Abstract
We evaluated the BURP (based upon repeat patterns) algorithm, which relies on sequencing of the Staphylococcus aureus protein A gene (spa), for its ability to infer clonal relatedness within a population of 110 wild-type strains. BURP clustering of the resulting 66 spa types was highly concordant with multilocus sequence typing (96.5% concordance) and pulsed-field gel electrophoresis (94.9%).
Staphylococcus aureus, the leading cause of nosocomial infections worldwide, causes a wide variety of infections (14, 18). In recent decades, the worldwide spread of methicillin-resistant S. aureus (MRSA) has become a major challenge in health care (5). More recently, community-acquired MRSA has intensified public health concerns (26). To understand these changes in epidemiology, different typing methods have been applied, including phage typing, pulsed-field gel electrophoresis (PFGE), and sequence-based typing methods. Sequence-based typing offers the advantage that results are easy to compare and communicate between different laboratories. Multilocus sequence typing (MLST) has become the “gold standard” for population analysis (15), but it has low discriminatory power and is expensive, so this method is mainly restricted to reference laboratories. As a result of the predominantly clonal evolution of S. aureus, sequencing of the repeat region of the protein A gene (spa) generates informative typing results and has quickly been established as a robust and highly discriminatory method (1, 3, 10, 16, 21, 24). The spa region consists of a variable number of 21- to 27-bp repeats with differing nucleotide compositions that result in different spa types. It has been observed that this region provides information not only about short-term epidemiology but also about long-term phylogeny and contains a reliable signal that could be utilized for the determination of clonal relatedness among individual strains (12).
Here we investigated a well-characterized collection of methicillin-sensitive S. aureus (MSSA) carriage strains by spa typing. To evaluate the ability of spa typing to determine the clonal relatedness of a natural population of S. aureus strains, we used the recently described grouping algorithm that is “based upon repeat patterns” (BURP) to cluster related spa types (17) and compared the results with MLST clonal complexes (CCs) and PFGE groups.
(This study was presented in part at the 107th General Meeting of the American Society for Microbiology, Toronto, Ontario, Canada, 21 to 25 May 2007.)
One hundred ten MSSA carriage strains that originated from a random and representative sample of nonhospitalized elderly individuals living independently in the Nottingham Health district (8) were analyzed. This collection was previously characterized by MLST, PFGE, phage typing, and analysis of randomly amplified polymorphic DNA (6). spa typing was performed as described elsewhere (4, 10, 16). BURP clusters (spa CCs) for these strains were determined, and clustering was portrayed with Ridom StaphType (version 1.5) software (Ridom GmbH, Würzburg, Germany). BURP offers two user-defined parameters that influence clustering: exclusion of spa types that are shorter than a certain number (x) of repeats and the maximum number of costs (y) for clustering spa types into the same group. Short spa types can be excluded from further analysis, because their information content is limited and no reliable evolutionary history can be inferred. The costs account for the genetic changes between two different spa types, whereas the algorithm attempts to minimize the genetic changes (“parsimony assumption”). The default parameters (x = 5; y = 4) were applied (17). For the grouping of MLST data, the available sequence types (ST) and the enhanced BURST (eBURST) (based upon related ST) tool were used (2). ST that shared at least five of seven identical alleles were grouped into a single CC. PFGE types and groups were assigned according to the criteria of Tenover et al. (25), which correspond to a maximum variation of three bands for types and fewer than seven bands for groups. The UPGMA (unweighted-pair group method using average linkages) dendrogram illustrating the similarity was built based on Dice coefficients of the SmaI macrorestriction profiles using MEGA (version 4) software (23). The index of diversity and simple concordance were calculated as previously described (7, 11, 19).
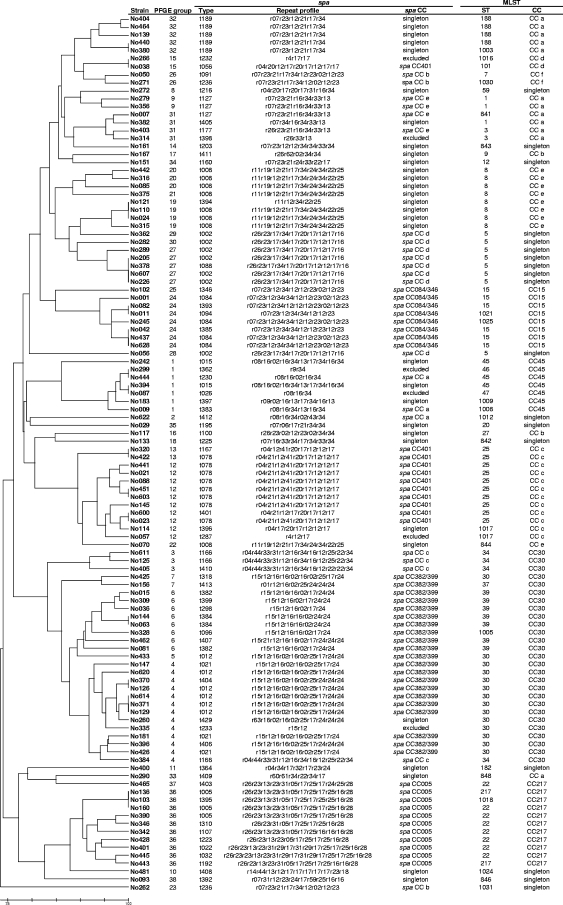
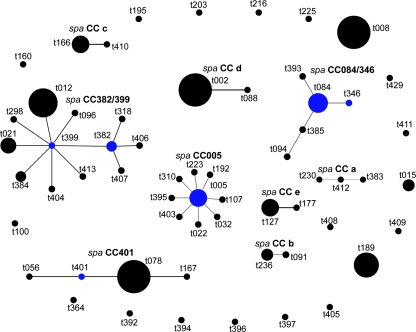
All 110 S. aureus strains were spa typeable, and they exhibited 66 different spa types (number of repeats, 2 to 16). spa types t008 and t078 were most frequent (eight isolates each). The index of diversity for spa typing was 0.98 (95% confidence interval, 0.97 to 0.99). Using the default analysis parameters of BURP, the resulting spa types were clustered into 9 spa CCs and 19 singletons. Six spa types with fewer than five repeats were excluded (spa types t026, t232, t233, t287, t362, and t398). Figure 1 shows the spa/BURP and corresponding MLST/eBURST and PFGE grouping results for each isolate. The clonal relatedness of all BURP-grouped spa types is illustrated in a population snapshot (Fig. 2). Four of the nine spa CCs had designated group founders (spa CC401, spa CC382/399, spa CC084/346, and spa CC005). The group founder within BURP clusters of at least three different spa types is defined as the spa type with the highest founder score (assigned to the spa type to which the relevant spa types and strains are most closely related). In two spa CCs (spa CC382/399 and spa CC084/346), spa types t382 and t399 and spa types t084 and t346 had identical founder scores, respectively.
FIG. 1.
UPGMA dendrogram illustrating the similarity (based on Dice coefficients) of SmaI macrorestriction profiles (PFGE) of 110 S. aureus strains carried by healthy individuals in the community. The UPGMA dendrogram was constructed using MEGA (version 4) software (23). The scale bar at the bottom shows percent similarity. The PFGE groups are arbitrarily numbered. CCs with a designated founder are named after the founder ST or spa type(s). CCs without an assigned founder are arbitrarily named with lowercase letters. spa types shorter than five repeats were excluded from BURP clustering. Abbreviations: r, repeat; t, type.
FIG. 2.
Population snapshot based on BURP analysis of 110 S. aureus strains. BURP grouping using default parameters resulted in 9 spa CCs, 20 singletons, and 6 excluded spa types (spa types t026, t232, t233, t287, t362, and t398). Each dot represents a unique spa type. The diameter of a dot is proportional to the quantity of the corresponding spa type. Blue dots represent group founders, defined as the spa type(s) with the highest founder score within a CC. Note that the spacing between linked spa types and between unlinked spa types and spa CCs provides no information concerning the genetic distance between them.
All STs belonging to CC217 and CC15 were grouped into spa CC005 and spa CC084/346. PFGE subdivided both spa CCs into two different groups. spa CC401 was clustered by eBURST into two different groups (CC c and CC d) that shared only one of the seven MLST loci (aroE). PFGE subdivided spa CC401 into three different groups. The 21 strains of spa CC382/399, exhibiting 12 different spa types, all clustered in MLST CC30 (ST30, ST37, ST39, and ST1005) by eBURST. Six other strains of CC30 were clustered by BURP in spa CC c, in one singleton (No260), and in one excluded spa type (No335). PFGE corroborated the group diversity (14 PFGE types). However, PFGE grouping resulted in five groups. Overall, grouping by BURP was highly concordant with that by eBURST (96.5%) and PFGE (94.9%). PFGE groups were 93.8% concordant with eBURST CCs. On the level of types instead of groups, concordance between spa types and ST or PFGE types was 96.8% or 97.1%, respectively, while PFGE types were 95.9% concordant with ST.
Two recent studies with strain collections that did not represent diverse natural populations of S. aureus (Fig. 2), but contained predominantly clinical isolates or mainly MRSA strains, showed a strong sampling bias (9, 22). In both studies, high concordances between BURP-grouped spa types and MLST and PFGE clusters were found, and only a few discrepancies were detected. In this study, similar discrepancies became apparent by using MLST-based grouping as a reference, e.g., in MLST CC30. MLST data are not greatly influenced by the effects of recombination, due to the use of BURST, which deduces CCs from allelic profiles. In contrast, spa and PFGE grouping algorithms lack any transformation of the original data and are therefore more sensitive to recombination events. Therefore, large chromosomal replacements, which affect macrorestriction patterns and spa typing substantially, are likely within CC30. Such events have already been shown in different clonal lineages, including CC30, by a previous study (20). These cases could be clarified by using an additional target from another genomic region (e.g., clumping factor B gene) (13). In some other instances, the use of BURP clustering is limited because short spa types are excluded from further analysis. However, analysis of the SpaServer content, comprising more than 42,000 isolates with more than 3,100 different spa types (10), shows that fewer than 7% of all spa types were affected by exclusion from BURP clustering.
In summary, BURP determines clonal relatedness, yielding results highly congruent with MLST and PFGE groupings, within an unbiased sampled population of MSSA strains based on spa data. spa typing and BURP should be considered for phylogenetic studies in addition to strain typing of MSSA.
Acknowledgments
A. Mellmann was funded by a grant from the Deutsche Forschungsgemeinschaft (ME 3205/1-1). D. Harmsen is one of the developers of the Ridom StaphType software mentioned in the article. The software is distributed and sold by the company Ridom GmbH, of which he is a partial owner. All other authors have declared no conflicts of interest.
We thank Phillip I. Tarr (Washington University School of Medicine, St. Louis, MO) for critical review of the manuscript.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. W. Friedrich, D. Harmsen, A. Holmes, X. Huijsdens, A. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frenay, H. M., A. E. Bunschoten, L. M. Schouls, W. J. van Leeuwen, C. M. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 1560-64. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich, A. W., W. Witte, D. Harmsen, H. de Lencastre, W. Hryniewicz, J. Scheres, and H. Westh. 2006. SeqNet.org: a European laboratory network for sequence-based typing of microbial pathogens. Euro Surveill. 11E060112.4. [DOI] [PubMed] [Google Scholar]
- 5.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 6.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 404544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 394190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann, H., A. Tami, S. Hori, M. Halwani, and R. Slack. 2002. Nottingham Staphylococcus aureus population study: prevalence of MRSA among elderly people in the community. BMJ 3241365-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallin, M., A. Deplano, O. Denis, R. De Mendonca, R. De Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 415442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn, G., P. Francioli, and D. S. Blanc. 2007. Double-locus sequence typing using clfB and spa, a fast and simple method for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 4554-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 15.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellmann, A., A. W. Friedrich, N. Rosenkötter, J. Rothgänger, H. Karch, R. Reintjes, and D. Harmsen. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellmann, A., T. Weniger, C. Berssenbrügge, J. Rothgänger, M. Sammeth, J. Stoye, and D. Harmsen. 2007. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Nosocomial Infections Surveillance (NNIS) System. 2003. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control 31481-498. [DOI] [PubMed] [Google Scholar]
- 19.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47222-229. [DOI] [PubMed] [Google Scholar]
- 20.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 1861060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 442533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 24.Tang, Y. W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 381347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]