Abstract
Information about norovirus (NoV) infections in Central America is limited. Through a passive community and hospital pediatric diarrhea surveillance program, a total of 542 stool samples were collected between March 2005 and February 2006 in León, Nicaragua. NoV was detected in 12% (65/542) of the children; of these, 11% (45/409) were in the community and 15% (20/133) were in the hospital, with most strains (88%) belonging to genogroup II. NoV infections were age and gender associated, with children of <2 years of age (P < 0.05) and girls (P < 0.05) being most affected. Breast-feeding did not reduce the number of NoV infections. An important proportion (57%) of NoV-infected children were coinfected with diarrheagenic Escherichia coli. A significant proportion (18/31) of NoV-positive children with dehydration required intravenous rehydration. Nucleotide sequence analysis (38/65) of the N-terminal and shell region in the capsid gene revealed that at least six genotypes (GI.4, GII.2, GII.4, GII.7, GII.17, and a potentially novel cluster termed “GII.18-Nica”) circulated during the study period, with GII.4 virus being predominant (26/38). The majority (20/26) of those GII.4 strains shared high nucleotide homology (99%) with the globally emerging Hunter strain. The mean viral load was approximately 15-fold higher in children infected with GII.4 virus than in those infected with other G.II viruses, with the highest viral load observed for the group of children infected with GII.4 and requiring intravenous rehydration. This study, the first of its type from a Central American country, suggests that NoV is an important etiological agent of acute diarrhea among children of <2 years of age in Nicaragua.
The importance of norovirus (NoV) infections in endemic community-based gastroenteritis is poorly understood, especially in low-income countries. NoV transmission is facilitated by the small dose of virus required for infection, the unusual stability of the virus outside the host, the prolonged period of virus shedding, and the high genetic variability of the virus (5, 20, 26, 33). Furthermore, poor sanitary conditions might also contribute to the spread of the virus. Since the introduction of PCR methodologies, several studies have shown that NoV is an important pathogen causing endemic gastroenteritis in young children, with infection rates from 7 to 22% (6, 14, 23, 26, 27). Although NoV infections are common, the disease does not seem to be severe enough to require frequent hospitalizations as is the case for rotavirus (24). Nevertheless, 7.1% of young children in the United States with NoV gastroenteritis required hospitalization between 1997 and 1999 (34).
NoV belongs to the family Caliciviridae and exhibits high genetic diversity. Five genogroups (genogroup I [GI] to GV) have been identified based on phylogenetic analysis of the complete capsid gene (33). The data have also revealed that the major human pathogen genogroup, GII, is divided into at least 17 clusters or genotypes (33). While the epidemiological and clinical implications of the genotypes are not fully understood, some reports suggest that a specific genotype resistant to the circulating immunity eventually emerges and increases the number of diarrhea cases in the community (2, 7, 15). Indeed, a striking increase in NoV outbreaks in Europe in 2002 coincided with the emergence of a new NoV GII.4 variant (15). Furthermore, in Australia in 2004, there was an increase in NoV GII activity associated with a novel GII.4 variant (termed “Hunter” virus) (2), and it has been suggested that some variants of NoV GII predominantly affect children (14). Given the emerging importance of NoV as a cause of sporadic diarrhea in children and the potential of this virus to affect populations lacking proper sanitary conditions, we have carried out a molecular epidemiology study to investigate a pediatric population in Nicaragua.
MATERIALS AND METHODS
Site description.
This study was carried out in the city of León, located in the northwest of Nicaragua and with an estimated population of 200,000 inhabitants, 12% of whom are children of less than 4 years of age. The climate is tropical; the rainy season starts in June and lasts until November, when the dry season starts. Sanitary conditions are insufficient in large sections of the city, especially in peripheral areas. The local health system (SILAIS; Local System for Integral Health Care) is comprised of three health centers (covering specific geographic territories and mainly attending outpatients) and the Oscar Danilo Rosales hospital school. Health units belonging to the health centers provide primary care in peripheral areas.
Study design.
Through, a community- and hospital-based study of sporadic acute diarrhea (defined as isolated cases with no known related cases) a total of 542 children of ≤5 years of age with acute diarrhea were enrolled in a longitudinal, prospective manner at five different health facilities from March 2005 to February 2006. After informed consent was given, epidemiological information from each case was obtained from parents or guardians and registered in a paper file. This study was approved by the local ethical committee for biometrics research (registration no. 61).
Clinical assessment.
The clinical information was obtained by reviewing the clinical records of the cases. The information was registered in a paper file containing answers to questions about symptoms such as fever (≥38°C), nausea, vomiting, loss of appetite, abdominal cramps, abdominal distension (gas), and number of loose stools during the past 24 h as well as information on dehydration status and treatment plan. All children were clinically evaluated by pediatricians or general practitioners following the WHO strategy for diarrhea management. This strategy is known as integrated management of childhood illness (IMCI, or AIEPI in Spanish) and was adopted by the Nicaraguan health system (MINSA). In brief, the IMCI strategy states that a child with diarrhea must be classified into one of the following categories depending on the dehydration status: “severe dehydration,” “some dehydration,” and “without dehydration.” Children falling into the “severe dehydration” category require hospitalization and immediate intravenous rehydration (plan C). Those children with “some dehydration” require active oral treatment with oral rehydration solution over a period of 4 hours in the health facility (plan B). After 4 hours, the child is reassessed and hospitalized if needed. Finally, children with no signs of dehydration were treated at home with extra fluids and continuing feeding to prevent dehydration (plan A).
Sampling and preliminary analysis.
Feces were collected in sterile plastic containers and transported at 4°C to the microbiology laboratory of UNAN-León. In hospitalized children, specimens were collected <24 h after admission. A 10% (wt/vol) suspension of the stools was prepared with phosphate-buffered saline (pH = 7.2), and two aliquots were frozen at −20°C for virus analysis (NoV, rotavirus, astrovirus, and adenovirus). Escherichia coli, Shigella spp., and Salmonella spp. were investigated by conventional bacterial culture procedures, and diarrheagenic E. coli (DEC) types (enterotoxigenic E. coli, enteropathogenic E. coli, enteroaggregative E. coli, enteroinvasive E. coli, and enterohemorrhagic E. coli) were studied by PCR-based methods (25). Parasites were investigated using conventional microscopy and staining methods. Furthermore, the presence of mucus or blood in stool was examined.
Viral antigen detection.
Two commercial enzyme immunoassays (enzyme-linked immunosorbent assay [ELISA] kits Idea K6043 and K6044 NoV; Dako Cytomation Ltd., United Kingdom) were used for NoV identification in fecal samples according to the manufacturer's instructions. The results were visually read and confirmed by absorbance measurements. Viral coinfections in the NoV-positive samples were investigated by use of the following three commercial enzyme immunoassays distributed by Dako Cytomation Ltd. (United Kingdom): Idea K6020 for rotavirus, Idea K6042 for astrovirus, and Idea K6021 for adenovirus. The procedures were carried out according to the manufacturer's instructions. The results were visually read and confirmed by absorbance measurements.
RNA extraction.
Viral RNA was extracted from stool suspensions of NoV-positive samples by use of a QIAmp viral RNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. A total of 60 μl of purified viral RNA was obtained and stored at −20°C until reverse transcription (RT) and PCR analysis.
RT.
RT was carried out as previously described (1). Briefly, 28 μl of the purified RNA was mixed with 50 pmol of random hexadeoxynucleotides [pd(N)6], and the mixture was denatured at 97°C for 5 min and quickly chilled on ice for 2 min, followed by the addition of one RT-PCR bead (Amersham Biosciences, United Kingdom) and RNase-free water to a final volume of 50 μl. The RT reaction was carried out for 30 min at 42°C to produce cDNA.
NoV confirmation by conventional PCR and real-time PCR.
The RT products were amplified by NoV PCR using a degenerated primer pool, p289hi/290hijk, which consisted of four different forward and two reverse primers (34) targeting the gene encoding the RNA-dependent RNA polymerase in ORF1, resulting in an amplicon of 319 bp for NoV. PCR conditions were used as described in reference 8. The presence of NoV was further confirmed with a genogroup-specific real-time PCR assay that uses primers NVG1f1b, NVG1rlux, NVG2flux1, and COG2R and that amplifies 88- and 89-bp fragments of the orf1-orf2 junction for NoV GI and GII, respectively (21).
Estimation of viral load.
In a subset of samples (n = 41), the viral load was quantified by real-time PCR in duplicate by use of sevenfold serial dilutions (101 to 107 copies) of external plasmid standards (21).
Selecting an approach for genotyping.
Two different PCR assays were evaluated to select an appropriate approach to define cluster or NoV genotypes. Fourteen samples randomly selected from NoV-positive specimens were analyzed by both assays. The first PCR assay, amplifying the N-terminal and shell (NS) region of the capsid gene (12, 21), could detect 9 of 14 samples (64%). The second PCR, amplifying region D downstream in the same gene (30), detected 7 of 14 samples (50%). Furthermore, more PCR product was observed with first PCR assay than with the second (data not shown). Based on these results, the PCR assay targeting the NS region was selected to be used for genotyping.
PCR amplifying the NoV NS region.
Five microliters of cDNA was added to a mix containing 5 μl of 10× high-fidelity PCR buffer (Invitrogen, Carlsbad, CA), 2 μl of 50 mM MgCl2, 1 μl of 10 mM deoxynucleoside triphosphate mix (Applied Biosystems, Warrington, United Kingdom), 1 μl of 10 pmol of each GI-specific primer (NVGIF1b and G1SKR) (12, 21) or GII primer (NVG2flux1 and G2SKR) (12, 21), 1 U of Platinum Taq DNA polymerase high-fidelity buffer (Invitrogen, Carlsbad CA), and RNase-free water to a final volume of 50 μl. PCRs in separate tubes for GI and GII were performed under the following conditions: 94°C for 4 min followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 7 min. The 381-bp and 390-bp amplicons obtained from GI and GII viruses, respectively, were visualized by 2% agarose gel electrophoresis followed by ethidium bromide staining.
PCR amplifying the NoV D region.
Five microliters of cDNA and 1 μl of 10 pmol of each GII-specific primer (forward, Cap D1 and Cap D3; reverse, Cap C) (30) were added to a mix containing the PCR reagents described for the PCR amplifying the NoV NS region. Thermocycling conditions were used as described elsewhere (30). A 253-bp PCR product was visualized as described above.
Nucleotide sequencing of the NS region.
The PCR products were purified using a reaction mixture containing 2.5 μl PCR product and 1 μl ExoSap-IT enzyme (GE Healthcare, Chalfont St. Giles, United Kingdom). This reaction was performed at 37°C for 15 min, followed by inactivation of the enzyme at 80°C for 15 min. Nucleotide sequencing was performed by using a DYEnamic dye terminator kit (GE Healthcare, Chalfont St. Giles, United Kingdom) with the primers for the NS region as previously described and a Mega BACE 500 automated sequencer (GE Healthcare, Chalfont St. Giles, United Kingdom). The PCR products were sequenced twice in both forward and reverse directions. Complete nucleotide sequences (accession numbers EU780735 to EU780768) were obtained by assembling overlapping contigs with DNASTAR (Madison, WI).
Genotyping by sequence analysis of the NS region.
Sequence alignment of the Nicaraguan strains and reference NoV genotypes was performed by using the Clustal W algorithm, version 1.83, and default parameters on the European Bioinformatics Institute server (EMBL-EBI). Phylogenetic analysis was performed using the MEGA 3.1 software package and the tree was constructed using the neighbor-joining and Kimura two-parameter methods (11, 28). The significance of the relationship was obtained by bootstrap resampling analysis (1,000 replications). The assignment of genotypes was done using pairwise nucleotide distance measurements as proposed in reference 10.
Statistical analysis.
NoV-positive specimens were compared in terms of gender, age group, and breast-feeding status by use of chi-square tests or Fisher's exact test. Mean viral loads were compared between genotypes and rehydration plan groups by use of analysis of variance, and tendencies were estimated by regression analysis. The software SPSS (Statistical Program for Social Science version 10.0.1 for Windows; Chicago, IL) and StatCalc from EpiInfo version 6 (CDC, Atlanta, GA) were used for statistical analysis.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this work are available under GenBank accession numbers EU780735 to EU780768.
RESULTS
NoV infections in Nicaragua are associated with age and gender but not with breast-feeding.
NoV was detected in 65 (12%) of the 542 stool samples analyzed with girls (15%), who were more frequently infected than were boys (10%) (P = 0.044), and children of less than 2 years of age were symptomatically infected more frequently than were children of 2 to 5 years of age (P = 0.002) (Table 1). However, in the group of hospitalized children no statistically significant association between gender and NoV infection was observed (P = 0.800), nor did breast-feeding reduce the number of NoV infections in this study (P = 0.107) (Table 1).
TABLE 1.
Epidemiological profile of pediatric norovirus diarrhea in children of ≤5 years of age from Leoń, Nicaragua
| Parameter | Value for:
|
||||||
|---|---|---|---|---|---|---|---|
| Both settings (n = 542)
|
Community (n = 409)
|
Hospital (n = 133)
|
|||||
| Total | No. of NoV strains (%) | P value | Total | No. of NoV (%) | Total | No. of NoV strains (%) | |
| Gender | |||||||
| Male | 321 | 31 (10) | 0.044a | 234 | 17 (7)b | 87 | 14 (16)c |
| Female | 221 | 34 (15) | 175 | 28 (16) | 46 | 6 (13) | |
| Age range (mo) | |||||||
| ≤6 | 96 | 12 (12) | 0.002d | 66 | 9 (14) | 30 | 3 (10) |
| 7-12 | 152 | 25 (16) | 100 | 15 (15) | 52 | 10 (19) | |
| 13-24 | 183 | 24 (13) | 141 | 17 (12) | 42 | 7 (17) | |
| 25-60 | 111 | 4 (4) | 102 | 4 (4) | 9 | 0 (0) | |
| Breast-feeding statuse | |||||||
| Exclusively breast-fed | 38 | 5 (13) | 0.107f | 30 | 3 (10) | 8 | 2 (25) |
| Nonexclusively breast-fed | 279 | 45 (16) | 216 | 33 (16) | 63 | 12 (18) | |
| Not breast-fed | 114 | 11 (10) | 61 | 5 (10) | 53 | 6 (11) | |
Chi-square test.
P = 0.005; chi-square test (female versus male in the community).
P = 0.80; Fisher's exact test (female versus male hospitalized children).
Chi-square test (≤24 versus 25 to 60 months of age).
Evaluated only for children of ≤24 months of age.
Chi-square test (breast-fed [exclusively and nonexclusively] versus not breast-fed).
Incidence of NoV in the community and in hospitalized children.
Forty-five (11%) of 409 children from the community were NoV positive, whereas NoV was detected in 20 (15%) of 133 hospitalized children (Table 1). NoV belonging to GII was the most common and was found in 88% (57/65) of the children, followed by GI, which was found in 11% (7/65). For one sample, the genogroup could not be determined (Table 2).
TABLE 2.
Seasonal distribution of pediatric norovirus infections and genogroups in Leon, Nicaragua
| Datea | No. of diarrheic samples | No. of NoV strains (%) | No. of NoV strains from indicated genogroupb
|
|
|---|---|---|---|---|
| GI | GII | |||
| Mar-05 | 22 | 1 (5) | 1 | |
| Apr-05 | 48 | 5 (10) | 5 | |
| May-05 | 52 | 8 (15) | 8 | |
| Jun-05 | 137 | 21 (15) | 5 | 16 |
| Jul-05 | 104 | 14 (13) | 2 | 12 |
| Aug-05 | 42 | 3 (7) | 3 | |
| Sept-05 | 20 | 1 (5) | 1 | |
| Oct-05 | 66 | 6 (9) | 6 | |
| Nov-05 | 28 | 1 (4) | 1 | |
| Dec-05 | 2 | 0 (0) | ||
| Jan-06 | 11 | 4 (36) | 3 | |
| Feb-06 | 10 | 1 (10) | 1 | |
| All dates | 542 | 65 (12) | 7 | 57 |
Mar, March; Apr, April; Jun, June; Jul, July; Aug, August; Sept, September; Oct, October; Nov, November; Dec, December; Jan, January; Feb, February; -05, 2005; -06, 2006.
Genogroup was not assigned for one NoV-positive sample that was confirmed NoV positive by ELISA and RT-PCR.
The seasonal peak of diarrhea is associated with the peak of NoV infections.
The highest percentage (36%; 4/11) of NoV infections was observed in January 2006, but the highest absolute number (43/65) of NoV infections occurred at the beginning of the rainy season (May to July 2005) and coincided with a nationwide epidemic peak of diarrhea (17) (Table 2). Most of the coinfections of NoV-positive children (30/37) occurred during this period. Furthermore, most of the hospitalized cases associated with NoV (14/18) occurred during the same rainy period, with GII virus being the predominant genogroup (10/14). GI viruses were detected only during the rainy season (Table 2).
Clinical features in single NoV infections.
To accurately describe a clinical profile of NoV disease in children, it was necessary to investigate all NoV-positive specimens for coinfections with other enteropathogens, including DEC, rotavirus, astrovirus, adenovirus, Entamoeba histolytica, Giardia lamblia, Ascaris lumbricoides, and Cryptosporidium spp. In 37 (57%) of the 65 NoV-positive cases, at least one more pathogen was found, with DEC being the most frequent coinfection, followed by adenovirus and Cryptosporidium. No coinfection with rotavirus was observed. For 28 (43%) of the 65 children, only NoV was detected, and these children presented with symptoms including appetite loss (79%), four to six loose stools in the previous 24 h (46%), vomiting (54%), dehydration (43%), fever of ≥38°C (29%), abdominal distension (14%), and mucus in the stools (14%); other signs, such as abdominal cramping, were reported for less than 4% (Table 3). The occurrence of coinfections modified the clinical pictures of NoV disease observed for singly infected children. Relevant modifications were occurrence of fever (43% versus 29%), mucus in the stool (22% versus 14%), >6 loose stools in the previous 24 h (54% versus 21%, P = 0,007), watery stools (78% versus 61%), and severe dehydration (35% versus 18%) (Table 3).
TABLE 3.
Clinical features of NoV-infected children with and without concomitant infections
| Clinical feature | No. (%) of NoV-positive children with:
|
OR (95% CI)b | P value | |
|---|---|---|---|---|
| Coinfectiona (n = 37) | “Single” infection (n = 28) | |||
| Fever of ≥38°C | 16 (43) | 8 (29) | 1.90 (0.59-6.21) | 0.225 |
| Vomiting | 18 (49) | 15 (54) | 0.82 (0.27-2.46) | 0.694 |
| Loss of appetite | 31 (84) | 22 (79) | 1.41 (0.34-5.84) | 0.591 |
| Abdominal cramping | 0 (0) | 1 (4) | NAe | NA |
| Abdominal distension (gas) | 5 (14) | 4 (14) | 0.94 (0.19-4.76) | 0.602f |
| Mucus | 8 (22) | 4 (14) | 1.66 (0.38-7.56) | 0.336f |
| No. of loose stools in the past 24 h | ||||
| 1-3 | 4 (11) | 9 (32) | 0.26 (0.06-1.09) | 0.034f |
| 4-6 | 13 (35) | 13 (46) | 0.63 (0.20-1.92) | 0.357 |
| >6 | 20 (54) | 6 (21) | 4.31 (1.26-15.35) | 0.007 |
| Stools | ||||
| Watery | 29 (78) | 17 (61) | 2.35 (0.69-8.06) | 0.121 |
| Loose | 8 (22) | 11 (39) | 0.43 (0.12-1.44) | 0.121 |
| Dehydration statusc | ||||
| Severe dehydrationd | 13 (35) | 5 (18) | 2.49 (0.68-9.62) | 0.123 |
| Some dehydration | 5 (14) | 2 (7) | 2.03 (0.31-16.63) | 0.344f |
| No dehydration | 19 (51) | 21 (75) | 0.35 (0.10-1.15) | 0.052 |
Coinfections were with DEC, adenovirus, or Cryptosporidium.
OR, odds ratio; 95% CI, 95% confidence interval.
Evaluated by following the EMCI strategy.
After clinical reassessment, 18 of the 31 NoV-positive children initially classified as having some dehydration were reclassified as having severe dehydration because they required hospitalization and intravenous rehydration.
NA, not applicable.
Fisher's exact test.
Clinical reassessment of NoV-positive children with some dehydration revealed that a significant proportion (18/31) required immediate intravenous rehydration (Table 3).
Genotypes and seasonality.
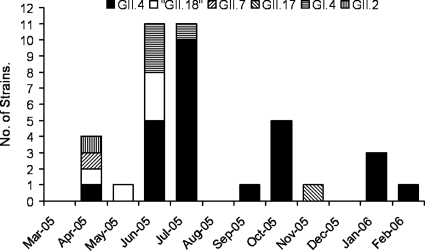
The highest diversity of NoV genotypes was observed in April, when at least four genotypes circulated; these four genotypes were GII.2, GII.4, GII.7, and “GII.18” (Fig. 1). Interestingly, one strain belonging to NoV GII did not match any of the 17 previously described genotypes (33) and was therefore tentatively assigned to “GII.18” (Fig. 2). This novel genotype also circulated in May and June 2005 (Fig. 1). During June, GI.4 and GII.4 were observed. In July, the genotype diversity was restricted, but more importantly, genotype GII.4 become predominant (10/11) (Fig. 1). During the following months, the NoV-positive specimens decreased, but in October, GII.4 reappeared once again. In November, the uncommon genotype GII.17 appeared, and during January 2006, the percentage of NoV-positive isolates increased up to 36%, and this increase was associated with the reemergence of GII.4 (Fig. 1).
FIG. 1.
Dynamics of NoV genotypes in children of ≤5 years of age with acute diarrhea in the period between March 2005 and February 2006. Of the genotyped strains, 97% were from children of ≤2 years of age. Feb, February; Mar, March; Apr, April; Jun, June; Jul, July; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December; Jan, January; -05, 2005; -06, 2006.
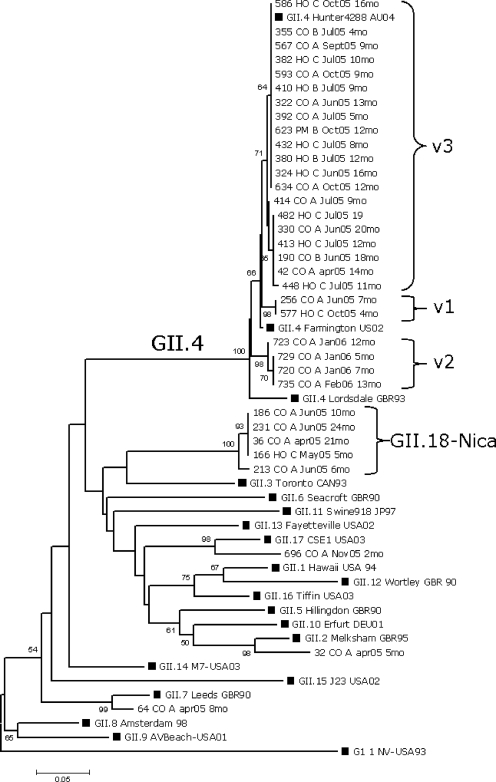
FIG. 2.
Phylogenetic analysis of the capsid NS region of GII NoV strains (n = 34). The tree was constructed based on the Kimura two-parameter and neighbor-joining methods by use of MEGA 3.1 software. Bootstrap values are shown at the branch nodes (values of <50% are not shown). GI.1 NV-USA 93 was used as the out-group. The black squares represent reference strains. For Nicaraguan strains, the number of the strain is given followed by patient hospitalization or community status (HO or CO, respectively), the patient rehydration plan (A, B, or C), the date of sample collection (see Fig. 1 legend for abbreviations), and the age of the child (mo, months).
Nucleotide sequence analysis of the NS and D regions of strains belonging to the novel cluster (“GII.18”) revealed low nucleotide homologies (84% and 77%, respectively) compared with what was seen for NoV sequences published in GenBank. Pairwise distance calculation of the NS region (mean, 0.314; range, 0.203 to 0.328) strongly suggests that the novel cluster represents a new genotype (10). Furthermore, a group of strains with >20% nucleotide variation in region D has been suggested to represent a novel genotype (30).
Molecular and epidemiological patterns of NoV GII.4.
Nucleotide and amino acid sequences of the GII.4 strains (n = 26) were compared with each other and with those of GII.4 reference strains. Thirteen substitutions at nucleotide positions 26, 99, 108, 117, 161, 162, 189, 192, 198, 222, 225, 226, and 234 were observed in the NS region of the GII.4 Nicaraguan strains. However, only substitutions at nucleotide positions 26 (G/A/C) and 161 (A/G) lead to amino acid changes. Based on these changes, GII.4 Nicaraguan strains were divided in three variants corresponding to the v1, v2, and v3 variants described by Gallimore and colleagues (7) (Fig. 2). These variant-defining point mutations encoded the following amino acid motifs at positions 6, 9, and 15, respectively: for v1, N6S9T15; for v2, N6N9T15; and for v3, N6T9T15. Most of the sequenced GII.4 strains (20/26) were classified as v3 and are highly related to “Hunter” virus, which was suggested to be a globally emerging strain in 2004 (2) (Fig. 2). The highest frequencies of v3 strains were observed in June, July, and October, months that are associated with a generally increased activity of NoV (Table 2). The v3 strains predominantly infected children of more than 6 months of age living in different areas of the city. Importantly, 7 out of 20 children infected with the v3 variant required hospitalization and intravenous rehydration. The v2 strains were found in January and February 2006, when the highest frequency (4/11) of NoV infections was observed. The v2 variant circulated in children who were living in the same neighborhood and who did not require rehydration. Variant 1 strains were isolated in June and October from hospital and community cases.
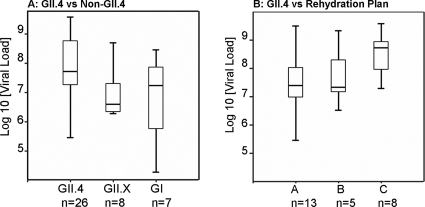
Viral load and dehydration.
To investigate if clinical severities were associated with virus concentration and genotype, a subset of randomly selected samples (n = 41) belonging to GI and GII were analyzed by real-time PCR (21). The geometric mean viral loads of NoV GI (n = 7) and GII (n = 34) were 5.7 × 106 and 3.8 × 107 genome equivalents per gram of fecal specimen, respectively (P = 0.305) (Fig. 3A). Furthermore, virus concentrations in specimens from children infected with NoV GII.4 were approximately 15-fold higher than those in specimens from children infected with other GII genotypes (7.2 × 107 versus 4.8 × 106) (P = 0.210) and 13-fold higher than those seen for GI genotypes (7.2 × 107 versus 5.7 × 106) (P = 0.235). The highest viral load was observed for the group of children infected with GII.4 and requiring intravenous rehydration (mean, 3.2 × 108) (Fig. 3B).
FIG. 3.
Viral load determined by genome equivalents per gram of feces. (A) GII.X represents GII.2, GII.7, GII.17, and “GII.18.” (B) Shedding in children infected with GII.4 virus and requiring either intravenous rehydration (plan C), active oral treatment with oral rehydration solution in the health center (plan B), or oral rehydration at home (plan A). Boxes represent interquartile ranges that contain 50% of the values. Lines across the boxes indicate medians.
DISCUSSION
This study describes the incidence and genetic diversity of NoV in pediatric diarrhea in a Central American country. The incidence found for this study (12%) is in the range found for other reports and confirms that infection rates in the studied population are similar to those for other high- and low-income countries (18). However, the rate (15%) of NoV diarrhea cases requiring hospitalization, which was high in comparison to those found in previous studies carried out in France, Australia, and the United States (13, 34), deserves special attention. The real hospitalization rate associated with NoV diarrhea is probably even higher, considering the sensitivity of the ELISA method used, which was 77% compared with that of RT-PCR assays (32). Attempts were made to estimate the frequency of false-negative samples. Reexamination of 33 randomly selected ELISA-negative samples revealed that 9% (3/33) of these were indeed NoV positive after RT-PCR and sequencing analysis. This suggests that the observed incidence of 12% is probably 10 to 20% higher in reality. However, due to practical circumstances, ELISA was used as the screening method instead of RT-PCR.
Several studies have suggested that NoV infection in young children occurs right after weaning or when NoV antibodies are not present at protective levels (6, 23, 26). In agreement with this, we found a marked trend toward higher rates of infection in children younger than 2 years of age (14% for those of ≤2 years of age versus 4% for those of >2 years of age [P < 0.01]), with children between 6 to 24 months of age being the most frequently affected (Table 1).
An association between breast-feeding and low rates of NoV infection, possibly due to the protective effect of maternal antibodies or the presence of fucosylated glycans in the breast milk of secretor-positive mothers, has previously been suggested (9, 16, 19). In the current study, no association was observed (the infection rate for breast-fed children was 16% and that for non-breast-fed children was 10% [P = 0.107]). One possible explanation is that the mothers may not have been exposed previously to the circulating strains and therefore lacked specific protective NoV antibodies, especially immunoglobulin A (16). Another possibility is that even though fucosylated glycans, such as the H antigen and Leb epitopes, are present in the breast milk of secretor-positive mothers, they did not play a protective role against the symptomatic infection.
From 1996 onward, a marked increase of gastroenteritis outbreaks has been associated with NoV GII infections and particularly with GII.4 infections. Specific variants within this genotype, such as the Lordsdale, Farmington Hills, and Hunter strains and, more recently, the 2006a and 2006b strains, have emerged in various geographic regions and spread around the world (2, 29, 31). Recently, Gallimore and coworkers found that within a 3-year period a single specific variant of GII.4 emerged in the United Kingdom and became predominant for a period until it was replaced by another variant from the pool (7). In the current study, we also observed emergence and selection of variants within a 1-year period. The most important increase of NoV infections was associated with v3 or “Hunter-like” strains, probably selected in a period of high genetic diversity, perhaps in April 2005 (Fig. 1). The described NoV increase may have been associated with the epidemic peak of diarrhea in Nicaragua between June and July 2005 (17). The observation that 90% (9/10) of the sequenced GII.4 strains from hospitalized children belonged to the v3 variant supports the hypothesis that the v3 variant is highly virulent (Fig. 2). While the v3 variant disappeared in January 2006, it was replaced by the v2 variant, which is likely to represent a spatial and temporal cluster. The highest percentage of NoV infection was observed in January 2006 and was associated with variant v2. An example of the sharp increase and rapid spread of certain NoV variants over large geographic areas was observed for the Farmington Hills strain in the United States (31). Our observation extends previous knowledge about the emergence and selection of GII.4 variants and suggests that particular variants with increased fitness are selected from a pool of cocirculating variants.
Recombination is a significant driving force for the evolution of NoV, and most recombination events occur in the ORF1-ORF2 junction located upstream of the capsid gene (3, 10). In this study, we have investigated the relationship between genotypes, as defined by partial capsid sequences, and disease severity. It is likely that some of the NoV strains investigated have undergone recombination events, but this probably did not affect the relationship between genotype and disease severity, since recombination does not usually take place within the capsid gene (3).
Interestingly, we found five cases of NoV belonging to a new genotype, tentatively termed GII.18-Nica. This was confirmed not only by analysis of sequences from the NS region but also by sequencing of region D (30). Furthermore, we sequenced 150 bp of the 3′ end of the polymerase gene for one GII.18 sample, which also clustered separately from other described genotypes (data not shown), further indicating that this strain is indeed distinct.
Chan and coworkers speculated that NoV GII strains have higher transmissibility than GI strains, as the viral load is 100-fold higher in patients infected with GII strains than in patients infected with GI strains (4). Our observations do not support such a general conclusion, because even though individuals infected with NoV GII shed higher amounts of viral genomes (7-fold), these amounts were was not as much as 100-fold higher. In agreement with our observation, Ozawa and coworkers reported that NoV GII has a mean viral load higher (13-fold) than that of NoV GI (22). The variance in titers of shed virus was high for all observations, and therefore no significant difference in the levels of shedding for GI and GII was observed (Fig. 3A).
Furthermore, the viral load was approximately 15-fold higher in children infected with GII.4 strains than in children infected with other GII strains. Moreover, increased virus shedding in children infected with specific genotypes might be associated with increased clinical severity. We observed that the mean viral load was higher among hospitalized children (plan C) than among nonhospitalized children infected with GII.4 virus (Fig. 3B). However, it must be taken into account that not only genotypes but also individual variations might be associated with high viral load. For instance, four individuals infected with GII.4 virus which was sampled 2 days after the onset of symptoms shed various amounts of virus, indicating that virus shedding is not only genotype dependent but also host dependent. The observed differences are also likely to depend on the time of sampling.
Interestingly, a trend toward increased virus shedding and lengths of illness was observed for children infected with GII.4 virus. In contrast, a trend toward decreased shedding and lengths of illness was observed for children infected with GI and other GII genotypes (data not shown).
Acknowledgments
This study was supported by grants from the Swedish International Development Cooperation Agency (grant 075007109), the Swedish Research Council (grant 10392), and the Swedish Council for Forestry, Environment and Spatial Planning (grant 245-2004-1821).
We thank Patricia Roiz, Soledad Calderon, Brenda Mora, and Silvia Altamirano for their valuable assistance with laboratory activities and fieldwork. We also thank Samuel Vilchez for DEC PCR analysis of NoV infection samples.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Bucardo, F., B. Karlsson, J. Nordgren, M. Paniagua, A. Gonzalez, J. J. Amador, F. Espinoza, and L. Svensson. 2007. Mutated G4P[8] rotavirus associated with a nationwide outbreak of gastroenteritis in Nicaragua in 2005. J. Clin. Microbiol. 45990-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, R. A., M. M. Tanaka, and P. A. White. 2007. Norovirus recombination. J. Gen. Virol. 883347-3359. [DOI] [PubMed] [Google Scholar]
- 4.Chan, M. C., J. J. Sung, R. K. Lam, P. K. Chan, N. L. Lee, R. W. Lai, and W. K. Leung. 2006. Fecal viral load and norovirus-associated gastroenteritis. Emerg. Infect. Dis. 121278-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duizer, E., P. Bijkerk, B. Rockx, A. De Groot, F. Twisk, and M. Koopmans. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 704538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas, T., X. Jiang, M. L. Guerrero, W. Zhong, N. Wilton, T. Berke, D. O. Matson, L. K. Pickering, and G. Ruiz-Palacios. 2000. Prevalence and genetic diversity of human caliciviruses (HuCVs) in Mexican children. J. Med. Virol. 62217-223. [PubMed] [Google Scholar]
- 7.Gallimore, C. I., M. Iturriza-Gomara, J. Xerry, J. Adigwe, and J. J. Gray. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 1521295-1303. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83145-154. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, X., P. Huang, W. Zhong, M. Tan, T. Farkas, A. L. Morrow, D. S. Newburg, G. M. Ruiz-Palacios, and L. K. Pickering. 2004. Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J. Infect. Dis. 1901850-1859. [DOI] [PubMed] [Google Scholar]
- 10.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299225-239. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide. J. Mol. Evol. 16111-120. [DOI] [PubMed] [Google Scholar]
- 12.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100107-114. [DOI] [PubMed] [Google Scholar]
- 13.Koopmans, M. 2003. Molecular epidemiology of human calicivirus, p. 523-554. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Elsevier, Amsterdam, The Netherlands.
- 14.Lindell, A. T., L. Grillner, L. Svensson, and B. Z. Wirgart. 2005. Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in children. J. Clin. Microbiol. 431086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363682-688. [DOI] [PubMed] [Google Scholar]
- 16.Makita, K., Y. Hayakawa, M. Okame, K. Homma, T. G. Phan, S. Okitsu, and H. Ushijima. 2007. First detection of IgA against norovirus in breast milk. Clin. Lab. 53125-128. [PubMed] [Google Scholar]
- 17.Ministerio de Salud de Nicaragua. 2005. Boletin epidemiologico del Ministerio de Salud, no. 42. Ministerio de Salud de Nicaragua, Managua, Nicaragua.
- 18.Moreno-Espinosa, S., T. Farkas, and X. Jiang. 2004. Human caliciviruses and pediatric gastroenteritis. Semin. Pediatr. Infect. Dis. 15237-245. [DOI] [PubMed] [Google Scholar]
- 19.Morrow, A. L., G. M. Ruiz-Palacios, X. Jiang, and D. S. Newburg. 2005. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 1351304-1307. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 7713117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordgren, J., F. Bucardo, O. Dienus, L. Svensson, and P. E. Lindgren. 2008. Novel light-upon-extension real-time PCR assays for detection and quantification of genogroup I and II noroviruses in clinical specimens. J. Clin. Microbiol. 46164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa, K., T. Oka, N. Takeda, and G. S. Hansman. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 453996-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang, X. L., J. Joensuu, and T. Vesikari. 1999. Human calicivirus-associated sporadic gastroenteritis in Finnish children less than two years of age followed prospectively during a rotavirus vaccine trial. Pediatr. Infect. Dis. J. 18420-426. [DOI] [PubMed] [Google Scholar]
- 24.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 382001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rockx, B., M. De Wit, H. Vennema, J. Vinje, E. De Bruin, Y. Van Duynhoven, and M. Koopmans. 2002. Natural history of human calicivirus infection: a prospective cohort study. Clin. Infect. Dis. 35246-253. [DOI] [PubMed] [Google Scholar]
- 27.Roman, E., A. Negredo, R. M. Dalton, I. Wilhelmi, and A. Sanchez-Fauquier. 2002. Molecular detection of human calicivirus among Spanish children with acute gastroenteritis. J. Clin. Microbiol. 403857-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 29.Tu, E. T., R. A. Bull, G. E. Greening, J. Hewitt, M. J. Lyon, J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII. 4 variants 2006a and 2006b. Clin. Infect. Dis. 46413-420. [DOI] [PubMed] [Google Scholar]
- 30.Vinje, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116109-117. [DOI] [PubMed] [Google Scholar]
- 31.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus-United States, 2002. J. Infect. Dis. 19027-36. [DOI] [PubMed] [Google Scholar]
- 32.Wilhelmi de Cal, I., A. Revilla, J. M. del Alamo, E. Roman, S. Moreno, and A. Sanchez-Fauquier. 2007. Evaluation of two commercial enzyme immunoassays for the detection of norovirus in faecal samples from hospitalised children with sporadic acute gastroenteritis. Clin. Microbiol. Infect. 13341-343. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]
- 34.Zintz, C., K. Bok, E. Parada, M. Barnes-Eley, T. Berke, M. A. Staat, P. Azimi, X. Jiang, and D. O. Matson. 2005. Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infect. Genet. Evol. 5281-290. [DOI] [PubMed] [Google Scholar]