR | |
| 11e | HC | RFLP with HaeIII/species-specific recA PCR | |
| 31e | HST | RFLP with HaeIII/species-specific recA PCR | |
| B. cenocepacia lineage IIIB | 1 | LMG 16654 | RFLP with HaeIII |
| 2 | HC | RFLP with HaeIII/species-specific recA PCR | |
| 1 | LMG 18870 | RFLP with HaeIII | |
| B. stabilis | 3 | HC | RFLP with HaeIII/species-specific recA PCR |
| 1 | LMG 10929 | RFLP with HaeIII | |
| B. vietnamiensis | 1 | LMG 21820 | RFLP with HaeIII |
| B. dolosa | 1 | LMG 19467 | RFLP with HaeIII |
| B. ambifaria | 1 | LMG 20983 | RFLP with HaeIII |
| B. anthina | 1 | LMG 14191 | RFLP with HaeIII |
| B. pyrrocinia | 2 | HC Sm 20/06, HC Sm 66/06 | PCR for 16S rRNA |
| S. maltophilia | 11 | HC | |
| 13 | HNLP | ||
| 2 | HC Ax 247/06, HC Ax 335/06 | PCR for 16S rRNA | |
| A. xylosoxidans | 9 | HC | |
| 10 | HNLP | ||
| 1 | ATCC 19606 | ||
| Acinetobacter baumannii | 1 | HC Ab 54/06 | PCR for 16S rRNA |
| 9 | HNLP | ||
| Acinetobacter spp. | 1 | ATCC 27511 | |
| Ralstonia pickettii | 1 | HC Rp 127 | PCR for 16S rRNA |
ATCC, American Type Culture Collection, Manassas, VA; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; LMG, Laboratorium Microbiologie Gent Culture Collection, Ghent, Belgium; HNLP, Hospital de Niños La Plata, La Plata, Argentina; HST, Hospital Santa Trinidad, Córdoba, Argentina; HC, Hospital de Clínicas, Buenos Aires, Argentina.
All clinical isolates were identified by use of the API or the Vitek system and the nine additional phenotypic tests indicated in Materials and Methods (19). BCC clinical isolates were further identified by recA PCR-RFLP analysis with primers BCR1 and BCR2 and the HaeIII restriction enzyme, followed by recA PCR with species-specific primers (50). Some clinical isolates belonging to genera other than BCC were randomly chosen to confirm their identities by16S rRNA gene sequencing.
Only nonmucoid P. aeruginosa strains were included.
All BCC reference strains were kindly provided by Laura Galanternik, Hospital Gutierrez, Buenos Aires, Argentina.
One strain was identified as B. cenocepacia lineage IIIA type D by recA PCR-RFLP analysis with HaeIII, and the rest of the isolates were atypical strains identified as B. cenocepacia lineage IIIA but had a recA PCR-HaeIII RFLP profile not compatible with the ones previously described for B. cenocepacia (50).
Clinical isolates and reference strains were stored at −70°C in brain heart infusion broth containing 10% glycerol. For FTIR analysis, they were unfrozen and subcultured once for 24 h on methylene blue (EMB) agar medium.
Isolation and phenotypic identification.
The isolation of clinical isolates was performed according to clinical microbiology practice recommendations for respiratory tract specimens from CF patients (11, 26, 27, 30, 50, 60). The isolates were first identified by using conventional biochemical methods (with the API 20NE system or the Vitek 2 card [bioMérieux]). Subsequently, additional phenotypic tests which allow the discrimination of B. cepacia-like and BCC bacteria were applied to obtain the definitive phenotype (11, 26, 60). Antibiotic susceptibilities were determined by the disk diffusion method on Mueller-Hinton medium (Difco Laboratories), according to the methods outlined by the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards; 1999) (40a). The identities of the strains identified as putative members of BCC by the aforementioned assays were further confirmed by molecular identification techniques.
Molecular identification of BCC isolates.
The total chromosomal DNA of each isolate, which had previously been cultured in tryptic soy agar (TSA) at 37°C for 24 to 48 h, was extracted by the boiling-lysis method described by Seo and Tsuchiya (51). The identification of the clinical isolates and the confirmation of the identities of the reference strains were performed as described by Mahenthiralingam et al. by PCR of the recA gene with primers BCR1 and BCR2 followed by RFLP analysis with the HaeIII restriction enzyme (33). A species-specific PCR was used to the confirm species of the clinical isolates (33). The specific RFLP patterns obtained for the different BCC species were compared with those reported previously (16, 33, 51, 56, 58).
Identification by 16S rRNA gene sequence analysis.
Total DNA was prepared as described by Sambrook et al. (48). Primers fD2 and rP2 (59), which are specific for the 16S rRNA gene, were used. Sequence analysis was performed as described by Bosshard et al. (3). The amplicons were purified and sequenced, and the fragments were analyzed with an automatic DNA sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems). These sequences were analyzed with the BLASTN program of the Genetics Computer Group. A similarity score of ≥99% with the reference sequence of a classified species was used to accept a species-level identification. If the score was between 98% and 95%, the clinical isolate was assigned to the corresponding genus.
FTIR, sample preparation, measurements, and data preprocessing.
For FTIR measurements, both clinical isolates received from hospitals on EMB medium and bacteria obtained from stocks that had been subcultured for 24 h were subcultured on TSA medium (Oxoid, Basingstoke, United Kingdom) for 5 ± 0.5 h at 37 ± 1°C. One loopful of these bacteria was directly harvested with a 1-mm-diameter platinum loop and suspended in 120 μl distilled water. The suspensions were vortexed at a high speed (2,000 rpm) for 15 min on an MS2 minishaker (IKA Works, Inc., Wilmington, NC) and centrifuged at 8,000 × g for 5 min, the supernatant was discarded, and the pelleted cells were suspended in 100 μl of distilled water. An aliquot of 80 μl was transferred to ZnSe optical plates and dried under moderate vacuum (0.1 bar) for 45 min to obtain transparent bacterial films (24).
FTIR absorption spectra between 4,000 and 650 cm−1 were acquired on a Spectrum One FTIR spectrometer (Perkin-Elmer Instruments) with 6-cm−1 spectral resolution and 64 scans. OPUS software (version 4.2; Bruker Optics GmbH, Ettlingen, Germany) was used for data preprocessing. The first derivatives were calculated using the Savitzky-Golay algorithm with nine smoothing points to increase the number of discriminative features present in the spectra and to minimize problems with baseline shifts. To avoid interference from the biomass variations among the different samples, the first derivatives were vector normalized in the full range (25, 38). At least 10 spectra of each strain from four independent bacterial cultures were included in the analysis to account for possible sources of variance in the sampling procedure. The levels of reproducibility among the replicates for every strain were calculated as the averages plus or minus 2 standard deviations of the so-called spectral distance (D), which is the measure of dissimilarity, where D is equal to (1 − r) × 1,000 and r is Pearson's correlation coefficient (24, 38). OPUS software (version 4.0; Bruker Optics GmbH) was used to calculate the average D values and standard deviations for the different spectral ranges.
A database was built with the vector-normalized first-derivative spectra of the clinical isolates and reference strains given in Table 1. Before the spectra were introduced into the library, raw spectral data were subjected to a quality test (QT) with OPUS software (38). Spectral quality was ensured by taking into account the intensity, water vapor level, signal-to-noise values, and signal-to-water vapor values. Additionally, a so-called α factor, which was calculated from the ratio of the intensities at wavelengths of 1,738 cm−1 (I1,738) and 1,540 cm−1 (I1,540) (i.e., α = I1,738/I1,540) with values of less than 0.25, was included in the QT. For spectral analysis, the database (which contained almost 2,000 vector-normalized first-derivative spectra) was divided into two subsets: (i) the training data set, which constituted the reference data of the first-derivative and vector-normalized spectra for the reference strains, the clinical isolates identified by 16S rRNA gene sequencing, and about 70% of the remaining clinical isolates randomly chosen from among representatives of all species studied (altogether 131 isolates), and (ii) the test data set, which contained the spectral data for the clinical isolates that were not included in the training data set (53 clinical isolates), which were used exclusively for external validation.
Semiquantitative determination of PHB.
In order to adjust the culture conditions so that poly-β-hydroxybutiric acid (PHB) did not interfere with FTIR discrimination analysis, the PHB content was evaluated from the IR absorbance spectra, as reported previously (25). Briefly, the I1,738 of the ester carbonyl peak, used as a marker band to estimate the PHB content, and the I1,540 of the amide II peak, used as an internal standard of the total biomass, were calculated from the vector-normalized spectra. The semiquantitative estimation of PHB was calculated according to the equation α = I1,738/I1,540 with OPUS software (version 4.0; Bruker Optics GmbH).
HCA.
Analysis for discrimination between the different groups of bacteria studied was performed by HCA with the first-derivative spectra of the training data set. Dendrograms were obtained by using Ward's clustering algorithm, and the D values, defined above, were calculated as a distance measure of similarity among the spectra (24, 29, 39).
ANN analysis.
The whole database, which contained almost 2,000 first-derivative spectra for 169 clinical isolates and 15 reference strains, was used for the development of ANNs. NeuroDeveloper software (version 2.3; Synthon GmbH, Heidelberg, Germany) was used to develop and optimize a modular classification system. The first-derivative spectra included in the training data set were used for (i) preprocessing, (ii) training, and (iii) internal validation of the networks. In the data preprocessing step, the selection of the best wavelengths (feature selection) was performed in the predefined spectral windows of 2,800 to 3,100 cm−1 and 1,650 to 900 cm−1 by using the COVAR algorithm (49). During the training procedure, the number of input neurons and the number of hidden neurons were optimized in the network topology in order to obtain good network performance. The internal validation was used to monitor the training process and to determine the optimal performance during the training process (49). To choose the spectra for use for internal validation, 10% of the spectra from each species in the training data set were randomly selected by the software. As an objective performance criterion of the ANNs, external validation was carried out with the test data set. This validation was used to establish the percentage of correct identifications as an objective measure to estimate the performance of the identification model.
TEM.
The removal of pili and/or other fiber appendages that adhered to the bacteria was evaluated by transmission electron microscopy (TEM). Bacteria that had been grown on TSA medium for 5 h were suspended in distilled water and vortexed for different times (5 to 30 min) at different vortexing intensities (high, medium, slow) with an MS2 minishaker (IKA). A drop of both the original and the sheared suspensions was transferred to Formvar-coated grids. Five minutes later, the fluid was removed by absorbing it with filter paper and the grids were negatively stained with 2% phosphotungstic acid (pH 5.2) for 40 s. The grids were observed under a JEM 1200 EX JEOL transmission electron microscope (Servicio Central de Microscopía Electrónica, Facultad de Ciencias Veterinarias, UNLP, Argentina) at 80 kV.
RESULTS AND DISCUSSION
Sample preparation for FTIR.
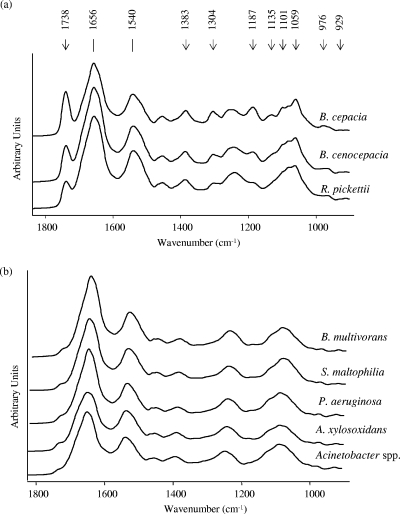
At the beginning of our studies, we noticed that even though we were working on the basis of strictly standardized culture conditions, sample preparation procedures, and spectral data acquisition parameters, the biological variance achieved for replicate measurements for the different clinical isolates was too high to allow accurate discrimination. In order to elucidate the reasons for this insufficient reproducibility, we focused our attention on those cellular components whose expression levels might be variable and might have relatively intense IR absorbance bands. It was previously reported that bacterial isolates recovered from CF patients could express PHB (28, 31). This intracellular component shows a strong ester carbonyl band at 1,738 cm−1, accompanied by a number of additional bands at 1,383, 1,304, 1,187, 1,135, 1,101, 1,059, and 976 cm−1 (38, 41). It was previously reported that these dominant IR absorption bands distributed throughout the whole mid-IR spectral range interfered with the discrimination of other bacteria by FTIR (25). Therefore, we hypothesized that the presence of PHB might be one of the sources of interference when FTIR was used to discriminate among nonfermenting gram-negative bacilli. In experiments in which we screened for bands characteristic of PHB among the spectra of the reference strains and local isolates, we noticed that B. cenocepacia, B. cepacia, and R. pickettii could be considered PHB producers, while no PHB signals were found for the other species under our working conditions (growth on TSA medium at 37°C for 24 h). Figure 1a shows the FTIR absorbance spectra obtained for three clinical isolates of PHB-producing bacteria after 24 h of growth and the PHB absorption bands that interfered with the discrimination of nonfermenting rods, while Fig. 1b shows the spectra for five clinical isolates of non-PHB-producing bacteria (P. aeruginosa, A. xylosoxidans, Acinetobacter spp., B. multivorans, and S. maltophilia).
FIG. 1.
FTIR absorbance spectra in the region from 1,800 to 900 cm−1 for intact nonfermentative gram-negative rods grown on TSA medium at 37°C for 24 h. (a) PHB-producing bacteria. The bands indicated with arrows correspond to the main absorption bands assigned to PHB. (b) Non-PHB-producing bacteria.
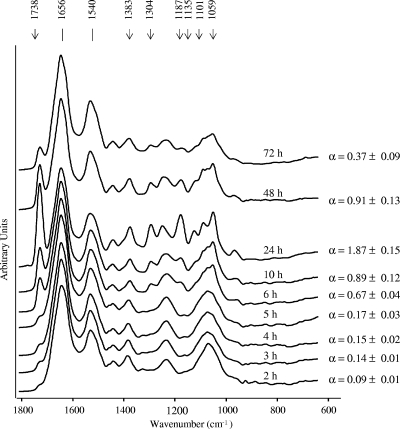
In order to determine the environmental conditions under which PHB accumulation can be reduced to levels that would allow the reproducible discrimination of bacteria, different growth conditions and incubation times were tested. Figure 2 shows the kinetics of PHB production for a clinical isolate of B. cenocepacia and the corresponding α values calculated from the FTIR spectra obtained for cells grown for different times on TSA medium at 37°C. For this clinical isolate, no significant PHB signals were detected during the first 5 h of incubation, with the corresponding α values being less than 0.17 ± 0.03. In the early exponential growth phase, the PHB signal started to increase, from an α value of 0.67 ± 0.04 at 6 h of incubation to the highest value of 1.87 ± 0.15 at 24 h of growth. Subsequently, the bacteria degraded PHB and its signal decreased again in the following hours, with an α value of 0.37 ± 0.09 reached after 72 h of growth. We performed the same analysis with other B. cenocepacia strains and found that the kinetics and the relative amounts of PHB accumulated by the cells were not the same for all the strains tested. Some of the clinical isolates showed a maximum of PHB accumulation at 48 h of growth; other isolates started producing PHB after 10 h of incubation. In addition, the highest α values calculated for the different isolates varied from 0.52 ± 0.15 to 2.10 ± 0.17, irrespective of the genera or the species considered (B. cenocepacia, B. cepacia, and R. pickettii). Nevertheless, for all the isolates tested (Table 1), no PHB production was observed before 6 h of growth on TSA medium at 37°C. From these results we concluded that the cultivation time should be as short as possible and should be less than 6 h to avoid PHB interference with species discrimination by FTIR. On the other hand, a sufficient amount of biomass should be available after the selected incubation time to obtain spectra with an adequate signal-to-noise ratio to pass the QT described above. When these requirements are taken into account, an incubation time of 5 h was selected to achieve a sufficient cell mass, and we found that the α value should be less than 0.25 to ensure the absence of interfering PHB signals. It is important to note that the growth kinetics and, thus, the level of PHB production must depend on additional variables, aside from the culture medium, incubation time, temperature, and inoculum size, which cannot easily be controlled experimentally. Therefore, we defined that all spectra which failed the QT because of signal-to-noise ratios that were too low and/or because they had α values of ≥0.25 must be rejected and the measurements must be repeated.
FIG. 2.
FTIR spectra (in the region from 1,800 to 600 cm−1) obtained during the growth of a B. cenocepacia clinical isolate as an example of the kinetics of PHB production of a nonfermentative rod growing on TSA medium at 37°C. The semiquantitative estimation of the PHB content, calculated from α (I1,738/I1,540) values, is indicated at each time of growth. The α values were calculated as the ratio of the I1,738 of the ester carbonyl peak, used as a PHB marker band, and the I1,540 of the amide II peak, used as an internal standard of the total biomass (37).
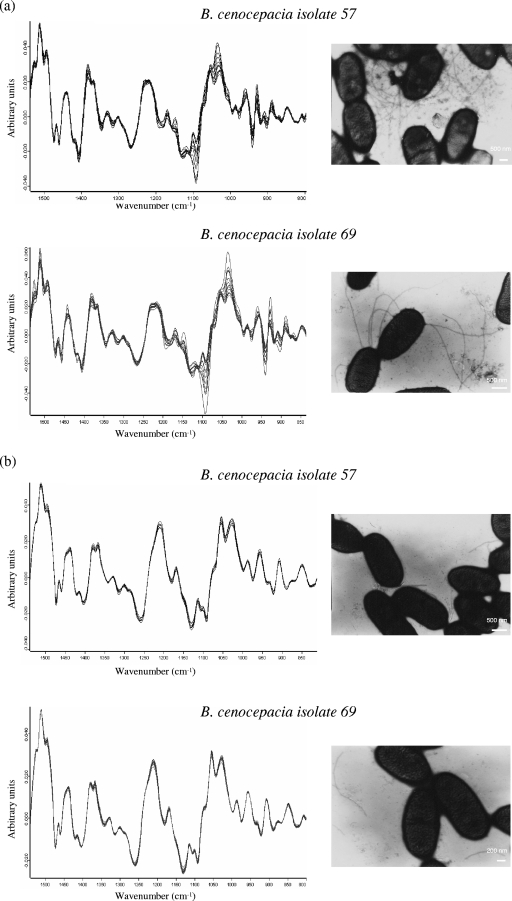
Once PHB interference was eliminated, the level of reproducibility for replicate measurements for different clinical isolates was studied with bacteria grown under the previously selected conditions (growth on TSA medium at 37°C for 5 h). Figure 3a shows the vector-normalized first-derivative spectra in the 1,500- to 800-cm−1 range for two B. cenocepacia clinical isolates (isolates 57 and 69) from five independent cultivation experiments (three replicate measurements each) and the micrographs of the corresponding cells obtained by TEM. As can be seen, significant heterogeneity among the replicate spectra for each strain was found in the 1,200- to 900-cm−1 region, which is mainly assigned to signals from carbohydrate vibrations, and in the spectral region of 1,500 to 1300 cm−1, known as the “mixed” region (24, 40). Cell components other than PHB that are also expressed at different levels and their chemical compositions may exhibit IR absorption bands in these spectral regions and might therefore constitute a possible source of variance among replicates. These components are appendage fibers, such as pili and flagella. It has been reported that these fibers are expressed or coexpressed by most of the gram-negative bacteria present in the lungs of CF patients (P. aeruginosa, Burkholderia spp., S. maltophilia, and Acinetobacter spp.) (15, 19, 21) and that a single pilus is a polymer formed by an ordered association of thousands of identical proteinaceous subunits (pilin), which might be glycosylated (4, 6, 42). Therefore, we studied the spectral heterogeneity obtained after the removal of these pilus or flagella fibers from bacterial cells by vigorous shaking and subsequent centrifugation (21, 44). Assays were performed with different times of vortexing (5 to 30 min) and different vortexing intensities (low, medium, high) with a minishaker (MS2; IKA), and the spectral heterogeneity was evaluated. We observed that a significant reduction in the variance among replicates could be obtained after vortexing of the cells at a high intensity for 15 min, centrifugation, and separation of the cells from the pili. Figure 3b shows the first-derivative spectra of clinical isolates B. cenocepacia 57 and 69 measured after application of the protocol described above and the corresponding micrographs obtained by TEM after vortexing of the cells.
FIG. 3.
Vector-normalized first-derivative spectra of two B. cenocepacia clinical isolates (isolates 57 and 69) in the 1,500- to 800-cm−1 range. (a) The heterogeneity of 15 replicate measurements for each strain in the spectral ranges of 1,200 to 900 cm−1 and 1,500 to 1,300 cm−1 and the corresponding micrographs obtained by TEM are shown. (b) Vector-normalized first-derivative spectra measured after vortexing of similar cells at the maximum intensity for 15 min and subsequent centrifugation at 8,000 × g for 5 min to separate the cells from free pilus appendages in the supernatants. Micrographs of the cells obtained by TEM after they were vortexed without centrifugation show the small fragments of pili or fibers suspended in the supernatants.
The variance among replicates in quantitative numbers was measured. We calculated the so-called D values for different IR spectral regions of the spectrum. By definition, the lowest D value indicates the highest reproducibility among replicates (24). Table 2 summarizes the spectral variances calculated for spectral windows of 3,000 to 2,800, 1,500 to 1,300, and 1,200 to 900 cm−1 before and after the application of shear stress to the cells. The D values calculated for clinical isolates B. cenocepacia 57 and 69 before shearing were relatively high for both strains in the spectral region assigned to carbohydrates compared to the values obtained, e.g., for the 3,000- to 2,800-cm−1 window. Table 2 also indicates the maximum and minimum D values (30.1 ± 28.7 and 13.2 ± 10.9, respectively) obtained in the carbohydrate region among all the B. cenocepacia clinical isolates assayed and the typical D values obtained for the species P. aeruginosa and S. maltophilia. In all cases tested, the D values calculated after removal of the fiber appendages decreased by a factor of nearly 10 for the carbohydrate region and more than five times in the 1,500- to 1,300-cm−1 region, while the D values for the spectral range from 3,000 to 2,800 cm−1 obtained after the removal of the detached fiber appendages from the cells by shearing and centrifugation decreased only by a factor of 2. The level of reproducibility obtained after shearing of the cells for 15 min at the maximum vortex intensity turned out to be suitable for the discrimination of bacteria even down to the species level.
TABLE 2.
Reproducibility levels obtained from cells before and after removing appendage structures calculated as D values for different spectral rangesa
| Species | Isolate |
D values for the following spectral ranges (cm−1):
|
|||||
|---|---|---|---|---|---|---|---|
| Before shearing
|
After shearing and centrifugation
|
||||||
| 3,000-2,800 | 1,500-1,300 | 1,200-900 | 3,000-2,800 | 1,500-1,300 | 1,200-900 | ||
| B. cenocepacia | 57 (HNLP) | 2.13 ± 1.62 | 6.19 ± 5.30 | 25.0 ± 23.2 | 0.75 ± 0.43 | 0.93 ± 0.66 | 1.94 ± 1.07 |
| 69 (HNLP) | 2.71 ± 1.03 | 6.35 ± 5.29 | 30.1 ± 28.7 | 1.11 ± 1.07 | 1.82 ± 1.55 | 2.75 ± 0.66 | |
| 15 (HST) | 1.94 ± 1.29 | 7.62 ± 6.64 | 13.2 ± 10.9 | 0.89 ± 068 | 0.95 ± 1.06 | 1.70 ± 0.88 | |
| P. aeruginosa | R2 (HST) | 2.73 ± 1.96 | 8.36 ± 7.19 | 35.9 ± 40.2 | 1.20 ± 0.95 | 2.19 ± 1.07 | 3.43 ± 1.95 |
| S. maltophilia | 50428 (HST) | 1.82 ± 1.69 | 10.12 ± 9.6 | 20.3 ± 23.2 | 0.31 ± 0.9 | 1.85 ± 1.34 | 2.99 ± 1.98 |
| 35668 (HST) | 1.9 ± 1.83 | 9.73 ± 8.25 | 15.3 ± 12.9 | 0.60 ± 0.40 | 0.96 ± 0.57 | 2.8 ± 2.1 | |
D values were calculated (36) for the B. cenocepacia clinical isolates before and after shearing in the spectral regions 1,200 to 900 cm−1 (assigned to carbohydrates), 1,500 to 1,300 cm−1 (mixed region), and 3,000 to 2,800 cm−1 (C—H stretching of >CH2 and —CH3). D values corresponding to one P. aeruginosa clinical isolate (isolate R2) and two S. maltophilia clinical isolates (isolates 50428 and 35668) are shown as examples of the typical D values obtained for genera other than Burkholderia. HNLP, Hospital de Niños La Plata, La Plata, Argentina; HST, Hospital Santa Trinidad, Córdoba, Argentina.
Therefore, to avoid interfering spectral signals due to various amounts of PHB accumulation and to fiber appendage expression in the discrimination of all strains of nonfermenting rods isolated from sputum samples from CF patients by FTIR, the bacteria had to be incubated for 5 ± 0.5 h on TSA medium at 37 ± 1°C, suspended in distilled water, vortexed at a high speed for 15 min, and centrifuged. The spectra of the suspended cells were then measured by FTIR, as described in the Materials and Methods section. This simple sampling protocol guaranteed a high level of reproducibility in a short time. It would be feasible to complete microbiological detection within a working day starting with the gram-negative nonfermentative rods obtained after 24 h of incubation on EMB agar, in view of the following requirements: 5 h of growth on TSA medium, 1 h for sample preparation, 15 min for the recording of spectral replicates, and a few minutes for data analysis.
Discrimination of gram-negative bacilli isolated from sputum samples by HCA.
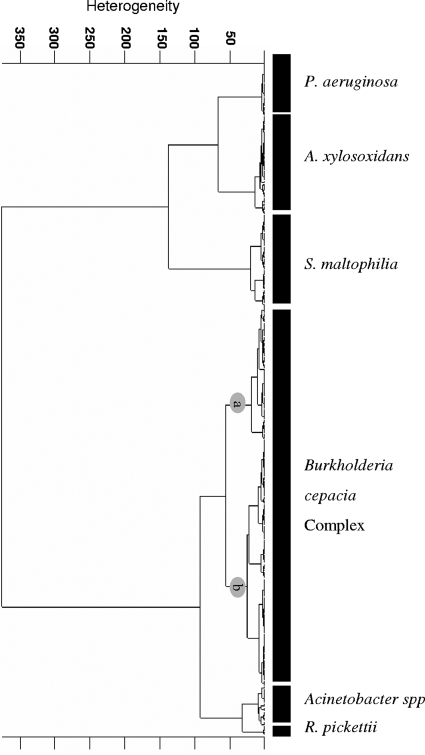
HCA was applied in an attempt to develop a discrimination model able to differentiate six groups of nonfermentative bacteria isolated from CF patients (P. aeruginosa, R. pickettii, A. xylosoxidans, Acinetobacter spp., S. maltophilia, and BCC bacteria). The selection of the best combination of spectral windows that produced the desired differentiation was performed with the training data set. The spectral windows 3,000 to 2,840, 1,200 to 900, and 1,400 to 1,250 cm−1 were found to contribute maximally to the discrimination of the aforementioned groups of bacteria. By using the information encoded in these spectral ranges as input data for the calculation of spectral distances and cluster analysis, a dendrogram was obtained (Ward's algorithm and OPUS software [Bruker Optics GmbH]), as shown in Fig. 4. This dendrogram shows a clear discrimination among the six different groups of bacteria analyzed. Although BCC bacteria were not discriminated to the species level, two main clusters (clusters a and b) were obtained. Interestingly, cluster a mainly contained the reference strains belonging to genomovars I to III, V to VII, and IX, while cluster b contained the clinical isolates and reference strains belonging to genomovars IV and VIII. These findings might be due to the fact that BCC bacteria, which are generally involved in chronic infection of the lungs of CF patients, where they live in biofilms, have a phenotypic evolution different from that of the reference strains (18, 22, 53). The performance of HCA for the discrimination of P. aeruginosa, R. pickettii, A. xylosoxidans, Acinetobacter spp., S. maltophilia, and BCC was tested with the external validation data (test data set), with the results for 97.3% of the strains being in the correct cluster. One A. xylosoxidans clinical isolate, one S. maltophilia clinical isolate, and three B. cenocepacia clinical isolates failed to cluster with the corresponding group and were thus misidentified. In the cases of A. xylosoxidans and S. maltophilia, this result could be due to the fact that the number of strains used was still not enough to cover the phenotypic variance within this group of bacteria. Similar results for optimization of the classification system were previously shown by Rebuffo et al. (45).
FIG. 4.
FTIR-based dendrogram of results of HCA of gram-negative rods obtained by using the first-derivative spectra in spectral windows of 3,000 to 2,840, 1,200 to 900, and 1,400 to 1,250 cm−1. The calculation was done on the basis of the spectral distances and the Ward algorithm. BCC bacteria are grouped in two clusters: cluster a mainly contained reference strains belonging to genomovars I to III, V to VII, and IX, while cluster b contained the clinical isolates and the reference strains belonging to genomovars IV and VIII.
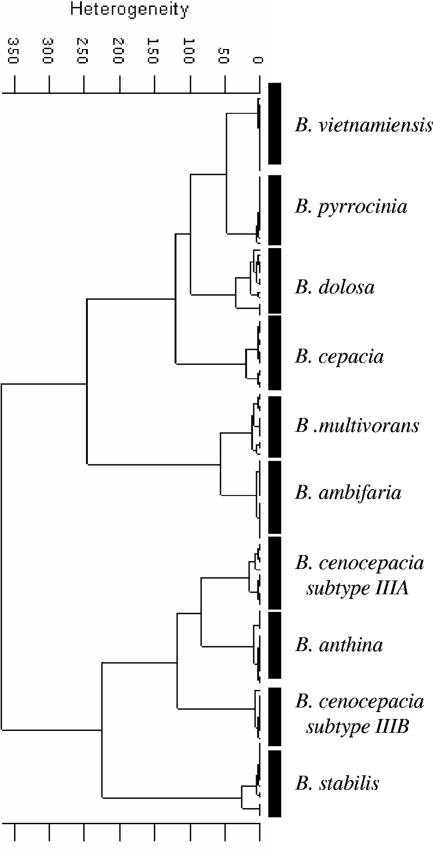
The next step in the identification procedure was to test if another specific combination of spectral windows for cluster analysis different from the one used for differentiation to the genus level would be more appropriate for discrimination of the BCC bacteria to the species level. With this aim, the 11 BCC reference strains listed in Table 1 were used in an HCA based on the spectral information contained in four different windows (1,200 to 900, 1,450 to 1,350, 1,330 to 1,250, and 3,000 to 2,800 cm−1) to discriminate among the nine species of BCC and the two subtypes of B. cenocepacia. The dendrogram displayed in Fig. 5 exhibits the clear discrimination of the nine species of BCC and the two subtypes of B. cenocepacia, which belonged to lineages IIIA and IIIB. However, when this cluster analysis was challenged by use of the well-identified clinical isolates, clustering in two different distinct groups of clinical and reference strains was obtained. We tried the use of other spectral window combinations, but in all of our trials the main result was the discrimination of the clinical and the reference strains (data not shown). Thus, as was previously reported for closely related bacteria in other genera (45), HCA does not seem to consider optimally the appropriate spectral information encoded in the spectra for the correct discrimination.
FIG. 5.
HCA showing the discrimination of the nine genomovars of BCC and two B. cenocepacia subtypes (subtypes IIIA and IIIB). The dendrogram was obtained by using the Ward algorithm, vector-normalized first-derivative spectra, and the information contained in spectral windows of 1,200 to 900, 1,450 to 1,350, 1,330 to 1,250, and 3.000 to 2,800 cm−1. HCA clearly discriminated among the nine species of BCC and two B. cenocepacia lineages (lineages IIIA and IIIB). Only reference strains were included in the analysis. Three replicate spectra from four independent experiments with each strain were used.
Identification by ANN analysis.
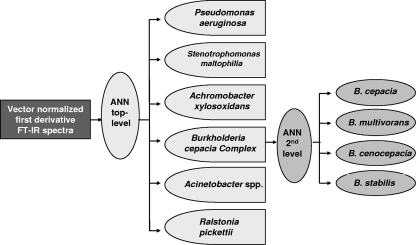
Taking into account the results described above, we applied ANN analysis, an advanced method based on so-called feature extraction techniques. With the aim of obtaining a clinically relevant identification system, this multivariate model was developed by considering the epidemiology of BCC. From the known occurrence of different BCC species in CF patients, it is evident that all species of this complex are potentially capable of colonizing the lungs of CF patients (11, 57). Nevertheless, studies of the distribution of BCC species in CF patients in more than 10 different countries, including the United States, Canada, France, Italy, and the United Kingdom, revealed that most isolates recovered form CF patients belong to the species B. cenocepacia, B. multivorans, B. cepacia, and B. stabilis (5, 10, 12, 16, 23, 54). The distribution of members of BCC in CF patients also often shows a highly disproportionate representation of species (13, 32). In this study, 84.3% of the isolates were B. cenocepacia, 9.4% were B. cepacia, 3.1% were B. stabilis, and 1.1% were B. multivorans (by species-specific recA PCR identification; Table 1). By taking into account the aforementioned BCC epidemiological data, a model based on ANNs was developed. The model was able to accurately differentiate not only the six groups of nonfermenting gram-negative rods normally isolated from sputum samples from CF patients but also the four most clinically relevant species of BCC bacteria recovered from patients (genomovars I to IV). The use of a modular model, composed of two ANNs, allowed the optimization of each network with individual data preprocessing, feature selection, and network topology. These two independently developed modules were then integrated into one ANN-based classification system (49) (Fig. 6).
FIG. 6.
Schematic diagram of the classification system based on a modular ANN for the identification of nonfermentative gram-negative rods isolated from sputum samples from CF patients on the basis of FTIR spectra. On the first level of the ANN architecture, six groups of nonfermenting gram-negative rods (P. aeruginosa, S. maltophilia, A. xylosoxidans, BCC, R. pickettii, and Acinetobacter spp.) are identified. In the second level of the classification scheme, the four most clinically relevant species of BCC bacteria (B. cepacia, B. multivorans, B. cenocepacia, and B. stabilis) are discriminated. These two independently developed modules were integrated into one ANN-based classification system.
As described in the Material and Methods section, the database used to develop this modular ANN system, which contained almost 2,000 first-derivative spectra for 169 clinical isolates and 15 reference strains, was divided into two subsets: (i) the training data set and (ii) the test data set, which contained the spectra for the clinical isolates that were not included in the training data set and that were exclusively used for external validation. The spectra for two isolates (isolates HST 8684 and HC BC05; Table 1) that were identified by molecular biology only to the genus level but that were identified as belonging to BCC were not used for training but were included in the test data set.
The optimal spectral features selected for use for the training of the top-level ANN was the most discriminative 60 wavelengths, two hidden units, and six output neurons. A fully connected feed-forward top-level ANN (Fig. 6) was trained by use of the Rprop algorithm (55). For the second-level ANN, 15 wavelengths with the highest potential to discriminate between the BCC species were selected, and two hidden neurons were optimized for the four output neurons. An internal validation of the ANN-based FTIR identification model developed was performed. For this, individual spectra of each strain were randomly selected from the training data set (131 isolates, including reference strains; Table 3) and tested for the correctness of the identification. In this internal validation, a 100% correct identification rate was obtained for all species tested (data not shown). When a spectrum belonging to a gram-negative nonfermentative rod isolated from a sputum sample was tested with this modular ANN model, a first step was used to check whether it was identified as a strain belonging to the species P. aeruginosa, S. maltophilia, A. xylosoxidans, BCC, R. pickettii, or Acinetobacter spp. by using the top-level ANN, as shown schematically in Fig. 6. If the identification output at this level resulted in a member of BCC, the second-level ANN of the modular system was used to assign the spectra to B. cepacia, B. multivorans, B. cenocepacia, or B. stabilis.
TABLE 3.
Identification results for modular ANNs obtained with the training data set and the test data set for external validation
| Net level and microorganism | No. (%) of isolatesa
|
||||
|---|---|---|---|---|---|
| Total | Training data set (training/internal validation)b | Test data set (external validation)c | Correct identification | No identification | |
| Top-level netd | |||||
| P. aeruginosa | 17 | 12 | 5 | 5 (100) | |
| BCC | 107 | 78 | 29 | 29 (100) | |
| S. maltophilia | 26 | 18 | 8 | 8 (100) | |
| A. xylosoxidans | 21 | 15 | 6 | 5 (83.3) | 1 (16.7) |
| R. pickettii | 2 | 2 | 2 (100) | ||
| Acinetobacter spp. | 11 | 8 | 3 | 3 (100) | |
| Total | 184 | 131 | 53 | 52 (98.1) | 1 (1.9) |
| Second-level nete | |||||
| B. cepacia | 10 | 7 | 3 | 3 (100) | |
| B. multivorans | 3 | 2 | 1 | 1 (100) | |
| B. cenocepacia | 83 | 58 | 25 | 23 (92) | 2 (8) |
| B. stabilis | 4 | 3 | 1 | 1 (100) | |
| BCCf | 2 | 2 | 2 (100) | ||
| Total | 102 | 70 | 32 | 30 (93.8) | 2 (6.2) |
There were no misidentifications.
The training data set contained 131 isolates (including reference strains) and was used for training and internal validation. The internal validation, which was performed with the 131 isolates (including reference strains) from the training data set, yielded 100% correct identification for all the species.
The test data set included 53 clinical isolates and was used for external validation.
NeuroDeveloper software (version 2.3; Synthon KG) was used to develop and optimize a modular system. The whole database used contained almost 2,000 first-derivative spectra obtained from 169 clinical isolates and 15 reference strains. The topology for the top-level net was 60 wavelengths, two hidden units, and six output neurons.
The topology for the second-level net was 15 wavelengths and two hidden neurons optimized for the four output neurons.
BCC corresponded to two clinical isolates (isolates HST 8684 and HC BC05) that were identified as belonging to BCC by recA PCR with primers BCR1 and BCR2 but that could not be identified by PCR with species-specific primers.
As an objective test of the performance of the model, an external validation was carried out with the first-derivative spectra for the 53 clinical isolates included in the test data set; the spectra for these isolates were not used for training or for internal validation. As indicated in Table 3, the top-level ANN did not identify only one A. xylosoxidans isolate. Interestingly, two strains (strains HST 8684 and HC BC05; Table 1) that could be identified only by molecular biology techniques as BCC strains were discriminated as belonging to BCC by the top-level network but could not be assigned to any of the BCC species by the second-level ANN (Table 3). The excellent overall performance of the modular ANN model was characterized by a 98.1% correct identification rate at the top level of the ANN and a 93.8% correct identification rate at the second level of the ANN. It should be emphasized that the ANN-based FTIR model developed here represents a system that can be used for the precise identification of P. aeruginosa, R. pickettii, A. xylosoxidans, Acinetobacter spp., S. maltophilia, and members of BCC. Additionally, it is able to discriminate BCC isolates to the species level and, in particular, can discriminate the four species that cause the majority of infections in CF patients (B. cepacia, B. multivorans, B. cenocepacia, and B. stabilis).
Chronic lung infections caused by the opportunistic pathogen that occur in CF patients are associated with extensive genetic adaptations and evolution of the infecting bacteria; they may evolve in response to selection pressure from the host environment and, probably, from other members present in the bacterial community (18, 22, 53). Therefore, our identification system, like other phenotypic and genotypic identification methodologies, requires continuous updating. However, the ANN-based FTIR technique has the advantage that it easily allows new isolates of the BCC species to be added to the database.
To extend phenotypic identification to most of the clinically relevant nonfermenting gram-negative bacilli isolated from CF patients, the present database will include further related species in the future, particularly those belonging to the genera Achromobacter, Acinetobacter, and Ralstonia, although they occur at very low frequencies (9).
It is important to remark that our preliminary results suggest that the use of FTIR in combination with ANNs is able to discriminate different phenotypes among isolates that are positive by B. cenocepacia-specific PCR. This difference in discrimination power would be assigned to the relatively low capacity of RLFP and BCC species-specific recA PCR to fully resolve the diversity within BCC compared with that of FTIR. We therefore consider that the use of the methodology based on FTIR developed in the present study, with the assistance of more powerful identification techniques, such as sequence-based identification schemes like MLST (1, 14), would represent an important tool that could be used to solve ambiguous results when newly emerging species of BCC are detected in CF patients.
Conclusions.
The accurate identification of pathogens from the respiratory tract of CF patients is crucial to ensure appropriate treatment and infection control. Among these organisms, the taxonomy and identification of members of the genus Burkholderia have been always very complex. In the present study, we illustrate for the first time the enormous potential of FTIR combined with ANN methodologies for the rapid and accurate identification of six of the most clinically relevant gram-negative nonfermenting bacilli isolated from sputum samples from CF patients (P. aeruginosa, R. pickettii, A. xylosoxidans, Acinetobacter spp., S. maltophilia, and bacteria belonging to BCC) and the discrimination of the four BCC species that occur at high frequencies in CF patients (B. cepacia, B. multivorans, B. cenocepacia, and B. stabilis).
We have put forward simple strategies that can be used to overcome experimental problems resulting from spectral interference with PHB and the different types of pilus and/or flagellar appendages produced by gram-negative rods, particularly when they are isolated from sputum samples from CF patients. The proposed approach represents a significant advance in the identification of gram-negative nonfermenting bacilli for routine analysis.
As opposed to other methods, the method developed in the present study requires small quantities of cells and minimal sample preparation, is inexpensive, and can lead to identification results within a few hours. In the future, it will be further developed to enlarge the database with data for a wider range of microbial species, as well as larger numbers of strains and isolates of each species. The new identification system proved to be reliable, rapid, and precise for practical use for the differentiation and identification of nonfermenting gram-negative bacteria isolated from sputum samples from CF patients collected for routine clinical detection.
Acknowledgments
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (grant BID 1728/OC-AR-PICT 34836), the Comisión de Investigaciones Científicas Gobierno de la Provincia de Buenos Aires (CIC-PBA), and Buenos Aires University (grant UBACYT B082). A. Bosch is member of the CIC PBA, and A. Miñán received a fellowship from CONICET.
We are grateful to Laura Galanternik for kindly providing us the BCC reference strains. We thank Julio Figari for excellent technical assistance.
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. J. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 434665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch, A., M. A. Golowczyc, A. Abraham, G. Garrote, G. De Antoni, and O. Yantorno. 2006. Rapid discrimination of lactobacilli isolated from kefir grains by FT-IR spectroscopy. Int. J. Food Microbiol. 111280-287. [DOI] [PubMed] [Google Scholar]
- 3.Bosshard, J. P. P., R. Zbinden, S. Abels, B. Böddinghaus, M. Altwegg, and E. C. Böttger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 441359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brimer, C. D., and T. C. Montie. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 1803209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisse, S., C. Cordevant, P. Vandamme, P. Bidet, C. Loukil, G. Chabanon, M. Lange, and E. Bingen. 2004. Species distribution and ribotype diversity of Burkholderia cepacia complex isolates from French patients with cystic fibrosis. J. Clin. Microbiol. 424824-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 27626479-26485. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., E. Falsen, B. Hoste, M. Ohlén, J. Goris, J. R. W. Govan, M. Gillis, and P. Vandamme. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pmomenusa sp. nov., Pandoraea sputorum sp. nov., and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50887-899. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., E. Falsen, M. Vancanneyt, B. Hoste, J. R. W. Govan, K. Kersters, and P. Vandamme. 1999. Classification of some Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int. J. Syst. Bacteriol. 49405-413. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., T. Spilker, R. Reik, P. Vandamme, and J. J. LiPuma. 2005. Use of PCR analyses to define the distribution of Ralstonia species recovered from patients with cystic fibrosis. J. Clin. Microbiol. 433463-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. [DOI] [PubMed] [Google Scholar]
- 11.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 393427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunha, M. V., J. H. Leitão, E. Mahenthiralingam, P. Vandamme, L. Lito, C. Barreto, M. J. Salgado, and I. Sa-Correia. 2003. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a seven-year study. J. Clin. Microbiol. 414113-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunha, M. V., A. Pinto-de-Olivera, L. Meirinhos-Soares, M. J. Salgado, J. Melo-Cristino, S. Correia, C. Barreto, and I. Sa-Correia. 2007. Exceptionally high representation of Burkholderia cepacia among B. cepacia complex isolates recovered from the major Portuguese cystic fibrosis center. J. Clin. Microbiol. 451628-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmastri, C., A. Baldwin, S. Tabacchioni, A. Bevivino, E. Mahenthiralingam, L. Chiarini, and C. Dowson. 2007. Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ. Microbiol. 91632-1639. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira-Garcia, D., M. B. Martinez, M. Dall'Agnol, M. Rosales, A. C. G. S. Azzuz, and J. A. Girón. 2002. Characterization of flagella produced by clinical strains of emerging opportunistic pathogen Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detsika, M. G., J. E. Corkill, M. Magalhäes, K. J. Glendinning, C. A. Hart, and C. Winstenley. 2003. Molecular typing of and distribution of genetic markers among Burkholderia cepacia complex isolates from Brazil. J. Clin. Microbiol. 414148-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferroni, A., I. Sermet-Gaudelus, I. E. Abachin, G. Quesne, G. Lenoir, P. Berche, and J. L. Gailard. 2002. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J. Clin. Microbiol. 403793-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fux, C. A., M. Shirtliff, P. Stoodley, and J. W. Costerton. 2005. Can laboratory reference strains mirror ‘real-world’ pathogenesis? Trends Microbiol. 1359-63. [DOI] [PubMed] [Google Scholar]
- 19.Ghol, O., A. Friedrich, M. Hoppert, and B. Averhoff. 2006. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl. Environ. Microbiol. 721394-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilligan, P. H. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 435-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein, R., L. Sun, U. Jiang, U. Sajjan, J. F. Forstiner, and C. Campanelli. 1995. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia. J. Bacteriol. 1771039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison, F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153917-923. [DOI] [PubMed] [Google Scholar]
- 23.Heath, D., G. Hohneker, C. Carriker, K. Smith, J. Routh, J. J. LiPuma, R. M. Aris, D. Weber, and P. H. Gilligan. 2002. Six-year molecular analysis of Burkholderia cepacia complex isolates among cystic fibrosis patients at a referral center for lung transplantation. J. Clin. Microbiol. 401188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helm, D., H. Labischinsky, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 13769-79. [DOI] [PubMed] [Google Scholar]
- 25.Helm, D., and D. Naumann. 1995. Identification of some bacterial cell components by FT-IR spectroscopy. FEMS Microbiol. Lett. 12675-80. [Google Scholar]
- 26.Henry, D. A., E. Mahenthiralingam, P. Vandamme, J. Coenye, and D. P. Speert. 2001. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 391073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isenberg, H. 2004. Clinical microbiology procedures handbook. ASM Press, Washington, DC.
- 28.Kessler, B., and N. J. Palleroni. 2000. Taxonomic implications of synthesis of poly-β-hydroxybutyrate and other poly-β-hydroxyalkanoates by aerobic pseudomonads. Int. J. Syst. Evol. Microbiol. 50711-713. [DOI] [PubMed] [Google Scholar]
- 29.Kirschner, C., K. Maquelin, P. Pina, N. A. Ngo Thi, L. P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Manfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopy study. J. Clin. Microbiol. 391763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiska, D., and P. H. Gilligan. 2003. Pseudomonas, p. 719-728. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 31.Lima, T. C. S., M. G. Breno, and C. M. Bonato. 1999. Bacteria isolated from sugarcane agroecosystem: their potential production of polyhydroxyalcanoates and resistance to antibiotics. Rev. Microbiol. (Brazil) 30214-224. [Google Scholar]
- 32.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 16492-96. [DOI] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 383165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3144-156. [DOI] [PubMed] [Google Scholar]
- 35.Maquelin, K., L. P. Choo-Smith, C. Kirschner, N. A. Ngo Thi, D. Naumann, and G. Puppels. 2002. Vibrational spectroscopic studies of microorganisms, p. 1-27. In J. M. Chalmers and P. R. Griffiths (ed.), Handbook of vibrational spectroscopy. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 36.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. A. Ngo Thi, T. van Vreeswijk, M. Stämbler, H. P. Endtz, H. A. Bruining, D. Naumann, and G. Puppels. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, M. B., and P. H. Gilligan. 2003. Laboratory aspects of management of chronic pulmonary infections in patients with cystic fibrosis. J. Clin. Microbiol. 414009-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naumann, D. 2000. Infrared spectroscopy in microbiology, p. 102-131. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry: applications, theory, and instrumentation. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 39.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy Nature 35181-82. [DOI] [PubMed] [Google Scholar]
- 40.Naumann, D. 2001. FT-infrared and FT-Raman Spectroscopy in biomedical research, p. 323-377. In H.-U. Gremlich and B. Yang (ed.), Infrared and Raman spectroscopy of biological material. Marcel Dekker, Inc., New York, NY.
- 40a.NCCLS. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 41.Nichols, P., J. Henson, J. Guckert, D. Nivens, and D. White. 1985. Fourier transform-infrared spectroscopic methods for microbial ecology: analysis of bacteria-polymer mixtures and biofilms. J. Microbiol. Methods 479-94. [DOI] [PubMed] [Google Scholar]
- 42.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 37832-38. [DOI] [PubMed] [Google Scholar]
- 43.Payne, G. W., P. Vandamme, S. H. Morgan, J. J. LiPuma, T. Coenye, A. J. Wightman, T. H. Jones, and E. Mahenthiralingam. 2005. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 713917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prieto, C. I., M. E. Rodriguez, A. Bosch, F. G. Chirdo, and O. M. Yantorno. 2003. Whole-bacterial cell enzyme-linked immunosorbent assay for cell-bound Moraxella bovis pili. Vet. Microbiol. 91157-168. [DOI] [PubMed] [Google Scholar]
- 45.Rebuffo, C., J. Schmitt, M. Wenning, F. von Stetten, and S. Scherer. 2006. Reliable and rapid identification of Listeria monocytogenes and Listeria species by artificial neural network-based Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 72994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saiman, L., Y. Chen, S. Tabbi, P. San Gabriel, J. Zhou, L. Liu, L. Lai, and S. Whittier. 2001. Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J. Clin. Microbiol. 393942-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 1757-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Schmitt, J., and T. Udelhoven. 2001. Use of artificial neural networks in biomedical diagnosis, p. 379-419. In H.-U. Gremlich and B. Yang (ed.), Infrared and Raman spectroscopy of biological material. Marcel Dekker, Inc., New York, NY.
- 50.Schreckenberger, P., M. Daneshvar, R. Weyant, and D. Hollis. 2003. Acinetobacter, Achromobacter, Chryseobacterium, Moraxella, and other nonfermentative gram-negative rods, p. 749-779. In P. R. Murray, E. J. Baron, J. M. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 51.Seo, S.-T., and K. Tsuchiya. 2005. Genotypic characterization of Burkholderia cenocepacia strains by rep-PCR and PCR-RFLP of the fliC gene. FEMS Microbiol. Lett. 24519-24. [DOI] [PubMed] [Google Scholar]
- 52.Serra, D., A. Bosch, D. Russo, M. E. Rodriguez, A. Zorreguieta, J. Schmitt, D. Naumann, and O. Yantorno. 2007. Continuous non-destructive monitoring of Bordetella pertussis biofilms by spectroscopy and other corroborative techniques. Anal. Bioanal. Chem. 3871759-1767. [DOI] [PubMed] [Google Scholar]
- 53.Smith, E. E., D. G. Buckley, Z. Wu, C. Saemphimmachak, L. R. Hoffman, D. A. D′Argenio, S. I. Miller, B. W. Ramsey, and D. P. Speert. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038305-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udelhoven, T., D. Naumann, and J. Schmitt. 2000. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 541471-1479. [Google Scholar]
- 56.Vandamme, P., B. Holmes, J. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 15491-96. [DOI] [PubMed] [Google Scholar]
- 57.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 471188-1200. [DOI] [PubMed] [Google Scholar]
- 58.Vermis, K., J. Coenye, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51937-940. [DOI] [PubMed] [Google Scholar]
- 59.Weisbourg, W., S. Barns, D. Pelletier, and D. Lane. 1991. 16S DNA amplification for phylogenetic study. J. Bacteriol. 173697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, J., E. Garber, M. Desai, and L. Saiman. 2006. Compliance of clinical microbiology laboratories in the United States with current recommendations for processing respiratory tract specimens from patients with cystic fibrosis. J. Clin. Microbiol. 441547-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]