Abstract
Previous epidemiological assessments of the prevalence versus special-pathogenicity hypothesis for urinary tract infection (UTI) pathogenesis in women may have been confounded by underlying host population differences between women with UTI and healthy controls and have not considered the clonal complexity of the fecal Escherichia coli population of the host. In the present study, 42 women with acute uncomplicated cystitis served as their own controls for an analysis of the causative E. coli strain and the concurrent intestinal E. coli population. Clonality among the urine isolate and 30 fecal colonies per subject was assessed by repetitive-element PCR and macrorestriction analysis. Each unique clone underwent PCR-based phylotyping and virulence genotyping. Molecular analysis resolved 109 unique clones (4 urine-only, 38 urine-fecal, and 67 fecal-only clones). Urine clones exhibited a significantly higher prevalence of group B2 than fecal-only clones (69% versus 10%; P < 0.001) and higher aggregate virulence scores (mean, 6.2 versus 2.9; P < 0.001). In multilevel regression models for predicting urine clone status, significant positive predictors included group B2, 10 individual virulence traits, the aggregate virulence score, fecal dominance, relative fecal abundance, and (unique to the present study) a pauciclonal fecal sample. In summary, within the fecal E. coli populations of women with acute cystitis, pauciclonality, clonal dominance, virulence, and group B2 status are closely intertwined. Phylogenetic group B2 status and/or associated virulence factors may promote fecal abundance and pauciclonality, thereby contributing to upstream steps in UTI pathogenesis. This relationship suggests a possible reconciliation of the prevalence and special-pathogenicity hypotheses.
Urinary tract infection (UTI), one of the most frequent types of bacterial infection in women, is usually due to Escherichia coli (1). Despite considerable study, the pathogenesis of UTI remains poorly understood. The host's fecal (and, in women, vaginal) flora is the most common immediate source for the infecting E. coli strain (22, 26). However, uncertainty remains regarding to what extent the causative strains represent simply the most prevalent fecal/vaginal E. coli clones within the host population or the affected individuals (the prevalence hypothesis) (24) or, instead, represent a distinctive, highly selected subset of the fecal/vaginal E. coli population with enhanced virulence potential (the special-pathogenicity hypothesis) (18).
Favoring the prevalence hypothesis, in acute E. coli UTI, the urine strain tends to be the host's predominant fecal strain (4, 18, 26). Favoring the special-pathogenicity hypothesis, acute-phase UTI isolates usually exhibit more virulence factors than fecal isolates from healthy hosts and derive predominantly from phylogenetic groups B2 and D (versus groups A and B1 for fecal strains) (6, 13, 27). However, such comparisons may be confounded by between-population host differences, since women who develop UTIs tend to differ genetically or behaviorally from UTI-free women (19, 20, 23) and, conceivably, could have intrinsically different fecal E. coli populations. Consequently, the ideal comparison group for acute-phase UTI isolates may be fecal isolates from the same hosts.
Our previous comparison of same-host urine and fecal isolates from women with acute E. coli cystitis (12) suggested that the UTI strain is usually both more virulent-appearing and more abundant than coisolated fecal strains. However, the very small number of subjects (n = 11) and fecal colonies (3 per host) examined in that study greatly limited the strength of these conclusions. Furthermore, the limited sampling of the fecal population in that study and previous similar studies involving girls (15, 18), women (11), men (7), and dogs (5) with UTIs precluded valid assessment of the clonal diversity of the host's fecal population, which might be expected to vary with the nature of the colonizing clones and to influence the risk of UTI. Accordingly, in the present study we sampled a larger population of women with acute uncomplicated E. coli UTIs and analyzed 30 fecal E. coli colonies per host (i.e., 4 times as many subjects and 10 times as many colonies per host as in our pilot study). We then assessed the relative abundance, molecularly inferred virulence, and phylogenetic background of each fecal clone in comparison with those of the UTI-causing E. coli strain of the host and assessed bacterial traits in relation to the clonal complexity of the source fecal sample.
MATERIALS AND METHODS
Patients.
Between January 2004 and March 2006, women presenting to the emergency department at Hospital Vall d'Hebron, Barcelona, Spain, with suspected acute uncomplicated cystitis were invited to participate in the study. Participation entailed providing informed consent, clinical data, and a self-collected rectal swab. Consecutive consenting women who met the following criteria were included: (i) age, 15 to 65 years; (ii) clinical and laboratory evidence of uncomplicated cystitis (as described previously [12]); (iii) no antimicrobial treatment within 21 days of presentation; (iv) E. coli as the sole urine microorganism; and (v) E. coli isolated from the rectal swab.
Urine microscopy and cultures.
Quantitative urine culture was done using chromogenic agar, followed by conventional identification (14). E. coli isolates were stored in 5% glycerol in Trypticase soy broth at −70°C until use.
Stool cultures.
Concurrent with urine collection, subjects collected a rectal swab using Amies transport medium. Rectal swabs were agitated in sterile saline, which was then diluted serially and spread onto MacConkey agar plates by using an automatic spiral spreading instrument. After 24 to 48 h of incubation (37°C), as many as 30 isolated colonies (as available) of putative E. coli were randomly picked. Colonies were identified using conventional methods (14). E. coli isolates were stored in 5% glycerol-supplemented broth at −70°C until use.
ERIC PCR.
Clonal relationships among the E. coli isolates from a given stool sample and the host's urine isolate were inferred from genomic enterobacterial repetitive intergenic consensus (ERIC) PCR profiles (25). Gel images were captured digitally (Gel Compar II, version 3.0; Applied Maths). Similarity relationships among profiles (within each subject) were estimated by using the Pearson correlation coefficient. Same-host isolates with ≥93% similar ERIC profiles were defined as clonally related. One representative of each putatively unique clone per fecal sample, plus each urine isolate, underwent phylotyping and virulence genotyping.
Phylotyping and virulence genotypes.
The E. coli phylogenetic group (A, B1, B2, or D) was determined by using a triplex PCR method (2). Sixteen virulence-associated genes (papA, papG [alleles I, II, and III], papC, fimH, afa/draBC, sfa/focDE, hlyA, cnf1, iutA, fyuA, kpsM II, traT, ibeA, and malX) were characterized by multiplex PCR, with appropriate positive and negative controls (8). The virulence score was the number of virulence genes detected, with pap elements counting collectively as a single trait. (Such in vitro testing predicts experimental virulence in vivo [8, 17].)
Pulsed-field gel electrophoresis.
E. coli isolates from a given subject that exhibited <93% similar ERIC profiles but identical results for the phylogenetic group and the virulence profile underwent pulsed-field gel electrophoretic analysis of XbaI-restricted total DNA (3). Isolates from a given subject that exhibited indistinguishable profiles (Dice similarity coefficient, 100%) were assumed to represent the same clone.
Definitions.
Each unique clone was assessed for its relative abundance in the host's fecal sample based on the proportion of fecal colonies it accounted for. The dominant clone was the sample's most abundant E. coli clone. If two clones were tied for the highest abundance in a sample, both were considered dominant. Pauciclonal fecal samples were arbitrarily defined as those with ≤4 E. coli clones; multiclonal samples were those with ≥5 E. coli clones.
Three distinct host ecological groups were defined on the basis of the relationship of each subject's fecal clone(s) with the urine clone (12). In group I, the stool culture yielded only the urine clone (designated a urine-fecal clone); in group II, it yielded the urine clone (urine-fecal clone) plus one or more nonurine (fecal-only) clones; and in group III, it yielded only one or more nonurine (fecal-only) clones, whereas the urine culture yielded a distinct (urine-only) clone.
Statistical methods.
Comparisons of proportions were tested using a chi-square test or Fisher's exact test (two-tailed), as appropriate. Comparisons of deviations from a theoretically expected distribution were tested by using the binomial test. Virulence scores were compared by using the Mann-Whitney U test. Due to the nonindependence of multiple isolates acquired from the same sample, mixed-model regression modeling (21) was used. The mixed-effects model is a two-stage (hierarchical) maximum-likelihood approach. The first stage describes the distribution of the outcomes within subjects (i.e., the distribution is conditional on random effects). The second stage describes the variability across subjects and allows for the comparison of other effects (e.g., urine versus fecal source). The number of isolates per sample is treated as a random variable. The significance criterion was a P value of <0.05.
Correlations among variables were assessed by using correspondence analysis, which uses a covariance matrix based on χ2 distances. The computation determines a plane defined by two principal axes of the analysis. The first axis, F1, accounts for most of the variance, and the second axis, F2, orthogonal to F1, accounts for the largest part of the variance not accounted for by F1. Correspondence analysis was conducted from a two-way table that had 109 rows, 1 per E. coli isolate, and 24 columns, 1 per variable.
Similarity relationships among the individual isolates with respect to virulence factor profiles were assessed by using principal-coordinates analysis (PCoA), a multivariate technique related to correspondence analysis that allows one to plot the major patterns within a multivariate data set, e.g., multiple loci and multiple samples (9). Using GenAlEx6 (16), PCoA was applied to the virulence factor data set as a way to collapse the multiple virulence factors for simplified among-group comparisons. Values for each clone from the first three PCoA axes, which capture most of the variance within the data set, were used in a one-way multivariate analysis of variance to test for differences between urine and fecal-only clones.
RESULTS
Subjects.
During the approximately 2-year study period, 85 women with suspected acute cystitis were enrolled. Of these, 42 fulfilled all clinical and bacteriological criteria and thus constituted the study population. The mean age was 30.4 years (range, 15 to 65 years).
Distribution of E. coli clones by source and host.
Overall, from the 42 subjects' fecal samples, 1,241 presumptive E. coli colonies were analyzed (30 colonies each for 38 subjects; 29, 28, 27, and 17 colonies each for 4 subjects, respectively). Of these, 1,117 colonies (90%) were confirmed as E. coli. According to molecular profiling, the 1,159 total E. coli colonies (1,117 fecal; 42 urine) represented 109 unique (by subject) clones. Of the 109 clones, 4 (4%) were found only in urine (urine-only clones), 38 (35%) in both urine and feces (urine-fecal clones), and 67 (61%) only in feces (fecal-only clones), for a total of 42 urine clones and 105 fecal clones.
Each subject had 1 urine clone and 1 to 11 (mean, 2.5) fecal clones. Comparison of each subject's urine clone and fecal clone(s) placed 14 subjects (33%) in ecological group I, 24 (57%) in group II, and 4 (10%) in group III (Table 1). Thus, in 90% (95% confidence interval [CI], 81% to 99%) of subjects (groups I and II), the urine clone was present in feces, with or without additional (fecal-only) clones.
TABLE 1.
Phylogenetic distribution of 109 Escherichia coli clones from women with acute cystitis in relation to ecological group and source
| Phylogenetic group | Prevalence (no. [%]) of the indicated phylogenetic group within the following ecological group and clone typea:
|
||||
|---|---|---|---|---|---|
| Group I (urine-fecal) (n = 14) | Group II
|
Group III
|
|||
| Urine-fecal (n = 24) | Fecal only (n = 55) | Urine only (n = 4) | Fecal only (n = 12) | ||
| A | 1 (7) | 3 (12) | 29 (53) | 1 (25) | 8 (66) |
| B1 | 0 | 2 (8) | 12 (22) | 0 | 0 |
| B2 | 11 (79) | 15 (62) | 5 (9) | 3 (75) | 2 (17) |
| D | 2 (14) | 4 (17) | 9 (16) | 0 | 2 (17) |
In group I, the urine clone is the sole fecal clone; in group II, the urine clone is present among the fecal clones; in group III, the urine clone is absent from the fecal clones.
Forty-six (42%) of the 109 total clones either were the most abundant clone in the host's fecal sample or were tied for being the most abundant and so were classified as dominant fecal clones. For 30 (71%; 95% CI, 57% to 85%) of the 42 subjects, the urine clone represented the host's dominant fecal clone.
Thirty-six (86%) of the 42 fecal samples yielded ≤4 clones each (i.e., were pauciclonal), collectively accounting for 66 (63%) of the 105 total fecal clones. The other six fecal samples (14%) yielded ≥5 clones each (i.e., were multiclonal), collectively accounting for the remaining 39 (37%) of the 105 fecal clones.
Phylogenetic group.
Table 2 shows the overall distribution of phylogenetic groups among the 109 clones, with a rank order, from the most to the least abundant, of A, B2, D, and B1. Because some subjects had multiple clones, a by-subject analysis yielded higher values than these for the proportion of the 42 women with at least 1 clone from a given phylogenetic group, i.e., group A was found in 42% of women versus 38% of clones, group B1 in 21% versus 13%, group B2 in 71% versus 33%, and group D in 31% versus 16%. Group B2 showed the largest (absolute and proportional) increase in estimated prevalence in the by-subject analysis, surpassing group A to become the most prevalent (by-subject) phylogenetic group overall.
TABLE 2.
Distribution of phylogenetic groups and virulence determinants according to source and the host's fecal clonal diversity among 109 Escherichia coli clones from women with acute cystitis
| Category and traita | Prevalence (no. [%]) among clones
|
P (urine vs fecal clones)b,c | Prevalence (no. [%]) among samples
|
P (pauciclonal vs multiclonal samples)b,d | |||
|---|---|---|---|---|---|---|---|
| Total (n = 109) | Total urine (n = 42) | Fecal only (n = 67) | Pauciclonal (≤4 clones) (n = 66) | Multiclonal (≥5 clones) (n = 39) | |||
| Phylogenetic group | |||||||
| A | 42 (38) | 5 (12) | 37 (55) | <0.001 | 19 (29) | 22 (56) | 0.005 |
| B1 | 14 (13) | 2 (5) | 12 (18) | 0.046 | 8 (12) | 6 (15) | |
| B2 | 36 (33) | 29 (69) | 7 (10) | <0.001 | 24 (36) | 9 (23) | |
| D | 17 (16) | 6 (14) | 11 (16) | 15 (23) | 2 (5) | 0.018 | |
| Adhesins | |||||||
| papC | 27 (25) | 17 (41) | 10 (15) | 0.003 | 23 (35) | 4 (10) | 0.005 |
| papA | 25 (23) | 17 (41) | 8 (12) | 0.001 | 20 (30) | 4 (10) | 0.018 |
| papG alelle II | 19 (17) | 12 (29) | 7 (10) | 0.015 | 18 (27) | 1 (3) | 0.001 |
| papG alelle III | 4 (4) | 4 (10) | 0 | 0.020 | 3 (5) | 0 | |
| fimH | 105 (96) | 41 (98) | 64 (96) | 63 (95) | 39 (100) | ||
| afa/draBC | 1 (1) | 1 (2) | 0 | 0 | 0 | ||
| sfa/focDE | 23 (21) | 17 (41) | 6 (9) | <0.001 | 17 (26) | 5 (13) | |
| Toxins | |||||||
| hly | 21 (19) | 15 (36) | 6 (9) | 0.001 | 16 (24) | 3 (8) | 0.033 |
| cnf1 | 17 (16) | 14 (33) | 3 (5) | <0.001 | 15 (23) | 1 (3) | 0.005 |
| Siderophores | |||||||
| fyuA | 60 (55) | 37 (88) | 23 (34) | <0.001 | 41 (62) | 15 (38) | 0.019 |
| iutA | 46 (42) | 26 (62) | 20 (30) | 0.001 | 34 (52) | 10 (26) | 0.009 |
| Protectins | |||||||
| kpsM II | 50 (46) | 34 (81) | 16 (24) | <0.001 | 37 (56) | 9 (23) | 0.001 |
| traT | 52 (48) | 24 (57) | 28 (42) | 34 (52) | 15 (38) | ||
| Invasin | |||||||
| ibeA | 12 (11) | 8 (19) | 4 (6) | 4 (6) | 7 (18) | ||
| Pathogenicity island marker | |||||||
| malX | 42 (39) | 29 (69) | 13 (19) | <0.001 | 29 (44) | 10 (26) | |
Virulence determinants: papC, pilus assembly; papA, P fimbrial structural subunit; papG alelles I (not detected), II, and III, P fimbrial adhesin molecule; fimH, type 1 fimbriae; afa/draBC, Dr-binding adhesions; sfa/focDE, S and F1C fimbriae; hlyA, hemolysin; cnf1, cytotoxic necrotizing factor 1; fyuA, yersiniabactin receptor; iutA, aerobactin; kpsM II, group II capsule synthesis; traT, serum resistance-associated outer membrane protein; ibeA, invasion of brain endothelium; malX, marker for a pathogenicity-associated island from strain CFT073.
P values are shown only for comparisons that yielded P values of <0.05.
For the 19 comparisons, the number of significant associations expected by chance alone (versus the number observed) was 1 (versus 14) at a P value of <0.05, 0.2 (versus 11) at a P value of <0.01, and 0.02 (versus 7) at a P value of <0.001.
For the 19 comparisons, the number of significant associations expected by chance alone (versus the number observed) was 1 (versus 10) at a P value of <0.05, 0.2 (versus 6) at a P value of <0.01, and 0.02 (versus none) at a P value of <0.001 (but 2 significant associations were observed at a P value of 0.001).
Virulence traits.
Of the 16 virulence genes sought, all but papG allele I were detected in ≥1 clone each, with overall prevalences ranging from 1% (afa/draBC) to 96% (fimH) (Table 2). Most individual virulence genes were significantly concentrated within groups B2 and D (data not shown). Accordingly, aggregate virulence scores were highest for group B2 (median, 7; range, 4 to 10), intermediate for group D (median, 5; range, 2 to 7), and lowest for groups A and B1 (median, 2; range, 0 to 4) (P < 0.001 for B2 versus each other group; P < 0.001 for group D versus group A or B1).
Bacterial characteristics versus source and colonization behavior.
Urine clones from ecological groups I, II, and III exhibited similarly high prevalences of group B2 and individual virulence genes, and similarly high aggregate virulence scores (see, e.g., Table 1). Likewise, fecal-only clones from groups II and III exhibited similarly low prevalences of group B2 (see, e.g., Table 1) and individual virulence genes, and similarly low aggregate virulence scores. Thus, all urine clones were combined for comparison with fecal-only clones (Table 2). Urine clones collectively exhibited significantly higher prevalences of group B2 and of multiple individual virulence genes, and significantly lower prevalences of groups A and B1, than the fecal-only clones. Likewise, the mean aggregate virulence scores of urine clones were significantly higher (mean scores, 6.2 [range, 1 to 10] versus 2.9 [range, 0 to 10]; P < 0.001).
Compared with nondominant fecal clones, dominant fecal clones more commonly represented the host's urine clone (i.e., were urine-fecal clones) (79% versus 24%; P < 0.001) and derived from group B2 (52% versus 15%; P < 0.001). They also exhibited higher virulence scores than other clones (5.2 versus 3.2; P < 0.001). Likewise, compared with clones from hosts with multiclonal fecal samples, clones from pauciclonal fecal samples exhibited a numerically higher prevalence of group B2, significantly higher prevalences of group D and of individual virulence genes (Table 2), a lower prevalence of group A, and higher aggregate virulence scores (mean score, 4.7, versus 3.0 for multiclonal fecal samples; P < 0.001).
Multilevel modeling.
Due to the nonindependence among multiple isolates acquired from the same sample, we next employed mixed-model regression modeling (21). In the resulting univariate models for predicting urine clone status among the 109 total clones, similar associations were documented as in the comparisons that considered the clones independent. Specifically, phylogenetic group A was a significant (and group B1 a borderline significant) negative predictor of urine clone status (data not shown), whereas group B2 was a significant positive predictor of urine clone status (odds ratio, 20.75; 95% CI, 7.08 to 60.74) and was the strongest univariate predictor overall. Other significant positive predictors included 10 individual virulence traits, the aggregate virulence score (a stronger predictor than any single virulence gene [odds ratio, 1.62; 95% CI, 1.34 to 1.97]), fecal dominance (odds ratio, 11.53; 95% CI, 4.52 to 29.46), relative fecal abundance (odds ratio, 18.37; 95% CI, 5.03 to 67.11), and a pauciclonal fecal sample (odds ratio, 5.03; 95% CI, 1.84 to 13.73).
Next, multivariable mixed models for predicting urine clone status were constructed using all possible combinations of group B2 (as the phylogenetic-group variable), the virulence score (as the virulence trait variable), pauciclonal sample status, and either dominant clone status or absolute fecal prevalence (as alternate fecal-abundance variables). Those models in which all predictor variables remained statistically significant are shown in Table 3. When group B2 was included in the model, the virulence score lost significance, but each of the three colonization variables individually retained statistical significance and improved the predictive power of the model (i.e., lowered the −2 log likelihood value). Without group B2 in the model, the virulence score was statistically significant and remained so in the presence of each of the colonization variables, individually or in pairwise combinations, with the colonization variables also remaining statistically significant.
TABLE 3.
Multivariable analysis of bacterial traits and colonization status as predictors of urine source according to multilevel modeling among 109 Escherichia coli clones from women with acute cystitis
| Model |
P value associated with the following variablea:
|
−2 LLb | ||||
|---|---|---|---|---|---|---|
| Group B2 | Virulence score | Pauciclonal sample | Dominant clone | Prevalence in feces | ||
| 1 | <0.001 | NS | 0.01 | — | — | 93 |
| 2 | <0.001 | NS | NS | 0.001 | — | 92 |
| 3 | <0.001 | NS | NS | — | 0.005 | 92 |
| 4 | — | <0.001 | 0.02 | — | — | 99 |
| 5 | — | <0.001 | — | 0.003 | — | 94 |
| 6 | — | <0.001 | — | — | 0.01 | 97 |
| 7 | — | <0.001 | 0.02 | <0.001 | — | 91 |
| 8 | — | <0.001 | 0.03 | — | 0.01 | 91 |
Models were constructed using all possible combinations of group B2, the virulence score, pauciclonal status of the fecal sample, and either dominant clone status or prevalence in feces. Only the eight models in which all candidate predictor variables yielded a P value of <0.05 are shown. —, variable not included in the particular model; NS, variable that, if added to the particular model, was not significant and did not change the significance of other variables in the model.
−2 LL (log likelihood) reflects the goodness of fit of the model, with lower values indicating a better fit.
Correspondence analysis.
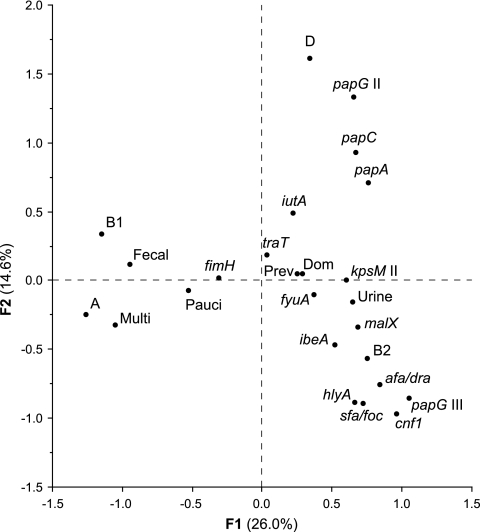
To explore overall relationships among the study variables, correspondence analysis was used. The resulting axes F1 and F2 accounted for 26% and 14.6% of total variance, respectively. In the F1-F2 plane, many virulence factors were clustered closely around urine clone status, as were group B2, dominant clone status, and prevalence (Fig. 1). Pauciclonal fecal-sample status was closer to urine clone status than was multiclonal sample status, whereas multiclonal sample status was closer to fecal-only clone status. Groups A and B1, both quite close to fecal-only clone status, were the variables most distant from urine clone status.
FIG. 1.
Correspondence analysis for bacterial traits and colonization status among 109 Escherichia coli isolates. The 16 virulence traits found among the 109 E. coli isolates, plus the four phylogenetic groups (A, B1, B2, and D), the clone source (urine or fecal), the pauciclonality (Pauci) or multiclonality (Multi) of the fecal sample, dominant (Dom) clone status, and prevalence (Prev) in feces, as computed from the correspondence analysis, are projected on the F1-F2 plane. The F1 axis accounted for 26% of total variance and the F2 axis for 14.6%.
PCoA.
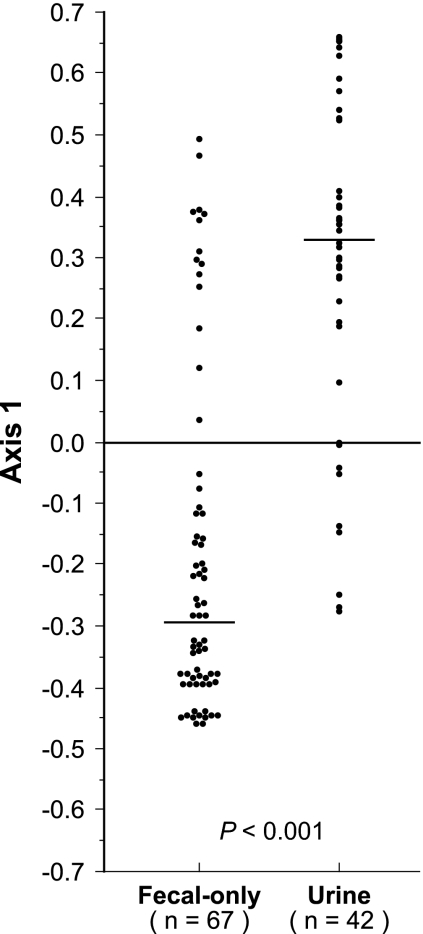
Next, PCoA was used to collapse the multidimensional data set for simplified between-group comparisons. In a PCoA that incorporated individual virulence factors, the phylogenetic group, fecal prevalence, dominant clone status, and pauciclonal sample status, the first three axes accounted for 46.6%, 17.6%, and 11.1% of total variance, respectively. Axis 1 significantly separated the urine and fecal-only isolates (P < 0.001), accounting for 42% of urine versus fecal-only clone status variance (Fig. 2). In contrast, axis 2 did not (P > 0.30), accounting for only 0.7% of urine versus fecal-only status variance. Axis 3 provided borderline significant separation of the urine and fecal-only isolates (P = 0.06) but accounted for only 3% of urine versus fecal-only status variance.
FIG. 2.
Distribution of 109 Escherichia coli isolates in relation to their source (urine or feces only) according to their values on the first axis of the PCoA.
DISCUSSION
In this study of 42 women with acute uncomplicated cystitis due to E. coli, we assessed the relative importance of fecal abundance versus the intrinsic virulence potential in determining which of the host's intestinal E. coli clones would cause the infection. We also undertook a novel assessment of the relationship between the clonal complexity of the fecal E. coli population and virulence-associated characteristics of the constituent strains. For each host, we studied the urine strain and 30 colonies from a pretherapy fecal sample, thereby detecting minority E. coli fecal populations (19).
This study is, to our knowledge, the first to assess a clone's quantitative fecal prevalence, or the pauciclonal versus multiclonal nature of the source fecal sample, in relation to the phylogenetic group, virulence factor profile, and UTI status, and to compare urine and fecal clones from the same hosts according to so broad an array of molecular traits by using sophisticated statistical methods such as multilevel modeling, correspondence analysis, and PCoA. Therefore, in addition to validating our previous findings (12) in a larger population and with greater methodological rigor, the present work also includes several novel features.
The study design allowed each woman to serve as her own control, thereby avoiding possible between-population differences that may confound comparisons of UTI isolates with fecal isolates from healthy hosts. This is an extremely powerful approach for removing the effects of known or unrecognized potential confounders, such as behavioral, environmental, physiological, or genetic differences between infected and uninfected hosts.
Our findings support three main conclusions. First, the urine clones, though almost always detectable in the host's fecal flora at the time of presentation, represented a highly biased subset of that flora, one enriched for traditionally recognized virulence traits and phylogenetic group B2, a result that supports the special-pathogenicity hypothesis. Second, however, the urine clone also tended to be the host's most abundant, or dominant, fecal clone, a result that supports the prevalence hypothesis. Finally, virulence traits and group B2 status were closely associated with fecal abundance, dominance, and pauciclonality. This suggests that prevalence and special pathogenicity are not alternative, mutually exclusive explanations for the occurrence of UTI. Instead, they may both be operational, perhaps contributing jointly to UTI pathogenesis, with virulence factors and other group B2-associated characteristics possibly promoting intestinal dominance (10) and thereby increasing the probability that subsequent steps in pathogenesis will occur.
In this regard, within the total clonal population, fecal abundance (and its close correlate, fecal dominance) was significantly associated with the virulence score (and its close correlate, group B2 status), and each of these domains individually was significantly associated with urine clone status. However, these domains were not precisely overlapping, since some independent predictive power for either domain was evident in the multivariable modeling. This suggests that virulence and abundance, although usually aligned, may sometimes substitute for one another in promoting UTI when either occurs alone. However, among the urine clones overall, dominance within the fecal reservoir did not appreciably reduce the requirement for virulence traits and group B2 status, evidence suggesting that the UTI-promoting effect of fecal abundance is generally insufficient of itself, in intact hosts, to allow low-virulence strains to cause UTI.
The observed association of fecal pauciclonality with group B2 status and virulence is novel and of potential biological and pathogenic significance. Why women with UTI who are colonized with many different E. coli clones tend to have primarily low-virulence, non-B2 clones, whereas those colonized with fewer E. coli clones tend to have high-virulence clones primarily from group B2, is unclear and warrants further study. It may be that certain women are particularly receptive to intestinal colonization with virulent clones, which then outcompete other clones and establish a low-diversity B2-dominated flora, whereas other women can support intestinal colonization with whatever clones they encounter, which probably are mostly low-virulence, non-B2 clones. Alternatively, this phenomenon could be a function of the strains themselves, with B2 strains tending to displace other phylogenetic groups through enhanced niche fitness or inhibition of other clones. Longitudinal studies could elucidate whether intestinal clonal complexity is a relatively stable trait specific to particular hosts or varies over time for a given host; whether expansion of minority B2 clones is typically accompanied by a reduction in overall intestinal clonal complexity; and whether antecedent intestinal clonal diversity predicts UTI risk.
Additional strengths of this study include the comparatively large number of subjects, the systematic approach to recruitment, and the clinical homogeneity of the study population. Limitations include the cross-sectional design, the use of multiple comparisons (which, however, was addressed by using multivariable analysis, correspondence analysis, and PCoA to supplement standard univariate analyses), and the absence of vaginal and periurethral sampling.
In summary, our findings suggest that the relationship between the structure of the fecal E. coli population and UTI pathogenesis is complex. Fecal pauciclonality, clonal dominance or relative abundance, molecularly inferred virulence, and group B2 status are closely intertwined. Phylogenetic group B2 status and/or virulence factors may promote fecal abundance and pauciclonality, thereby contributing to upstream steps in UTI pathogenesis. This relationship, if confirmed, suggests a possible reconciliation of the prevalence and special-pathogenicity hypotheses. Furthermore, pauciclonality of the intestinal E. coli population may represent a previously unrecognized correlate of pathogenicity deserving of further investigation.
Acknowledgments
This work was supported by grants from the Fondo de Investigación Sanitaria (FIS 01/1353 and FIS 02/1887), Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III; the Spanish Network for Research in Infectious Diseases (REIPI C03/14 and REIPI RD06/0008); and the Office of Research and Development, Medical Research Service, Department of Veterans Affairs. All authors declare no conflict of interest.
We thank the study participants for their contributions.
Footnotes
Published ahead of print on 21 May 2008.
REFERENCES
- 1.Andreu, A. 2005. Pathogenesis of urinary tract infections. Enferm. Infecc. Microbiol. Clin. 23(Suppl. 4)15-21. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 2.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 664555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 352977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruneberg, R. N. 1969. Relationship of infecting urinary organism to the faecal flora in patients with symptomatic urinary infection. Lancet ii766-768. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, J. R., T. T. O'Bryan, D. A. Low, G. Ling, P. Delavari, C. Fasching, T. A. Russo, U. Carlino, and A. L. Stell. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect. Immun. 683327-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, J. R., K. Owens, A. Gajewski, and M. A. Kuskowski. 2005. Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J. Clin. Microbiol. 436064-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson, J. R., F. Scheutz, P. Ulleryd, M. A. Kuskowski, T. T. O'Bryan, and T. Sandberg. 2005. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J. Clin. Microbiol. 433895-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181261-272. [DOI] [PubMed] [Google Scholar]
- 9.Kruskal, J. B., and M. Wish. 1978. Multidimensional scaling. Sage Publications, Beverly Hills, CA.
- 10.Le Gall, T., O. Clermont, S. Gouriou, B. Picard, X. Nassif, E. Denamur, and O. Tenaillon. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 242373-2384. [DOI] [PubMed] [Google Scholar]
- 11.Manges, A. R., J. R. Johnson, and L. W. Riley. 2004. Intestinal population dynamics of UTI-causing Escherichia coli within heterosexual couples. Curr. Issues Intest. Microbiol. 549-57. [PubMed] [Google Scholar]
- 12.Moreno, E., A. Andreu, T. Perez, M. Sabate, J. R. Johnson, and G. Prats. 2006. Relationship between Escherichia coli strains causing urinary tract infection in women and the dominant faecal flora of the same hosts. Epidemiol. Infect. 1341015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno, E., I. Planells, G. Prats, A. M. Planes, G. Moreno, and A. Andreu. 2005. Comparative study of Escherichia coli virulence determinants in strains causing urinary tract bacteremia versus strains causing pyelonephritis and other sources of bacteremia. Diagn. Microbiol. Infect. Dis. 5393-99. [DOI] [PubMed] [Google Scholar]
- 14.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken. 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 15.Nowrouzian, F. L., I. Adlerberth, and A. E. Wold. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8834-840. [DOI] [PubMed] [Google Scholar]
- 16.Peakall, R., and P. E. Smouse. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plos, K., H. Connell, U. Jodal, B. I. Marklund, S. Marild, B. Wettergren, and C. Svanborg. 1995. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J. Infect. Dis. 171625-631. [DOI] [PubMed] [Google Scholar]
- 19.Schlager, T. A., T. S. Whittam, J. O. Hendley, J. L. Bhang, C. L. Wobbe, and A. Stapleton. 2003. Variation in frequency of the virulence-factor gene in Escherichia coli clones colonizing the stools and urinary tracts of healthy prepubertal girls. J. Infect. Dis. 1881059-1064. [DOI] [PubMed] [Google Scholar]
- 20.Scholes, D., T. M. Hooton, P. L. Roberts, A. E. Stapleton, K. Gupta, and W. E. Stamm. 2000. Risk factors for recurrent urinary tract infection in young women. J. Infect. Dis. 1821177-1182. [DOI] [PubMed] [Google Scholar]
- 21.Singer, J. D., and J. B. Willett. 2003. Applied longitudinal data analysis. Oxford University Press, New York, NY.
- 22.Stamm, W. E., T. M. Hooton, J. R. Johnson, C. Johnson, A. Stapleton, P. L. Roberts, S. L. Moseley, and S. D. Fihn. 1989. Urinary tract infections: from pathogenesis to treatment. J. Infect. Dis. 159400-406. [DOI] [PubMed] [Google Scholar]
- 23.Stapleton, A., T. M. Hooton, C. Fennell, P. L. Roberts, and W. E. Stamm. 1995. Effect of secretor status on vaginal and rectal colonization with fimbriated Escherichia coli in women with and without recurrent urinary tract infection. J. Infect. Dis. 171717-720. [DOI] [PubMed] [Google Scholar]
- 24.Turck, M., and R. G. Petersdorf. 1962. The epidemiology of nonenteric Escherichia coli infections: prevalence of serological groups. J. Clin. Investig. 411760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 196823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto, S., T. Tsukamoto, A. Terai, H. Kurazono, Y. Takeda, and O. Yoshida. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J. Urol. 1571127-1129. [PubMed] [Google Scholar]
- 27.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 403951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]