Abstract
BK virus (BKV) is the infectious cause of polyomavirus-associated nephropathy. Screening guidelines for renal-transplant recipients define levels of viremia and viruria that are actionable for additional testing or intervention. However, standardized real-time PCR primers, probes, and standards are unavailable, and the extent of agreement among published assays is unknown. We compared seven TaqMan real-time PCR primer/probe sets (three designed at this institution, three described in the literature, and one purchased) in conjunction with two different standards to prospectively measure BKV titers in 251 urine specimens submitted to our clinical laboratory. We observed substantial disagreement among assays attributable both to features of primer and probe design and to choice of reference material. The most significant source of error among individual specimens was primer or probe mismatch due to subtype-associated polymorphisms, primarily among subtype III and IV isolates. In contrast, measurement of the most abundant subtypes (Ia, V, and VI) were typically uniform among all seven assays. Finally, we describe and validate a new clinical assay designed to reliably measure all subtypes encountered in our study population (Ia, Ic, III, IV, and VI). Consideration of available BKV sequence information in conjunction with details of subtype distribution allowed us to develop a redesigned assay with markedly improved performance. These results suggest that both accurate BKV measurement and the uniform application of BKV screening guidelines could be significantly improved by the use of standardized reference materials and PCR primers and probes.
BK virus (BKV) is a small, nearly ubiquitous DNA virus in the family Polyomaviridae that causes chronic, usually asymptomatic infection in immunocompetent individuals. Since the advent of potent antirejection therapy, BKV has emerged as the primary infectious cause of polyomavirus-associated nephropathy (PVAN), affecting 1 to 10% of renal-transplant recipients, and with frequencies of graft loss between 10% and 80% (7).
Quantitative assays for BKV in urine and serum are used as both screening and diagnostic tests for PVAN in renal-transplant recipients. Prospective studies of renal-allograft patients have demonstrated that high levels of BKV viruria precede sustained viremia, which in turn precedes evidence of allograft dysfunction (2). Furthermore, screening blood or urine for high levels of BKV replication in conjunction with interventions, such as reduction of immunosuppression (2) or treatment with cidofovir (12), triggered at defined levels of viremia or viruria has been associated with improved clinical outcomes.
Renal-transplant programs have instituted BKV screening protocols, and quantitative BKV testing is increasingly performed in molecular virology laboratories. In 2005, an expert panel recommended the use of either urine cytology or nucleic-acid-based testing to screen renal-transplant recipients during the first 2 years after transplantation who present with evidence of allograft dysfunction and accompanying renal biopsy (7). These guidelines included quantitative cutoffs for BKV loads that indicate the need for additional testing: urine DNA loads of >107 copies/ml or plasma DNA loads of >104 copies/ml that persist for more than 3 weeks constitute a diagnosis of “presumptive PVAN” and should be followed up with a renal biopsy.
BKV isolates have been grouped into subtypes, initially serologically (13), later using VP1 sequence data and restriction enzyme analysis (11), and most recently by phylogenetic analysis of full-genome viral DNA sequences (17, 27). Subtype designations have evolved over time. In this report, we refer to subtypes Ia, Ic, III, IV, V, and VI as described previously (27). According to this scheme, subtypes Ib1 and Ib2 (17) have been reclassified as VI and V, respectively. Subtype II isolates are rare and are not represented in this study.
The distribution of subtypes among human populations has a strong geographic component, with regions of highest prevalence for each subtype as follows: Ia, Africa; Ic, Northeast Asia, particularly Japan; III, Africa; IV and V, Europe and Asia; and IV, Southeast Asia (14, 34). The distribution of subtypes outside of Europe, Africa, and Asia has not been well described. No correlation between subtype and either virologic or clinical characteristics has been demonstrated to date.
A wide variety of primers, probes, and standards for quantitative-PCR assays are in use in both clinical and research laboratories. The absence of standardized BKV assays constitutes a major obstacle to the comparison of BK titers measured at different institutions and may severely limit the usefulness of generalized quantitative viral-load cutoffs in screening and diagnostic protocols. In addition, we have had anecdotal experience with clinical specimens associated with unexpectedly low BKV concentrations by existing clinical assays.
In this study, we describe the extent and sources of variability between seven different real-time PCR assays for the measurement of BKV in urine specimens. We report that interassay variability is substantial and that the variability is to a large extent dependent on the viral subtype. Our findings highlight the importance, not only of appropriate primer and probe design, but also of the consideration of viral population variability in the selection of an appropriate reference material for each assay. Finally, we describe improved performance of an assay using primers and probes redesigned using additional BKV sequence information now available.
MATERIALS AND METHODS
Specimen collection and DNA extraction.
A total of 251 consecutive urine specimens with volumes of ≥0.5 ml submitted to our clinical laboratory for BKV testing were prospectively collected between August and November 2006. The study population consisted of 151 males, 96 females, and 4 persons of unknown gender, with a median age of 50 years (range, 5 to 80 years). Approximately 56% of the specimens were submitted from either a renal-transplant clinic, a nephrology clinic, or an inpatient renal-transplant unit within our institution; others were submitted from elsewhere in the University of Washington health care system or other outside locations. Viral DNA was extracted in nine batches of 32 wells (28 patient specimens; a mixed-patient standard [MPS] [see below]; and negative, high, and low positive controls) from 200-μl urine aliquots and eluted into 100 μl using the Roche MagNaPure instrument and the DNA Isolation Kit I. After extraction, each sample was diluted with 500 μl of MagNaPure elution buffer, frozen, and stored at −20°C in three 200-μl aliquots until it was used.
Standards and controls.
Three different standard materials were used, including a plasmid containing the entire genome of the Dunlop strain (ATCC 45025, referred to here as Dunlop), purified MM strain BKV DNA (ATCC 45026) purchased from Advanced Biotechnologies Inc. (Columbia, MD), and a mixed urine pool (the mixed-patient standard [MPS]). Because the MM strain contains a deletion in the large T region of the genome, the T3 and T4 primer/probe sets did not amplify this material. The MPS, prepared to reflect viral genetic variation within our patient population, was made by pooling randomly selected urine samples from 30 different patients containing BKV titers of >9 × 105/ml. The pooled urine samples were frozen in 250-μl aliquots. Control material included in each extraction and PCR assay was prepared by pooling 30 positive samples in a manner similar to that of the MPS material; both a high control and low control (a 1:400 dilution of the high control) were prepared.
The concentration of the MM standard was provided in the package insert. The concentration of the Dunlop plasmid standard (DPS) was determined both by UV spectrophotometry at 260 nm and by quantitative real-time PCR as the average of values calculated using the V1, T1, and T2 assays with the MM standard material as a reference. Results from the V2 and V3 assays were not included because of sequence differences between the Dunlop and MM strains in these primer/probe sites. The concentration of the Dunlop plasmid calculated using spectrophotometry agreed well with results obtained from quantitative PCR. The concentration of the MPS was calculated similarly using quantitative PCR. Serial 100-fold dilutions of each set of standards were run in duplicate and included in each PCR experiment at the following concentrations: Dunlop, 0.7, 2.7, 4.7, 6.7, and 8.7 log10 templates/50-μl reaction mixture; MPS, 0.35, 1.35, 3.35, and 5.35 log10 templates/50-μl reaction mixture; and MM, 1.0, 2.0, 3.0, 4.0, and 5.0 log10 templates/50-μl reaction mixture.
Primers and probes.
Seven different primer/probe sets were used in parallel comparison studies and are named according to the first letter of the BKV genomic region (V or T) followed by a numeral corresponding to the relative genomic location. PCRs containing the V1 and T1 primer sets were performed according to previously published methods (8, 32). The V2 primer set is the BK version 1.0 analyte-specific reagent supplied by Nanogen, Inc., and reactions were performed according to conditions from Associated Regional and University Pathologists laboratories (J. Stevenson, A. Turiak, and D. Hillyard, poster presented at the 21st Annual Meeting of the Pan American Society for Virology, 2005, poster TA33). Two assays were designed at the University of Washington and performed as previously described. They were T2, including the Pep-1/Pep-2 primers (1) with a TaqMan probe (16), and T4 (5).
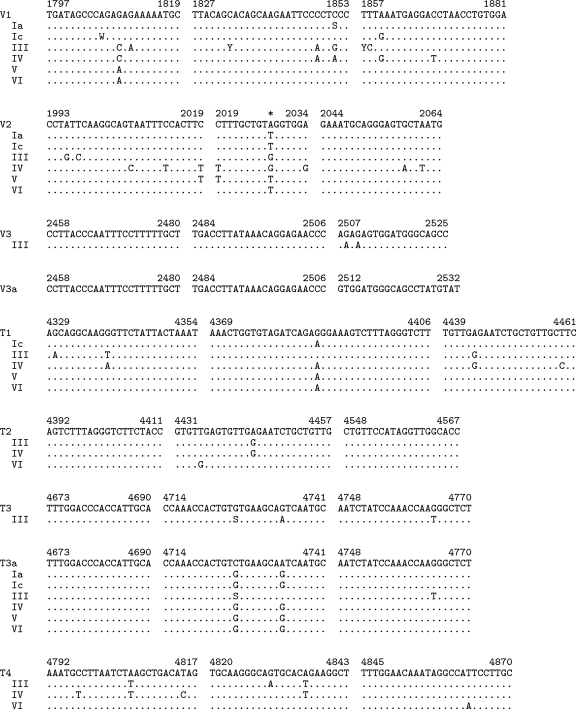
Two previously unpublished primer/probe sets, named V3 and T3, were designed for this study (see Fig. 3 for sequences). Both probes were labeled with 6-carboxyfluorescein and 6-carboxytetramethylrhodamine on the 5′ and 3′ ends, respectively. After results with the original seven primer sets were reviewed, an additional assay, named V3T3, was designed, consisting of a mixture of the V3 and T3 primer/probe sets with a modified V3 reverse primer (5′-ATACATAGGCTGCCCATCCAC-3′) and a second, genotype III-specific T3 probe (6-carboxyfluorescein-CCAAACCACTGTCTGAAGCAATCAATGC-6-carboxytetramethylrhodamine).
FIG. 3.
Alignments of primers and probes with consensus sequences of BKV subtypes. Sequences corresponding to forward primers (left), probes (center), and reverse primers (right) are shown for each primer/probe set, along with corresponding regions of consensus sequences representing BKV subtypes Ia, Ic, III, IV, V, and VI. V3a and T3a indicate sequences of the modified primers and probes derived from the V3 and T3 sequences, respectively, used in the V3T3 assay. The numbers indicate positions in the BKV genome based on the Dunlop reference strain (GenBank accession no. V01108). Subtypes with consensus sequences identical to those of a given primer/probe set are omitted. Nucleotides identical to the primer sequence at each position are shown as periods. IUPAC ambiguity codes are used where the consensus sequence could not be defined as a simple majority of sequences in a subtype. The codes used are defined as follows: S, G/C; Y, C/T; W, A/T. The position in the V2 probe appearing below the asterisk is occupied by a modified base (Nanogen, Inc., personal communication). Sequences of all reverse primers and T1, V2, and T2 probes are shown as the reverse complements of the synthesized oligonucleotides.
Quantitative PCRs.
The amplification conditions for the V3, T3, and V3T3 assays were as follows: 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 20 seconds and 60°C for 1 min. The final concentrations of the V3 primers and probe were 400 nM and 200 nM, respectively, and the final concentrations of the T3 primers and probes were 150 nM and 100 nM, respectively, for both the individual and the combined reactions. The MgCl2 final concentrations for V3, T3, and V3T3 were 7 mM, 4 mM, and 5.5 mM, respectively. The remaining reagents in the master mixture were included as previously described (16).
PCRs were performed in duplicate after previously extracted and diluted samples were thawed. Experiments were repeated if disagreements between duplicates were suggestive of experimental error. A total of nine 96-well plates were run with each primer/probe mixture, with each plate containing dilutions of the DPS, the MPS, the high and low controls, and 28 patient samples. A subset of experiments also included the MM standard. All initial real-time PCR assays were run on an Applied Biosystems Inc. (ABI) 7900 instrument. Reaction mixtures containing the V3T3 mixture were performed on an ABI 7700. Cycle thresholds (CT) were determined using the automated baseline threshold analysis mode with the version 2.2.2 SDS software (ABI 7900) or version SDS 1.9.1 (ABI 7700). CT data were exported to Microsoft Excel spreadsheets and used to populate a database (SQLite; http://www.sqlite.org) from which data for calculations was extracted using custom software written in either Python (http://www.python.org/) or R (20).
Calculations and statistics.
Linear least-squares regression lines were calculated for amplifications of serially diluted reference materials for each primer/probe set with 18 CT points (nine runs in duplicate) at each standard curve concentration. For some of the primer/probe mixes, the lowest standard concentration did not amplify; in those cases, only the positive wells were used. BKV titers were calculated using each assay by determining the number of copies/reaction mixture corresponding to the average of duplicate CTs using both the MPS and DPS curves.
An “expected” value for each specimen was defined as the average of the highest two viral-load measurements. Samples were considered discrepant if at least one primer/probe mix gave a quantitative result 1.0 log10 template/reaction mixture lower than the average of the top two primer/probe mix results (i.e., the expected value) for that sample. Analyses performed using an expected value calculated using either the highest single value or the average of the highest three values produced nearly identical results.
The significance of the difference in variance between values calculated for the positive control using DPS and MPS was demonstrated using the nonparametric Fligner-Killeen test of homogeneity of variances (function “fligner.test”) implemented in R 2.6 (4, 20). Comparison of positive control concentrations was performed using pairwise t tests calculated with nonpooled standard deviations with Bonferroni adjustment for multiple comparison, using the function “pairwise.t.test” in R 2.6 (4, 20). The significance of differences in measurement of viral loads in clinical specimens attributable to the assay was assessed using the Kruskal-Wallis rank sum test using the function “kruskal.test” in R 2.6.
Comparisons between expected values for discrepant specimens and measurements using the V3T3 assay and between the T2 and V3T3 assays for prospectively collected urine and serum specimens were performed by Deming regression using MedCalc v9.3.5.0 (MedCalc Software, Belgium); plots were generated with R 2.6 (20).
BKV sequence analysis.
Records containing sequences spanning the BKV coding region (Dunlop positions 388 to 5153) were downloaded from GenBank; 104 records were available as of March 2007. A single isolate was arbitrarily selected when multiple sequences appeared to originate from a single BKV-infected subject based on descriptions in the corresponding references (3, 10, 17, 27, 29). The remaining 69 sequences were assigned to BKV subtype Ia, Ic, III, IV, V, or VI as defined previously (27). A multiple-sequence alignment was constructed from individual pairwise alignments of each sequence to the Dunlop reference strain using needle (21) and custom software written in the Python scripting language (http://www.python.org). Phylogenetic trees were built from positions 388 to 5153 of this alignment using DNADIST and NEIGHBOR (6) with default parameters. A subset of sequences from GenBank were classified as described previously (17, 27); unclassified sequences were then assigned to a subtype based on their positions in the phylogenetic tree relative to the classified sequences. GenBank accession numbers and subtype assignments were as follows: Ia (10 sequences), AB263912, AB263913, AB263914, AB263926, AB263928, AB263938, DQ305492, NC_001538, V01108, and V01109; Ic (15 sequences), AB211372, AB211375, AB211376, AB211377, AB211378, AB211380, AB211382, AB211383, AB211384, AB213487, AB217918, AB217920, AB217921, AB263930, and AB263931; III (4 sequences>), AB211386, AB263916, AB263920, and PLYCGAS; IV (7 sequences), AB211387, AB211388, AB211389, AB211390, AB217919, AB260033, and AB260034; V (19 sequences), AB211370, AB260028, AB260029, AB260030, AB260031, AB260032, AB263915, AB263917, AB263918, AB263919, AB263921, AB263923, AB263924, AB263925, AB263935, AB263936, AY628234, DQ989794, and DQ989795; and VI (14 sequences), AB211369, AB211371, AB211373, AB211374, AB263922, AB263927, AB263929, AB263932, AB263933, AB263934, AB263937, AY628231, AY628237, and DQ989813. Consensus sequences were generated from sequences representing each subtype. For positions at which no single nucleotide was present in the majority of sequences, IUPAC ambiguity codes were used to represent mixtures of bases.
DNA sequencing.
PCR primers identical to Dunlop nucleotides 4186 to 4211 (forward primer) and 4899 to 4917 (reverse primer) were designed to amplify a 731-base pair region of the BKV large T region. PCR amplifications were performed on an ABI 7000 instrument. Reaction mixtures contained Herculase Hotstart DNA polymerase (Stratagene, La Jolla, CA) at 0.1 U/μl, 1× reaction buffer, 2% dimethyl sulfoxide, 200 μM deoxynucleoside triphosphates, 0.4 μM primers, and a 1:500 dilution of Sybr green stock material (Invitrogen, Carlsbad, CA). Conditions for amplification consisted of 95°C for 20 s, followed by 45 cycles of 95°C, 55°C, and 68°C for 20 s, 30 s, and 1 min, respectively. PCR products were prepared for sequencing using the DNA Clean and Concentration kit (Zymo Research Corp., Orange, CA) according to the manufacturer's instructions and eluted in 35 μl water. DNA sequencing was performed at DNA sequencing facilities at either the Department of Biochemistry or the Department of Genome Sciences High Throughput Genomics Unit, University of Washington, using the Applied Biosystems Big Dye Terminator kit under standard conditions. Sequencing reactions were submitted with 8 pmol primer and approximately 100 to 200 ng template per reaction. Each PCR product was sequenced in both the forward and reverse directions using the amplification primers.
Sequencing of subtype III and IV isolates proved difficult due to long poly(A) and poly(T) tracts, and additional sequencing reactions using primers identical to Dunlop nucleotides 4290 to 4316 (forward) and 4878 to 4897 (reverse), as well as T2-reverse and T3-reverse primers, were performed as necessary to generate sufficient reads over difficult regions to permit confident identification of polymorphisms.
Sequences were trimmed, analyzed, and assembled using either Sequencher 4.6 (Gene Codes Corporation, Ann Arbor, MI) or SeqScape 2.1 (Applied Biosystems, Foster City, CA). Sequence alignments and neighbor-joining trees were created using CLUSTAL W 1.83 with default parameters (30). Subtype assignments were made by grouping unknown sequences and reference sequences for each subtype by phylogenetic analysis.
Nucleotide sequence accession numbers.
BKV sequences were deposited in GenBank and have accession numbers EU681732 to EU681775.
RESULTS
Choice of reference material strongly influences agreement among assays.
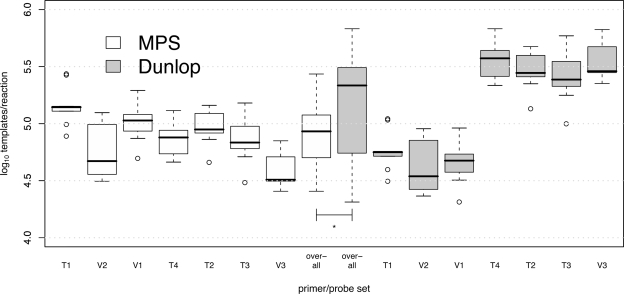
Before comparing the performances of different assays for measurement of the BKV concentrations in clinical specimens, we considered the effect of the choice of reference material on interassay variability of the positive control material. All calculations of BKV DNA copy numbers were made using standard curves generated with two different reference materials: MPS, DNA extracted from a pool of urine specimens containing BKV at high titer, and Dunlop, a bacterial plasmid containing the full genome of the Dunlop reference strain. To compare assay characteristics using both reference materials, we combined results for the positive controls across all experiments (the averages of duplicate reactions in each 96-well plate for a total of nine values for each primer/probe set).
Figure 1 shows the location of each of the seven primer/probe sets in the BKV genome. Figure 2 shows quantitative-PCR results for the positive control material with concentrations calculated according to standard curves constructed using either MPS or Dunlop. Values calculated using MPS aggregated across all primer/probe sets had a significantly lower variance than those calculated using the Dunlop plasmid (P < 0.0001). In addition, each of the V1, V2, and T1 assays resulted in concentrations significantly (approximately 10-fold) lower than each of the other four assays when Dunlop was used as a reference (the corrected P value was <0.0001 for each comparison in pairwise t tests). Using MPS as a reference, this pattern was not reproduced, although a few assays did yield significantly different concentrations for the positive control (T1 versus T4, and V3 versus V1, T1, T2, and T4; P < 0.05 for all). Therefore, a distinct assay-specific bias was directly attributable to the choice of reference material, and the MPS was associated with improved interassay consistency.
FIG. 1.
Map of BKV coding regions with the positions of primers and probes used in quantitative-PCR assays. The numbering is based on the Dunlop reference strain.
FIG. 2.
Measurement of positive control with each primer/probe set. Each box plot labeled with the name of a primer/probe set represents the distribution of the averages of each set of duplicate reactions in each of nine PCR experiments. Box plots labeled “overall” show the overall distribution of values pooled from all seven assays. The values were calculated using standard curves constructed using either MPS or the Dunlop plasmid. The black horizontal bar, box, whiskers, and open circles indicate the median, interquartile range (IQR), points 1.5 × IQR beyond the first and third quartiles, and outliers, respectively. The asterisk indicates that the overall variance for pooled data from all assays differed significantly, depending on the reference material used in the calculation (P < 0.0001).
BKV measurement in clinical specimens is not uniform across assays.
Next, we considered the relative performances of all seven real-time PCR assays in a set of prospectively collected clinical urine specimens. For each specimen, an expected value was calculated as the average of the two highest values produced by the seven primer/probe sets (expressed as log10 templates per reaction). Expected values were calculated separately using both the MPS and DPS. Of 251 urine specimens analyzed, 98 showed an expected value of at least 10 templates/reaction calculated using the MPS. We restricted all subsequent analyses to this set of specimens to avoid sampling effects at low numbers of templates. The expected values calculated using MPS ranged from 1.0 (by definition) to 7.8 (median, 4.1 log10 templates/reaction); concentrations of the same specimens calculated using the DPS ranged from 1.2 to 8.7 (median, 4.4 log10 templates/reaction). We found significant assay-specific differences in viral-load measurements using Dunlop as a reference (P < 0.0001; chi-square = 29.8; 6 degrees of freedom; Kruskal-Wallis rank sum test) but no significant difference among assays using MPS (P = 0.98).
To provide a preliminary description of the performance of each of the seven assays for clinical specimens, we tallied the number of viral-load measurements that were concordant, discrepant, or negative for all seven primer/probe sets. Discrepancy was defined as a difference between observed and expected values exceeding 1.0 log10 template/reaction. The overall performance of all seven assays using either MPS or Dunlop for viral-load calculation is shown in Table 1.
TABLE 1.
Comparison of quantitative performances for each primer/probe set using MPS and DPS among specimens with an expected value of at least 10 tem plates/reaction
| Primer | MPS
|
DPS
|
||||
|---|---|---|---|---|---|---|
| No. concordant | No. discrepanta | No. negativeb | No. concordant | No. discrepant | No. negative | |
| V1 | 85 | 10 | 3 | 15 | 80 | 3 |
| V2 | 92 | 6 | 0 | 68 | 30 | 0 |
| V3 | 88 | 9 | 1 | 91 | 6 | 1 |
| T1 | 97 | 0 | 1 | 43 | 54 | 1 |
| T2 | 90 | 4 | 4 | 85 | 9 | 4 |
| T3 | 93 | 0 | 5 | 91 | 2 | 5 |
| T4 | 91 | 6 | 1 | 92 | 5 | 1 |
| Total | 75 | 10 | 13 | 6 | 79 | 13 |
The difference between observed and expected values exceeded 1.0 log10 for one or more primer/probes among specimens that contained amplifiable BKV using all seven assays.
The CT was >45 in duplicate.
Using MPS as a reference, a total of 10 specimens were discrepant and 13 were negative for at least one assay. The T1 assay (none discrepant and a single negative) appeared to have the most favorable performance overall using MPS. However, the numbers of discrepant values were strikingly different using Dunlop as a reference, with 79 of 98 specimens at least 10-fold lower than expected for at least one assay. The majority of discrepancies occurred with the V1, V2, and T1 assays (54, 30, and 80 discrepant specimens, respectively). These are the same three assays that demonstrated a negative bias for the control material using the DPS as a reference (Fig. 2). As a group, the V1, V2, and T1 assays produced viral-load measurements in urine specimens that were significantly lower than those of the other four assays using Dunlop (but not MPS) as a reference (P < 0.0001; Kruskal-Wallis rank sum test). Thus, the predominant source of the discrepancies among clinical specimens using the DPS was the lower-than-expected concentrations calculated using the V1, V2, and T1 assays in a pattern that paralleled the negative bias in the measurement of the positive control using the same three assays.
Alignments of BKV full-genome sequences predict primer/probe mismatches.
We hypothesized that one likely explanation for assay discrepancy or failure would be reduced PCR primer or probe binding due to the presence of nucleotide polymorphisms in the target BKV sequences. BKV isolates can be grouped into subtypes defined by phylogeny that have different geographic distributions (27, 34). Polymorphisms affecting assay performance might be either subtype associated, and therefore shared by most or all members of a subtype, or associated with a more recent mutational event (i.e., the divergence of BKV subtypes). To predict subtype-associated polymorphisms in the primer and probe regions, consensus sequences corresponding to BKV subtypes Ia, Ic, III, IV, V, and VI were constructed using sequences downloaded from GenBank. Alignments of consensus sequences of BKV subtypes corresponding to each of the primers and probes are shown in Fig. 3. All primers and probes are predicted to contain mismatches to subtype III sequences, and all but T3 and V3 have mismatches to subtype IV sequences. Otherwise, the primer/probe sets can be divided into two classes on the basis of these alignments: those that are predicted to have few mismatches with the remaining subtypes (T4, T2, T3, and V3) and those predicted to have numerous mismatches with multiple subtypes (T1, V2, and V1).
Quantitative discrepancies are associated with subtypes III and IV.
The distribution of BKV subtypes found infecting the population that provided specimens to our laboratory (serving primarily the Seattle area but also the rest of Washington, as well as Alaska, Montana, Wyoming, and Idaho) has not been defined. To determine the approximate relative abundances of BKV subtypes in our reference population, and to identify non-subtype-associated polymorphisms affecting assay performance, we sequenced a PCR-amplified region spanning 732 base pairs in 45 specimens. The specimens selected for sequencing included 14 discrepant specimens with expected viral titers of >2.0 log10 templates/reaction, as well as 31 nondiscrepant specimens chosen to represent the full range of viral loads of >2 log10 templates/reaction (all values were calculated using MPS). The region sequenced spans the T1, T2, T3, and T4 amplicons. Each BKV sequence was assigned to a subtype by its position in a phylogenetic tree that included reference sequences representing subtypes Ia, Ic, III, IV, V, and VI.
The subtype distribution and number of discrepant specimens within each subtype are shown in Table 2. Statistical analyses comparing the distribution of discrepancies or viral loads among subtypes were not attempted because of potential sampling bias in the selection of specimens for sequencing. However, the average viral loads were similar across subtypes by inspection. The most prevalent subtypes identified were Ia, V, and VI (together accounting for over 75% of sequenced specimens). Notably, all subtype III and IV specimens were discrepant by at least one assay.
TABLE 2.
Subtype distribution among sequenced specimens
| Subtype | Count | No. discrepanta (%) | Viral loadb |
|---|---|---|---|
| Ia | 8 | 3 (37.5) | 5.7 |
| Ic | 1 | 0 (0.0) | 4.1 |
| III | 6 | 6 (100.0) | 3.7 |
| IV | 3 | 3 (100.0) | 6.0 |
| V | 17 | 2 (11.8) | 4.9 |
| VI | 10 | 0 (0.0) | 5.4 |
| Total | 45 | 14 | 5.0 |
Discrepant specimens are those for which the difference between the expected and observed values exceeds 1.0 log10 for one or more primer/probes.
The viral load is the average of expected values in log10 templates/reaction.
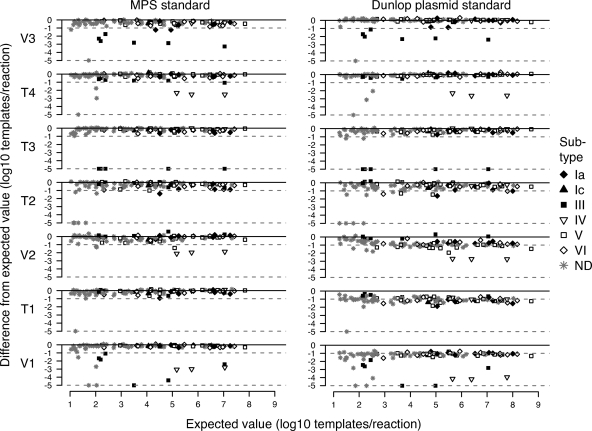
Importantly, we found that although the performance of specific assays did depend on the subtype, subtype-dependent discrepancies were not dependent on the viral load. Details of viral-load measurements using each assay, as well as subtype assignments among discrepant specimens, are presented in Table 3. These relationships are also highlighted in Fig. 4, which plots the expected values for BKV copy numbers against deviations from the expected values for each assay. BKV copy numbers of subtype III specimens were uniformly underestimated by the V1, V3, and T3 assays across the full range of viral loads measured. Similarly, viral loads among subtype IV specimens were uniformly underestimated by the T4, V1, and V2 assays.
TABLE 3.
Quantitative-PCR results for measurement of BKV among urine specimens discrepant for one or more assays, calculated using the MPS standard
| Patient | Subtypea | Expected resultb | Result for primer/probec:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | T1 | T2 | T3 | T4 | |||
| 062 | IV | 7.0 | 7.2 | 4.5 | 5.2 | 6.9 | 4.3 | 6.9 | 6.7 |
| 152 | III | 7.0 | 6.8 | 6.0 | 7.3 | 6.4 | 4.6 | 0 | 3.8 |
| 065 | IV | 5.8 | 5.8 | 3.2 | 3.8 | 5.4 | 2.8 | 5.7 | 5.4 |
| 174 | IV | 5.2 | 5.2 | 2.8 | 3.1 | 5.0 | 2.1 | 5.1 | 4.6 |
| 009 | V | 5.1 | 5.1 | 4.7 | 3.7 | 4.8 | 5.1 | 4.5 | 4.6 |
| 139 | Ia | 4.9 | 4.8 | 5.0 | 4.8 | 4.7 | 4.9 | 4.7 | 3.7 |
| 015 | III | 4.8 | 4.2 | 4.0 | 5.4 | 4.2 | 0.4 | 0 | 2.0 |
| 242 | Ia | 4.5 | 3.6 | 4.2 | 4.6 | 3.1 | 4.4 | 3.9 | 4.4 |
| 055 | Ia | 4.3 | 4.2 | 4.2 | 4.0 | 3.9 | 4.4 | 4.3 | 3.1 |
| 090 | V | 4.1 | 3.5 | 4.3 | 3.6 | 3.1 | 3.9 | 3.7 | 3.9 |
| 013 | III | 3.5 | 3.3 | 2.7 | 3.6 | 3.4 | 0 | 3.2 | 0.7 |
| 112 | III | 2.4 | 2.4 | 1.7 | 2.4 | 2.4 | 1.3 | 0 | 0.6 |
| 010 | III | 2.2 | 2.3 | 1.7 | 2.1 | 2.1 | 0.4 | 0 | −0.4 |
| 175 | III | 2.1 | 2.0 | 1.5 | 2.3 | 1.9 | 0.5 | 0 | −0.2d |
| 034 | ND | 2.0 | 2.1 | −0.9 | 0.7 | 0.7 | 0 | 2.0 | 1.6 |
| 032 | ND | 2.0 | 1.6 | 0.3 | 0.8 | 1.7 | −0.7 | 2.1 | 1.9 |
| 026 | ND | 1.7 | 1.2 | 1.4 | 2.1 | 1.1 | 0 | 1.2 | 0 |
| 162 | ND | 1.6 | 0.7 | 1.6 | 1.5 | 0 | 1.1 | 0.8 | 1.6 |
| 094 | ND | 1.3 | 1.2 | 0 | 0.4 | 0 | −0.2 | 1.3 | 1.3 |
| 031 | ND | 1.2 | 0 | 1.1 | 1.3 | 0.0d | 0.7 | 0.6 | 0.9 |
| 188 | ND | 1.2 | 0.8 | 0.8 | 1.3 | 0 | 0.6 | 0.2 | 1.0 |
| 158 | ND | 1.1 | 1.2 | 1.0 | 1.0 | 0 | 1.0 | 0.6 | 0.5 |
| 016 | ND | 1.0 | 0.7† | 0.9 | 0.5 | 1.1d | 0.5 | 0.4d | −0.2 |
ND, not determined.
The average of the highest two of seven values obtained for each specimen in units of log10 templates per reaction.
Averages of duplicate results are shown; results for which the difference between the expected and observed values exceeds 1.0 log10 for one or more primer/probes are shown in boldface; the value is set to zero when both reactions have CT of >45 (negative numbers describe values less than 1.0).
The value shown is for a single positive reaction.
FIG. 4.
Deviation of viral loads from expected values using each primer/probe set, calculated using MPS (left) and Dunlop (right) as references. The expected value of each specimen (x axis) is plotted against the difference between the expected value and the value obtained in each assay (y axis). Only the 98 specimens with expected values of >10 templates/reaction are shown. Negative specimens and those with deviations from expected values of >5 log10 templates/reaction are plotted on the dotted line at the base of each plot. Specimens with an undetermined subtype (ND) are plotted in gray.
Interestingly, measurement of one specimen using the T3 assay was affected by nucleotide substitution at a position that is partially conserved among subtype III sequences. The BKV concentration of specimen 013 calculated using T3 (Table 3 and Fig. 4) was the same as the expected value, despite the assay's failure to amplify other subtype III specimens. Position 4726 is polymorphic among published subtype III BKV sequences and is either G (identical to the primer sequence) or C. Specimen 013 contains a G at this position, leaving only a single A/G mismatch with the T3 probe at position 4734. Therefore, the single subtype III-associated polymorphism in the T3 probe region was tolerated with no effect on assay accuracy, while the presence of two polymorphisms caused assay failure.
Few other polymorphisms underlying primers or probes were identified in the sequenced specimens. Specimen 015 (subtype III) contained G4380C within the T1 probe. Specimens 016, 018, and 189 (all subtype V) contained A4336G within the T1 forward primer. None of these specimens was discrepant using the T1 assay with MPS as a reference (Table 3). Therefore, among the limited number of specimens considered in this study, BKV assay failure was primarily caused by subtype-associated polymorphisms, with a limited contribution by polymorphisms not tightly linked to a subtype.
Improved primer design eliminates subtype-associated assay failure.
Given the results of the multiassay comparison, we hypothesized that an assay combining slightly modified versions of the V3 and T3 primer/probe sets should accurately measure the viral loads of all BKV subtypes using the Dunlop reference material. Accordingly, we modified the V3 assay by shifting the reverse primer to correspond to Dunlop positions 2512 to 2532, resulting in a primer predicted to be identical to the consensus sequence for each subtype. In addition, we included a second T3 probe designed to detect subtype III BKV isolates in unimolar concentrations to the original probe (Fig. 3). Slight modifications were also made to the reaction conditions to permit multiplexed amplification of the combined V3 and T3 primer/probe sets; this assay is referred to as V3T3.
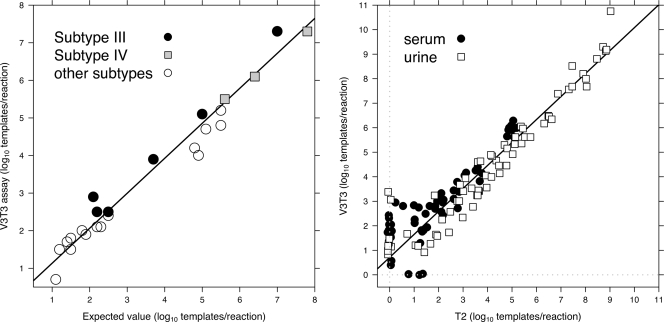
Reamplification of the previously discrepant specimens using V3T3 resulted in calculated concentrations that corresponded well to the expected values, with a correlation coefficient of approximately 0.97; the data were fitted to a line with a slope of 0.93 (0.85 to 1.01) and a y intercept of 0.20 (−0.09 to 0.50) log10 templates/reaction (95% confidence intervals are in parentheses) (Fig. 5, left). Among the specimens tested, the difference between the expected value and the concentration calculated using the V3T3 assay was at most 0.9 log10 templates/reaction. A single specimen, number 016, failed to amplify using V3T3; this failure was likely due to degradation of this low-titer material (expected value, 10 templates/reaction) following an additional freeze-thaw cycle and storage for approximately 1 year.
FIG. 5.
(Left) Comparison of expected values versus concentrations calculated using the V3T3 assay for specimens defined as discrepant. Subtype III and IV specimens are highlighted as indicated in the legend. (Right) Comparison of the T2 and V3T3 assays for 230 consecutive clinical specimens. Only specimens with a CT of <45 cycles for at least one of the two assays appear in the plot (60/96 and 69/134 serum and urine specimens, respectively); those with a CT of ≥45 were assigned a value of zero and are shown slightly offset from the origin of each axis (gray dotted line) to avoid overplotting. The regression line was calculated after excluding points that failed to amplify within 45 cycles in either of the two assays.
Finally, to compare the performance of the V3T3 assay with that of T2, the assay previously used in our clinical laboratory, we used both assays to measure BKV titers in 230 consecutively collected serum or urine specimens using the Dunlop reference material as a standard. A comparison of the measurements of each specimen using these two assays is shown in Fig. 5, right. Among all specimens tested, 60/96 (63%) serum specimens and 69/134 (52%) urine specimens were amplified within 45 cycles with at least one of the two assays. Among those that were positive using the V3T3 assay, 14 serum and 9 urine specimens failed to amplify using the T2 assay. Conversely, only three serum specimens that were positive with T2 failed to amplify using the V3T3 primer/probe set; they had concentrations of 0.8, 1.2, and 1.4 log10 templates/reaction according to the T2 assay. If a “positive” specimen is defined as one with a CT of <45 for at least one assay, the approximate sensitivities were 82% for the T2 assay and 98% for V3T3. The R2 value for the comparison of T2 and V3T3 was 0.92, and the data were fitted to a line with a slope of 0.97 (0.91 to 1.04) and a y intercept of 0.58 (0.26 to 0.89). Therefore, the improved assay agreed well with the T2 assay among specimens positive for both but gave results approximately fourfold higher on average using identically prepared standard curves of the Dunlop reference material.
DISCUSSION
This study was undertaken to document the extent of interassay variability in the measurement of BKV loads by real-time PCR. We demonstrated that both differences in primer and probe design and the combination of certain primers and probes with specific reference materials could lead to substantial disagreement between assays. Furthermore, the extent of variability of certain assays was substantially influenced by the BKV subtype. Viral-load measurement by real-time PCR is dependent on the amplification efficiency of the primers, the efficiency of annealing and enzymatic cleavage of the probes, and the relative amplification efficiency of the target versus the reference material; we described examples of assay variability attributable to each of these three assay components.
Primer design.
Only in the last few years have a substantial number of full-genome BKV DNA sequences been published (3, 10, 17, 27, 29); thus, the extent of BKV genetic diversity is not necessarily reflected in the design of commonly used PCR reagents. The importance of locating real-time PCR primers and probes within well-conserved genomic regions is well known (15, 19). The experiments in this study illustrate some potential manifestations of primer/probe mismatches. For example, false-negative results (that is, the failure to detect virus present at high copy number) were consistently seen when the T3 assay was used to amplify most subtype III isolates. Similarly, the V2, T4, and V1 assays generated viral-load measurements that were consistently low for all three subtype IV specimens (Table 3 and Fig. 4). These failures should perhaps have been anticipated given the extent of nucleotide mismatches between the affected primers and probes and their viral targets. There were two predicted nucleotide mismatches between the T3 probe and the subtype III consensus sequence and extensive mismatches between the subtype IV consensus sequence and the T4, V2, and V1 primers and probe (Fig. 3). However, this study and others (33) illustrate the difficulty of predicting the extent to which primer and probe mismatches will affect assay performance. For example, a predicted mismatch of 2 nucleotides between the T4 probe and the subtype III consensus sequence did not result in consistently inaccurate viral-load measurement using that assay (Table 3 and Fig. 3).
Reference material.
It is increasingly recognized that the availability of suitable reference material is necessary to provide quantitative consistency in virologic assays performed in different laboratories (26, 31). For example, international efforts have led to the availability of international standards for human immunodeficiency virus, hepatitis A virus, hepatitis B virus, hepatitis C virus, and human parvovirus B19 (9, 22, 23, 24, 25). It is important to note that while synthesis of primers and probes used in published assays may be trivial, the standards used are frequently either undescribed or difficult to obtain; indeed, this was the case with the T1 and T4 assays (8, 32). In this study, we used the Dunlop plasmid, which was the only readily available clone of a full-length BKV genome that could be amplified by all assays. Here, we found that the choice of reference material strongly influenced the agreement of viral-load measurements of the same specimen using different sets of primers and probes.
Our specific observation was that when primers and probes were an exact match with corresponding sequences in the reference material but had mismatches with extracted viral DNA, the calculated viral load was lower than expected. Figure 2 and Fig. 4 graphically illustrate the consistently lower viral-load measurements resulting from the use of the Dunlop standard with the V1, V2, and T1 primers and probes. We hypothesize that the negative bias, when it appears, is the result of a mismatch in amplification efficiency between the DPS (relatively high in the absence of primer and probe mismatches) and the most prevalent BKV strains represented in the study population (relatively low due to the presence of mismatches, as shown in Fig. 3).
In contrast, the expected viral-load values were obtained when the V1, V2, and T1 primer sets were used with the MPS. Because this reference material is a complex mixture derived from pooled urine specimens, the overall extent of primer/probe mismatches with MPS is expected (on average) to be similar to the extent of mismatches with clinical specimens. Expected values were observed when the other four primer/probe sets, none of which are predicted to have extensive mismatches with predominant subtypes of BKV (i.e., Ia, V, and VI), were used with both Dunlop and MPS.
These findings highlight the importance of choosing standards that reflect the circulating viral genetic diversity in a population (predicted, in this case, by the BKV subtype distribution) for accurate and reliable viral-load determination using PCR-based assays.
BKV subtype distribution and assay performance.
The current study is the first to describe the distribution of BKV subtypes in a North American population. Most of the discrepancies that we observed are attributable to subtype-associated nucleotide polymorphisms, specifically among subtype III and IV isolates, which are the most divergent of the subtypes (27). Pairwise distances between the consensus nucleotide sequences spanning the BKV coding regions are in the 1 to 2% range among subtypes Ia, Ic, V, and IV, whereas distances between both the subtype III and IV consensus sequences and each of the other subtypes are in the 4 to 5% range (data not shown). The divergence of subtypes III and IV is reflected in the extent of mismatch with a number of the primers and probes used in this study (Fig. 3). Mutations acquired more recently in BKV evolutionary history (that is, since the time of divergence of the major subtypes) played little role in assay failure. This observation is consistent with the limited intrasubtype variability that has been described among published sequences (17, 27), as well as the reported stability of BKV sequences within renal-transplant recipients over time (29). Thus, most of the variability among BKV isolates is subtype associated. We hypothesize that anecdotal experiences with “sporadic” failure of clinical assays to accurately measure BKV may actually be attributable to minority subtypes that would be predicted to be associated with reduced assay performance, given the extent of subtype-associated polymorphisms in the primer or probe binding region.
Despite the fact that subtype III viruses are often described as rare in the literature (18) and are poorly represented among published sequences, this subtype represented approximately 13% (6/45) of the isolates sequenced in this study. As a result, any assay including primers and probes with extensive mismatches to the subtype III consensus performed poorly. Another specific manifestation of the subtype distribution was the effect of mismatches between the DPS (subtype Ia) and isolates belonging to the most abundant subtypes, V and VI (see the discussion above). It is likely that a standard containing sequences from one of the more abundant subtypes would not have been associated with the same extent of variability among assays as we observed using the Dunlop strain.
New assay design and validation.
In this report, we describe a new clinical assay, called V3T3, composed of a mixture of primers and probes originally used in the V3 and T3 assays (both designed for this study), with slight modification intended to improve amplification of subtype III viruses. Both primer/probe sets are suitable for use with the Dunlop reference strain, which is available from ATCC as a plasmid containing the entire viral genome. Calculation of BKV concentrations in urine specimens that produced discrepant results among the original seven assays agreed well with expected values, and viral-load measurement was insensitive to the subtype. Furthermore, while V3T3 agreed quantitatively with our current clinical assay (T2) among specimens for which any amplification was detected, the V3T3 assay yielded positive results in 23 of 230 specimens that failed to amplify using T2. We suggest that this new assay demonstrated improved performance characteristics primarily because it incorporates sequence information and subtype distribution data not available previously.
Significantly, these results suggest that the discrepancies revealed by repeated measurement of specimens with seven different assays would likely not have been detected in the course of routine assay validation commonly performed in clinical laboratories. Clinical assay validation studies typically emphasize accuracy and precision in the measurement of control material; these types of validation studies are by definition difficult to apply to a large number of clinical specimens containing virus in unknown quantity, since they rely on the measurement of reference material containing a known concentration of virus. The difficulty and expense of performing large-scale comparisons of multiple assays underscores the need for a common set of reagents if clinical guidelines for the interpretation of BKV load data are to be applied uniformly across institutions.
Acknowledgments
We thank Amalia Magaret for her review of the statistical analysis and editorial suggestions.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Arthur, R. R., S. Dagostin, and K. V. Shah. 1989. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J. Clin. Microbiol. 271174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, D. C., I. Agha, D. L. Bohl, M. A. Schnitzler, K. L. Hardinger, M. Lockwood, S. Torrence, R. Schuessler, T. Roby, M. Gaudreault-Keener, and G. A. Storch. 2005. Incidence of BK with tacrolimus versus cyclosporine and impact of preemptive immunosuppression reduction. Am. J. Transplant. 5582-594. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y., P. M. Sharp, M. Fowkes, O. Kocher, J. T. Joseph, and I. J. Koralnik. 2004. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J. Gen. Virol. 852651-2663. [DOI] [PubMed] [Google Scholar]
- 4.Conover, W. J., M. E. Johnson, and M. M. Johnson. 1981. A comparative study of tests for homogeneity of variances, with applications to the outer continental shelf bidding data. Technometrics 23351-361. [Google Scholar]
- 5.Erard, V., B. Storer, L. Corey, J. Nollkamper, M.-L. Huang, A. Limaye, and M. Boeckh. 2004. BK virus infection in hematopoietic stem cell transplant recipients: frequency, risk factors, and association with postengraftment hemorrhagic cystitis. Clin. Infect. Dis. 391861-1865. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 2007. PHYLIP (Phylogeny Inference Package), version 3.4. Department of Genome Sciences, University of Washington, Seattle.
- 7.Hirsch, H. H., D. C. Brennan, C. B. Drachenberg, F. Ginevri, J. Gordon, A. P. Limaye, M. J. Mihatsch, V. Nickeleit, E. Ramos, P. Randhawa, R. Shapiro, J. Steiger, M. Suthanthiran, and J. Trofe. 2005. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation 791277-1286. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch, H. H., M. Mohaupt, and T. Klimkait. 2001. Prospective monitoring of BK virus load after discontinuing sirolimus treatment in a renal transplant patient with BK virus nephropathy. J. Infect. Dis. 1841494-1495. [DOI] [PubMed] [Google Scholar]
- 9.Holmes, H., C. Davis, A. Heath, I. Hewlett, and N. Lelie. 2001. An international collaborative study to establish the 1st international standard for HIV-1 RNA for use in nucleic acid-based techniques. J. Virol. Methods 92141-150. [DOI] [PubMed] [Google Scholar]
- 10.Ikegaya, H., P. J. Saukko, R. Tertti, K. P. Metsarinne, M. J. Carr, B. Crowley, K. Sakurada, H.-Y. Zheng, T. Kitamura, and Y. Yogo. 2006. Identification of a genomic subgroup of BK polyomavirus spread in European populations. J. Gen. Virol. 873201-3208. [DOI] [PubMed] [Google Scholar]
- 11.Jin, L., P. E. Gibson, J. C. Booth, and J. P. Clewley. 1993. Genomic typing of BK virus in clinical specimens by direct sequencing of polymerase chain reaction products. J. Med. Virol. 4111-17. [DOI] [PubMed] [Google Scholar]
- 12.Kadambi, P. V., M. A. Josephson, J. Williams, L. Corey, K. R. Jerome, S. M. Meehan, and A. P. Limaye. 2003. Treatment of refractory BK virus-associated nephropathy with cidofovir. Am. J. Transplant. 3186-191. [DOI] [PubMed] [Google Scholar]
- 13.Knowles, W. A., P. E. Gibson, and S. D. Gardner. 1989. Serological typing scheme for BK-like isolates of human polyomavirus. J. Med. Virol. 28118-123. [DOI] [PubMed] [Google Scholar]
- 14.Krumbholz, A., R. Zell, R. Egerer, A. Sauerbrei, A. Helming, B. Gruhn, and P. Wutzler. 2006. Prevalence of BK virus subtype I in Germany. J. Med. Virol. 781588-1598. [DOI] [PubMed] [Google Scholar]
- 15.Lengerova, M., Z. Racil, P. Volfova, J. Lochmanova, J. Berkovcova, D. Dvorakova, J. Vorlicek, and J. Mayer. 2007. Real-time PCR diagnostics failure caused by nucleotide variability within exon 4 of the human cytomegalovirus major immediate-early gene. J. Clin. Microbiol. 451042-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limaye, A. P., K. R. Jerome, C. S. Kuhr, J. Ferrenberg, M. L. Huang, C. L. Davis, L. Corey, and C. L. Marsh. 2001. Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J. Infect. Dis. 1831669-1672. [DOI] [PubMed] [Google Scholar]
- 17.Nishimoto, Y., T. Takasaka, M. Hasegawa, H.-Y. Zheng, Q. Chen, C. Sugimoto, T. Kitamura, and Y. Yogo. 2006. Evolution of BK virus based on complete genome data. J. Mol. Evol. 63341-352. [DOI] [PubMed] [Google Scholar]
- 18.Nukuzuma, S., T. Takasaka, H.-Y. Zheng, S. Zhong, Q. Chen, T. Kitamura, and Y. Yogo. 2006. Subtype I BK polyomavirus strains grow more efficiently in human renal epithelial cells than subtype IV strains. J. Gen. Virol. 871893-1901. [DOI] [PubMed] [Google Scholar]
- 19.Nye, M. B., A. R. Leman, M. E. Meyer, M. A. Menegus, and P. G. Rothberg. 2005. Sequence diversity in the glycoprotein B gene complicates real-time PCR assays for detection and quantification of cytomegalovirus. J. Clin. Microbiol. 434968-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team. 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 21.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16276-277. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha, J., W. Gerlich, N. Lelie, P. Dawson, K. Heermann, and A. Heath. 2001. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 8063-71. [DOI] [PubMed] [Google Scholar]
- 23.Saldanha, J., A. Heath, N. Lelie, G. Pisani, and M.-Y. Yu. 2005. A World Health Organization international standard for hepatitis A virus RNA nucleic acid amplification technology assays. Vox Sang. 8952-58. [DOI] [PubMed] [Google Scholar]
- 24.Saldanha, J., N. Lelie, A. Heath, et al. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76149-158. [DOI] [PubMed] [Google Scholar]
- 25.Saldanha, J., N. Lelie, M. W. Yu, and A. Heath. 2002. Establishment of the first World Health Organization international standard for human parvovirus B19 DNA nucleic acid amplification techniques. Vox Sang. 8224-31. [DOI] [PubMed] [Google Scholar]
- 26.Schirm, J., A. M. van Loon, E. Valentine-Thon, P. E. Klapper, J. Reid, and G. M. Cleator. 2002. External quality assessment program for qualitative and quantitative detection of hepatitis C virus RNA in diagnostic virology. J. Clin. Microbiol. 402973-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, P. M., G. Gupta, A. Vats, R. Shapiro, and P. Randhawa. 2006. Phylogenetic analysis of polyomavirus BK sequences. J. Virol. 808869-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Takasaka, T., N. Goya, H. Ishida, K. Tanabe, H. Toma, T. Fujioka, S. Omori, H.-Y. Zheng, Q. Chen, S. Nukuzuma, T. Kitamura, and Y. Yogo. 2006. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J. Gen. Virol. 87303-306. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentine-Thon, E. 2002. Quality control in nucleic acid testing—where do we stand? J. Clin. Virol. 25(Suppl. 3)S13-S21. [DOI] [PubMed] [Google Scholar]
- 32.Vats, A., R. Shapiro, P. Singh Randhawa, V. Scantlebury, A. Tuzuner, M. Saxena, M. L. Moritz, T. J. Beattie, T. Gonwa, M. D. Green, and D. Ellis. 2003. Quantitative viral load monitoring and cidofovir therapy for the management of BK virus-associated nephropathy in children and adults. Transplantation 75105-112. [DOI] [PubMed] [Google Scholar]
- 33.Whiley, D. M., and T. P. Sloots. 2005. Sequence variation in primer targets affects the accuracy of viral quantitative PCR. J. Clin. Virol. 34104-107. [DOI] [PubMed] [Google Scholar]
- 34.Zheng, H.-Y., Y. Nishimoto, Q. Chen, M. Hasegawa, S. Zhong, H. Ikegaya, N. Ohno, C. Sugimoto, T. Takasaka, T. Kitamura, and Y. Yogo. 2007. Relationships between BK virus lineages and human populations. Microbes Infect. 9204-213. [DOI] [PubMed] [Google Scholar]