Abstract
Real-time PCR is a powerful tool for the detection and typing of herpes simplex virus (HSV). HSV types 1 and 2 can be distinguished by using the differences in the melting temperatures (Tms) of the hybridization probes. The efficacy of Tm profiling with low DNA concentrations was evaluated in this study.
Herpes simplex virus (HSV) type 1 (HSV-1) and HSV-2 are common viral pathogens that cause superficial mucocutaneous lesions that can progress to systemic infections. Typically, the clinical manifestations of HSV-1 infection include orofacial blisters and dermal lesions, whereas both HSV-1 and HSV-2 are associated with genital lesions (10, 11). Accurate genotyping can facilitate the counseling of patients with HSV infections, as the two subtypes behave differently in terms of their shedding and reoccurrence (9, 10). HSV-1 and HSV-2 are also known to disseminate to the central nervous system, leading to encephalitis and meningitis (10, 11). Since the prompt administration of antiviral drugs can improve the prognosis, the rapid detection in cerebrospinal fluid (CSF) is of utmost importance (3, 8, 10).
The isolation and discrimination of HSV-1 from HSV-2 was historically performed by using virus culture and immunofluorescence; however, molecular typing of HSV by real-time PCR has become the new “gold standard” due to its speed, sensitivity, and accuracy (1, 2). For example, commercial kits designed for use on the Roche Diagnostics (Mannheim, Germany) LightCycler platform can simultaneously detected and differentiate HSV-1 from HSV-2. By exploiting nucleotide polymorphism differences between genes encoding HSV-1 and HSV-2 DNA polymerase I, melting-curve analysis of fluorogenic hybridization probes can be used to distinguish HSV-1 from HSV-2. Recently, the detection and subtyping of HSV from CSF on the LightCycler platform have been validated for clinical use (3). However, following the implementation of LightCycler PCR in the molecular epidemiology laboratory at the Queen Elizabeth II Health Sciences Center (Halifax, Nova Scotia, Canada), atypical melting-curve profiles were occasionally observed for specimens containing low viral titers. In 2007, approximately 3,000 clinical specimens were analyzed, with 121 displaying atypical melting-curve profiles. The majority were linked to known sequence variations in the probe binding site (3a, 7). However, 14 had wild-type HSV-2 sequences yet still displayed atypical melting-temperature (Tm) profiles. Furthermore, their crossing-point (Cp) values ranged from 33 to 40, suggesting that atypical melting-curve profiles may result from low viral titers. In this study, the performance characteristics of Tm profiling with low concentrations of HSV-1 and HSV-2 DNA was investigated.
The detection of HSV was performed with a LightCycler HSV 1/2 detection kit (Roche Diagnostics, Mannheim, Germany). PCR amplifications were performed on a LightCycler (version 2.0) thermocycler, and Tm values were determined with the software provided by the manufacturer. The template DNA consisted of an HSV-1- and HSV-2-positive control (a mixture containing HSV-1 and HSV-2) provided with the kit, in-house-generated plasmid-borne HSV-1- and HSV-2-positive controls (described below), or DNA extracted from HSV-1- and HSV-2-positive clinical specimens. Extractions were performed with 200-μl samples and a MagNA Pure total nucleic acid isolation kit on an automated MagNA Pure LightCycler instrument (Roche Diagnostics, Mannheim, Germany), and RNA/DNA was eluted in a final volume of 100 μl.
Plasmid controls were generated by PCR amplification with template HSV-1 and HSV-2 DNA extracted from clinical isolates of HSV-1 and HSV-2 that had previously been identified with the LightCycler HSV 1/2 detection kit, viral culture and immunofluorescence, and a combination of PCR and restriction endonuclease digestion (5). PCR amplification was on an MJ Research PTC 200 thermal cycler (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada) by using the following parameters: 94°C for 5 min and 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 60 s, with a final extension at 72°C for 5 min. Reactions with 50-μl reaction mixtures were performed with an Expand high-fidelity PCR system (Roche Diagnostics, Laval, Quebec, Canada), according to the instructions provided by the manufacturer. A modified primer pair, which consisted of primer FESPYXHO (5′-CTGCTCGAGTGCGAAAAAACGTTC-3′) and primer RESPYBAM (5′-CTGGGATCCGGGGCGCTCGGCTAAC-3′), which target the DNA polymerase gene (2), was synthesized by Sigma Genosys (Oakville, Ontario, Canada). The 226-bp amplicon was digested with BamHI and XhoI and ligated into similarly digested pBlueScript KS (Strategene), and the resulting constructs were transferred into Escherichia coli DH5α by electroporation (6). After overnight growth at 37°C on Luria-Bertani agar containing ampicillin (100 μg/ml), transformants were confirmed by real-time PCR and conventional PCR with the primer pair described above. Plasmids were purified from PCR-positive transformants by using a QIAprep Spin Miniprep kit (Qiagen, Mississauga, Ontario, Canada) and were used to estimate the copy number in clinical specimens. Briefly, BamHI-digested plasmid-borne HSV-1- and HSV-2-positive controls were subjected to 1% agarose gel electrophoresis and purified in 10-μl volumes with a MinElute PCR purification kit (Qiagen). Linearized plasmids were quantified by spectrophotometry and were serially diluted 10-fold in the elution buffer provided in the MagNA Pure total nucleic acid extraction kit. Diluted DNA was subjected to real-time PCR analysis with the LightCycler HSV 1/2 detection kit (Roche Diagnostics, Mannheim, Germany); this allowed the Cp values to be related to their corresponding copy numbers. The standard curve spanned a dynamic range of 1 to 107 copies.
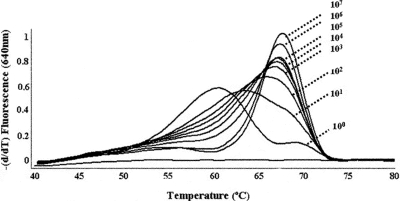
Following real-time PCR amplification of DNA extracted from diluted HSV-1 clinical specimens (oral, genital, and CSF specimens), no significant differences in Tm (ΔTm) were observed (Table 1, sample 1). Similarly, HSV-2-positive specimens with DNA concentrations of ≥66 copies displayed melting-curve profiles and Tm values that resembled those for the positive control. In contrast, a reproducible and significant decrease in Tm values was observed with lower DNA concentrations (≤55 copies) for both plasmid-borne HSV-2 (Fig. 1) and HSV-2-positive clinical specimens (Table 1, samples 2 to 6). A consistent trend was observed among the HSV-2-positive specimens obtained from various sources (CSF, genital, and oral specimens). When 10-fold serial dilutions of DNA extracted from several HSV-2-positive clinical specimens were evaluated (Table 1, samples 2 to 6), all specimens with Cp values above 33 (corresponding to ≤55 copies of viral DNA) displayed a marked decrease in Tm values compared to the Tm value for the positive control provided with the kit. Furthermore, HSV-2 dilutions with Cp values of ≥36 not only displayed a shift in Tm values but also no longer displayed the characteristic bell-shaped melting curve; therefore, the genotype could not be resolved by the instrument's software. At these concentrations of HSV-2 DNA (1 to 10 copies), the Tm values fell outside the acceptable range (±2.5°C) recommended by the manufacturer. It should be noted that a similar range in sensitivity (20 genomic copies) was reported by others (2); however, no mention of a shift in Tm values at low concentrations of HSV-2 DNA were reported. The data provided in this report demonstrate that low viral titers (≤55 copies; Cp values, ≥33) result in atypical melting-curve profiles. This may be of particular concern for specimens of poor quality or specimens collected during a period with low levels of viral shedding.
TABLE 1.
Representative effects of low viral titers on Tm values of HSV-specific probes
| Sample no. (type) | Dilution | Copy no.a | Cp | Genotype | Tm | ΔTm |
|---|---|---|---|---|---|---|
| 1 (CSF) | 100 | 50,679 | 21.70 | HSV-1 | 53.33 | −0.90 |
| 10−1 | 3,320 | 25.67 | HSV-1 | 53.21 | −1.02 | |
| 10−2 | 369 | 28.87 | HSV-1 | 53.29 | −0.94 | |
| 10−3 | 42 | 32.04 | HSV-1 | 53.37 | −0.86 | |
| 10−4 | 3 | 35.79 | HSV-1 | 53.46 | −0.77 | |
| 2 (CSF) | 100 | 27283 | 23.97 | HSV-2 | 67.03 | 0.81 |
| 10−1 | 2,775 | 27.54 | HSV-2 | 66.47 | 0.25 | |
| 10−2 | 282 | 31.11 | HSV-2 | 66.18 | −0.04 | |
| 10−3 | 53 | 33.72 | UKNb | 65.70 | −0.52 | |
| 10−4 | 3 | 38.57 | UKN | 61.52 | −4.70c | |
| 3 (Genital, penis) | 100 | 4,602 | 26.75 | HSV-2 | 66.67 | 0.45 |
| 10−1 | 423 | 30.48 | HSV-2 | 66.40 | 0.18 | |
| 10−2 | 35 | 34.37 | HSV-2 | 65.91 | −0.31 | |
| 10−3 | 4 | 37.84 | UKN | 63.67 | −2.55c | |
| 10−4 | 1 | >40.00 | UKN | 61.02 | −5.20c | |
| 4 (Genital, vulva) | 100 | 4,235 | 26.88 | HSV-2 | 66.56 | 0.34 |
| 10−1 | 386 | 30.62 | HSV-2 | 66.16 | −0.06 | |
| 10−2 | 46 | 33.95 | UKN | 65.77 | −0.45 | |
| 10−3 | 9 | 36.48 | UKN | 63.01 | −3.21c | |
| 5 (Oral, lip) | 100 | 2,176 | 27.92 | HSV-2 | 66.40 | 0.18 |
| 10−1 | 323 | 30.90 | HSV-2 | 66.10 | −0.12 | |
| 10−2 | 56 | 33.63 | UKN | 65.69 | −0.53 | |
| 10−3 | 1 | >40.00 | UKN | 60.07 | −6.15c | |
| 6 (Oral, tongue) | 100 | 381 | 30.64 | HSV-2 | 66.17 | −0.05 |
| 10−1 | 66 | 33.37 | HSV-2 | 65.36 | −0.86 | |
| 10−2 | 9 | 36.48 | UKN | 63.09 | −3.13c |
The copy number was estimated in relation to the copy number on a standard curve generated with plasmid-borne controls.
UKN, unknown; the software was unable to genotype the specimens.
Compared to the Tm values for the HSV-1- or HSV-2-positive control, the ΔTm values were above the acceptable variation (±2.5°C) recommended by the manufacturer.
FIG. 1.
Effect of low DNA concentrations on the melting-curve profile of HSV-2. PCRs were performed with template DNA that consisted of linearized plasmid containing the LightCycler HSV-2 target. The copy numbers ranged from 100 to 107. The profiles are representative not only of plasmid-borne HSV-2 but also of diluted HSV-2 clinical specimens.
To ensure that the decreased Tm values were not caused by mutations in the probe binding site (3a, 7), HSV-2 dilutions displaying a shift in Tm were concentrated with the MinElute PCR purification kit (Qiagen) and subjected to conventional and real-time PCRs. The amplicons generated by conventional PCR were submitted for DNA sequencing (Dalgen Microbial Genetics Centre, Halifax, Nova Scotia, Canada). The DNA sequences were identical to the DNA sequence of wild-type HSV-2. Interestingly, when concentrated HSV-2 DNA was subjected to real-time PCR analysis, specimens regained the typical bell-shaped profiles and the Tm values were restored to levels comparable to the level for the control provided with the kit. These data suggest that the shifts in Tm not attributed to sequence variations in the HSV-2 target were attributed to low concentrations of viral DNA.
Several hypotheses were proposed to explain the decreased Tm values obtained with low concentrations of HSV-2. First, since HSV-2 has a higher Tm than HSV-1, the probe binding sites likely have a higher G+C content, and therefore, more hydrogen bonds would be required for complete annealing. With low concentrations of HSV-2, the time required for the probes to find their complementary target may be insufficient under the conditions suggested by the manufacturer. The shifted Tm values could thus be explained by incomplete annealing, in which case the probes would melt at a lower temperature. To evaluate whether the shift in Tm of diluted HSV-2 was attributed to improper annealing, an additional step of annealing for 1 min at 40°C was added prior to melting-curve analysis. The increased annealing period did not resolve the changes in the Tm values.
Second, pH or salt concentrations may have a greater influence on probe binding to HSV-2 DNA than their influence on binding to HSV-1 DNA. The effect of the salt concentration or pH may be masked at high concentrations of HSV-2 DNA but may be significant at low concentrations. The composition of the elution buffer provided in the MagNA Pure LightCycler total nucleic acid isolation kit is proprietary; however, the kit insert states that DNA/RNA can be eluted with an alternative buffer consisting of 10 mM Tris, pH 8.0. When the results for HSV-2-positive specimens diluted in MagNA Pure elution buffer were compared to those for the same samples diluted in Tris buffer or water alone (Table 2), Tm shifts and genotyping failures were once again observed with low concentrations of HSV-2. Therefore, three commonly used elution buffers demonstrated the same problematic HSV-2 profiles at low viral titers.
TABLE 2.
Effects of diluent and UNG on Tm values of HSV-2-specific probes
| Diluent | Dilution | Copy no.a | Cp | Genotype | Tm | ΔTm |
|---|---|---|---|---|---|---|
| H2O (0.5 U UNG) | 10−1 | 107 | 32.63 | HSV-2 | 66.33 | −0.20 |
| 10−2 | 5 | 37.54 | UKN | 59.55 | −6.98c | |
| 10−3 | 2 | 39.44 | UKN | 56.46 | −10.07c | |
| H2O (no UNG) | 10−1 | 93 | 32.81 | HSV-2 | 66.15 | −0.38 |
| 10−2 | 18 | 35.42 | UKN | 61.07 | −5.46c | |
| 10−3 | 2 | 39.36 | UKN | 56.38 | −10.15c | |
| 10 mM Tris, pH 8.0 (0.5 U UNG) | 10−1 | 95 | 32.82 | HSV-2 | 66.82 | 0.29 |
| 10−2 | 8 | 36.59 | UKN | 60.70 | −5.83c | |
| 10−3 | 1 | >40.00 | UKN | 59.25 | −7.28c | |
| 10 mM Tris, pH 8.0 (no UNG) | 10−1 | 75 | 33.15 | HSV-2 | 66.63 | 0.10 |
| 10−2 | 13 | 35.92 | UKN | 61.24 | −5.29c | |
| 10−3 | 1 | >40.00 | UKN | 54.91 | −11.62c | |
| EB,b pH 8.0 (0.5 U UNG) | 10−1 | 93 | 32.85 | HSV-2 | 66.63 | 0.10 |
| 10−2 | 8 | 35.40 | UKN | 60.98 | −5.55c | |
| 10−3 | 1 | >40.00 | UKN | 54.91 | −11.62c | |
| EB, pH 8.0 (no UNG) | 10−1 | 77 | 32.55 | HSV-2 | 66.83 | 0.30 |
| 10−2 | 3 | 35.79 | UKN | 60.87 | −5.66c | |
| 10−3 | 2 | 39.22 | UKN | 59.65 | −6.88c |
The copy number was estimated in relation to the copy number on a standard curve generated with plasmid-borne controls.
EB, elution buffer provided with the MagNA Pure LightCycler total nucleic acid kit.
Compared to the Tm values for the HSV-1- or HSV-2-positive control, the ΔTm values were above the acceptable variation (±2.5°C) recommended by the manufacturer.
A final hypothesis to explain the Tm shifts for diluted HSV-2 could be the influence of uracil-N-glycosylase (UNG). Although UNG is known to decrease the Tm of HSV-2, its influence here is unlikely since UNG also decreases the Tm of HSV-1 (4). At low concentrations of viral DNA, only the HSV-2-positive specimens displayed altered Tm profiles. Furthermore, the effect of UNG was observed irrespective of the DNA concentration (4). Nonetheless, to exclude the influence of UNG, this enzyme was omitted from the assay (Table 2). No significant difference was observed with or without UNG at the concentrations suggested by the manufacturer.
In summary, Tm analysis may not be suitable for the genotyping of HSV-2 when the viral copy numbers are equivalent to or less than 56 (Cp value, ≥33). Failure to genotype HSV-2 due to low viral DNA concentrations would be problematic in a clinical setting, since additional diagnostic methods would be required, thus increasing sample turnaround times and costs. Although the cause of the apparent shifts in Tm remains elusive, melting-curve profiles and Tm values were restored to an acceptable range when the viral DNA was concentrated. It remains to be determined whether the observations from this study are target specific or are applicable to all hybridization probes or real-time PCR chemistries that permit melting-curve analyses.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Burrows, J., A. Nitsche, B. Bayla, E. Walker, G. Higgins, and T. Kok. 2002. Detection and subtyping of herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical setting by LightCycler PCR. J. Clin. Microbiol. 38795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson, K. E., B. D. Alexander, C. Woods, C. Petti, and L. B. Reller. 2007. Validation of laboratory screening criteria for herpes simplex virus testing of cerebrospinal fluid. J. Clin. Microbiol. 45721-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Issa, N. C., M. J. Espy, J. R. Uhl, and T. F. Smith. 2005. Sequencing and resolution of amplified herpes simplex virus DNA with intermediate melting curves as genotype 1 or 2 by LightCycler PCR assay. J. Clin. Microbiol. 431843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBlanc, J., J. Pettipas, S. Campbell, R. J. Davidson, and T. F. Hatchette. 2008. Uracil-DNA glycosylase (UNG) influences the melting temperature (Tm) herpes simplex virus (HSV) hybridization probes. J. Virol. Methods 151158-160. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg, F., and P. Lebon. 1991. Amplification and characterization of herpesvirus DNA in cerebrospinal fluid from patients with acute encephalitis. J. Clin. Microbiol. 292412-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 7.Stevenson, J., W. Hymas, and D. Hillyard. 2005. Effect of sequence polymorphisms on performance of two real-time PCR assays for detection of herpes simplex virus. J. Clin. Microbiol. 432391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyring, S. K., D. Baker, and W. Snowden. 2002. Valacyclovir for herpes simplex virus infection: long-term safety and sustained efficacy after 20 years' experience with acyclovir. J. Infect. Dis. 186(Suppl. 1)S40-S46. [DOI] [PubMed] [Google Scholar]
- 9.Warren, T., and C. Ebel. 2005. Counseling the patient who has genital herpes or genital human papillomavirus infection. Infect. Dis. Clin. N. Am. 19459-476. [DOI] [PubMed] [Google Scholar]
- 10.Whitley, R. J., C. A. Alford, M. S., Hirsch, R. T. Schooley, J. P. Luby, F. Y. Aoki, D. Hanley, A. J. Nahmias, and S. J. Soong. 1986. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N. Engl. J. Med. 314144-149. [DOI] [PubMed] [Google Scholar]
- 11.Whitley, R. J. 1990. Viral encephalitis. N. Engl. J. Med. 323242-250. [DOI] [PubMed] [Google Scholar]