Abstract
As a positive-strand RNA virus, hepatitis E virus (HEV) produces an intermediate negative-strand RNA when it replicates. Thus, the detection of negative-strand viral RNA is indicative of HEV replication. The objective of this study was to develop a negative-strand-specific reverse transcription-PCR (RT-PCR) assay for the identification of extrahepatic sites of HEV replication. Briefly, a 494-bp fragment within the orf1 gene of a chicken strain of HEV (designated avian HEV) was amplified and cloned into a pSK plasmid. A synthetic negative-strand viral RNA was generated from the plasmid by in vitro transcription and was used to standardize the assay. A nested set of primers was designed to amplify a 232-bp fragment of the negative-strand viral RNA. The assay was found to detect up to 10 pg and 10−5 pg of negative-strand HEV RNA in first- and second-round PCRs, respectively. The standardized negative-strand-specific RT-PCR assay was subsequently used to test 13 conveniently obtained tissue specimens collected sequentially on different days postinoculation from chickens experimentally infected with avian HEV. In addition to the liver, the negative-strand-specific RT-PCR assay identified replicative viral RNA in gastrointestinal tissues, including the colorectal, cecal, jejunal, ileal, duodenal, and cecal tonsil tissues. The detection of replicative viral RNA in these tissues indicates that after oral ingestion of the virus, HEV replicates in the gastrointestinal tract before it reaches the liver. This is the first report on the identification of extrahepatic sites of HEV replication in animals after experimental infection via the natural route. The assay should be of value for studying HEV replication and pathogenesis.
Hepatitis E virus (HEV) is a small, nonenveloped, positive-sense, single-strand RNA virus belonging to the genus Hepevirus in the family Hepeviridae (5, 7, 25). The genome of HEV is approximately 7.2 kb and contains three open reading frames (ORFs). ORF1 codes for the nonstructural proteins. ORF2 encodes the immunogenic capsid protein; and ORF3, the smallest of the three ORFs, is believed to encode a cytoskeleton-associated phosphoprotein that may be involved in virus replication (5, 6, 25-27). HEV does not grow efficiently in cell culture, and the lack of an established in vitro cell culture system has been a major impediment in the study of HEV (7, 20).
HEV causes acute hepatitis E in humans, which is a major public health concern in many developing countries of Asia and Africa as well as in Mexico. Sporadic cases of acute hepatitis E not related to traveling to regions where HEV is endemic have also been reported in numerous industrialized countries, including the United States (1, 4, 7, 24, 26, 27, 30, 31, 33, 35). Antibodies to HEV have been detected in a significant proportion of healthy individuals in the United States and other industrialized countries, although the incidence of disease is only sporadic in these countries (8, 31, 35). The fecal-oral route is the primary mode of transmission (7, 18), and waterborne epidemics due to contamination of drinking water or water supplies are characteristic of hepatitis E outbreaks (25). The disease mainly affects young adults and causes a 15 to 25% mortality rate in infected pregnant women (19, 25), even though the overall mortality rate is less than 1%.
In 1997, Meng et al. (21) discovered and characterized the first animal strain of HEV, swine HEV, from commercial pigs in the United States. Since then, swine HEV has been isolated from pigs in different geographical regions of the world (4, 20, 24, 30, 33, 35). Recently, a chicken strain of HEV, designated avian HEV, was isolated from chickens with hepatitis-splenomegaly (HS) syndrome in the United States and was characterized (12-14). The complete genomic sequence of avian HEV was determined and was found to be similar to that of mammalian HEVs. Despite an approximately 50 to 60% nucleotide sequence identity with mammalian HEVs, avian HEV shares many significant structural and functional features with human and swine HEVs (3, 15, 28). Avian HEV also contains antigenic epitopes in its capsid protein that are both common to and unique from both human and swine HEVs (9, 10, 36), suggesting that avian HEV is similar to human HEV genetically and antigenically. The discovery of avian HEV and its association with a hepatic disease provided a unique homologous animal model for the study of HEV, since chickens are the natural host of avian HEV. The clinical and pathological findings associated with avian HEV infection in chickens infected by the natural route have been reported previously (2).
Due to the lack of an efficient cell culture system and a practical animal model, little is known about the mechanism of HEV replication or pathogenesis. Extrahepatic sites of HEV replication were demonstrated in pigs experimentally infected via the intravenous route of inoculation (34), but the reproduction of HEV infection after inoculation by the natural oral route in pigs and nonhuman primates has been very difficult (18). The objective of this study was to develop a negative-strand-specific reverse transcription (RT)-PCR assay for the identification of extrahepatic sites of HEV replication. By using convenient tissue samples collected from chickens experimentally infected with avian HEV by the natural route of infection in a previous study (2), we identified extrahepatic sites of HEV replication by a negative-strand-specific RT-PCR assay.
MATERIALS AND METHODS
Virus.
The avian HEV isolate used in this study was originally recovered from a bile sample from a 56-week-old chicken with HS syndrome (13). The avian HEV stock has an infectious titer of 5 × 104.5 50% chicken infectious doses per ml (2, 29).
Tissue samples used for development and validation of negative-strand-specific RT-PCR assay.
Conveniently obtained tissue samples were collected from chickens experimentally infected with avian HEV by the natural route in a previous study (2). Thirteen different tissues, including the tissues of the colorectum, jejunum, ileum, duodenum, cecum, cecal tonsils, thymus, spleen, lung, heart, kidney, pancreas, and liver, were collected at each necropsy from all the chickens (n = 28) infected with avian HEV by the nature (oronasal) route and from the negative control chickens (n = 28). The tissue samples were collected from two chickens in each group that were necropsied at 1, 3, 5, 7, 10, 13, 16, 20, 24, 28, 35, 42, and 56 days postinoculation (dpi). The tissues were stored at −80°C.
Samples of the tissues were each homogenized in 10% sterile phosphate-buffered saline and stored at −80°C. At the time of RNA extraction, a portion of the tissue homogenate was clarified by centrifugation at 3,000 rpm for 15 min at 4°C (centrifuge 5810, rotor A-4-44; Eppendorf) and was subsequently used for the detection of positive-strand avian HEV RNA by a regular RT-PCR assay.
RT-PCR detection of positive-strand avian HEV RNA.
RT-PCR was performed to detect avian HEV RNAs in tissue homogenates, as described previously (28-29). Briefly, total RNA extracted from 100 μl of the 10% tissue homogenate was resuspended in 12.25 μl of DNase-, RNase-, and proteinase-free water (Invitrogen). RT was performed at 42°C for 60 min with 1 μl (10 pM) of reverse primer P2 (5′-ACAGTTTCACCTCAGGCTCG-3′) (28, 29). Five microliters of the resulting cDNA was amplified in a 50-μl reaction mixture with Platinum high-fidelity supermix (Invitrogen) by using a nested PCR. The first round of PCR with forward primer P1 (5′-ACAACATCCACCCCTACAAG-3′) and reverse primer P2 produced an expected fragment of 595 bp. For the second round of PCR, forward primer P3 (5′-AGAACAATGGTTGGCGGTCC-3′) and reverse primer P4 (5′-GAGGGCAAGCCACCTAAAAC-3′) were used to amplify an expected fragment of 394 bp. The PCR parameters consisted of an initial incubation at 94°C for 9 min, followed by 39 cycles of denaturation at 94°C for 0.5 min, annealing at 52°C for 0.5 min, and extension at 72°C for 1 min and a final extension at 72°C for 7 min. The PCR conditions for the second round of PCR were the same as those for the first round, except that an annealing temperature of 56°C was used.
Generation of synthetic negative-strand avian HEV RNA as a positive control for development of a negative-strand-specific RT-PCR assay.
To develop and standardize a negative-strand-specific RT-PCR assay, a negative strand of viral RNA had to be generated for use as a positive control. Briefly, total RNA was extracted from an infectious stock of avian HEV. RT was performed at 42°C for 60 min with 1 μl (10 pM) of reverse primer N2 (5′-CCGGGCTGATGGTCTCGATTAG-3′). Five microliters of the resulting cDNA was amplified in a 50-μl reaction mixture with Platinum high-fidelity supermix (Invitrogen). A 494-bp orf1 fragment of avian HEV was amplified by PCR with primer F3088 (5′-CGCTGTAGTGGGATCCATGTTGGTG-3′) and primer R3559 (5′-TGTCTCGAGGGGTTGATTGGTCC-3′). Forward primer F3088 has an introduced BamHI restriction site and reverse primer R3559 has an introduced XhoI site to facilitate the subsequent cloning steps (the restriction sites are underlined). The resulting PCR product was excised from the agarose gel and purified with a GeneClean II kit. The purified PCR product was ligated into a TA vector with T4 DNA ligase (Stratagene). The ligation mixture was transformed into One Shot TOP 10 chemically competent Escherichia coli cells (Invitrogen), 100 μl was spread on LB agar plates containing ampicillin, and the plates were incubated overnight. After identification of the plasmid containing the insert and confirmation by restriction digestion, the insert was subcloned into plasmid pBluescript II SK(+) (PSK II; Stratagene) by directional cloning with the restriction enzymes BamHI and XhoI. Recombinant plasmid PSK II containing the orf1 insert was isolated and confirmed by sequencing.
The recombinant PSK II plasmid was linearized and purified by phenol-chloroform extraction, and a synthetic negative-strand RNA was transcribed in vitro with T7 polymerase by using an mMessage mMachine T7 transcription kit (Ambion). To remove the plasmid DNA, the transcribed negative-strand vial RNA was separated in a 1% agarose gel, and the RNA band was excised from the gel and purified by use of an RNaid isolation kit (QBiogene). The purified RNA was further treated with DNase for 60 min at 37°C and extracted with the TRI reagent (Molecular Research Center, Inc.) to further eliminate plasmid DNA contamination. A nested PCR (without the RT step) was done with two sets of primers (primers EF1 and ER1 and primers EF2 and ER2) to rule out any potential plasmid DNA contamination.
Standardization of a negative-strand-specific RT-PCR assay.
The synthetic negative-strand viral RNA transcribed in vitro was quantified and serially diluted from 102 to 10−15 ng in 100 μl phosphate-buffered saline, and a negative-strand-specific RT-PCR was performed with each serial dilution. RT was performed as described above with primer EF1 (5′-ATGTTGGTGGGGTGCTGGTCGAGATTG-3′). Primers EF1 and ER1 (5′-GGGTTGATTGGTCCGATATGATGCCAG-3′) were used in the first round and primers EF2 (5′-TTGTTGGACATACCCCCGGCCCAC-3′) and ER2 (5′-TAATCACCGCAAGACGGCTAGTGG-3′) were used in the second round of nested PCR for the detection of negative-strand viral RNA. The 232-bp product obtained from the second round of the nested PCR was sequenced for confirmation.
Once the negative-strand-specific RT-PCR assay was standardized, we applied the assay and retested all the tissue and serum samples that were positive for positive-strand viral RNA to identify replicative viral RNA and the sites of HEV replication in various tissues.
DNA sequencing.
The PCR products amplified from various tissues were excised from 0.8% agarose gel and purified with a GeneClean III kit (Qbiogene), and both strands were sequenced at the Virginia Bioinformatics Institute Core Laboratory Facility with an automated DNA sequencer (Applied Biosystems).
RESULTS
Sensitivity of negative-strand-specific RT-PCR assay for detection of replicative HEV RNA.
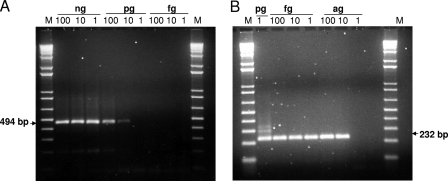
A negative-strand-specific RT-PCR assay was standardized by using a synthetic negative-strand viral RNA that was transcribed in vitro from a plasmid containing an avian HEV insert as a positive control. The 232-bp product from the second round of PCR was sequenced to confirm the specificity. The negative-strand-specific RT-PCR assay detected up to 10 pg negative-strand viral RNA in the first round and approximately 10−5 pg negative-strand viral RNA in the second round of nested PCR (Fig. 1). The level of detection of 10−5 pg corresponds to approximately 36 copies of viral RNA.
FIG. 1.
Development and standardization of a negative-strand-specific RT-PCR assay. The synthetic negative-strand HEV RNA transcript was serially diluted, as indicated (ng, pg, fg, ag), and tested by a nested PCR. The expected products in the first round (A) and the second round (B) of PCR are indicated. Lanes M, 1-kb plus ladder.
Distribution of positive-strand HEV RNA in various tissues.
Before the tissue samples were tested for replicative negative-strand viral RNA, all tissue samples were first tested by a regular RT-PCR assay to detect the positive-strand viral RNA. Positive-strand avian HEV RNA was detected in various tissues at times that varied from 1 to 56 dpi. However, viral RNA was more frequently detected in the liver and gastrointestinal (GI) tissues and was much less frequently found in non-GI tissues (Table 1). Viremia was transient at 20 and 35 dpi, and viral RNA was detected in most of the tissues during the period of viremia (2). However, positive-strand viral RNA was detected in a number of tissues in the absence of viremia. Liver tissue was positive for viral RNA from 3 dpi (2). Avian HEV RNA was consistently detected in colorectal tissue from 1 dpi to 24 dpi and then at 35 and 56 dpi. Jejunal tissue was positive from 7 dpi, and ileal tissue was positive from 1 dpi. Viral RNA was detected in duodenal tissue only from approximately the third week but was consistently detected in cecal tissue during almost the entire duration of the experiment. Cecal tonsil tissues were transiently positive. Positive-strand viral RNA was also detected in thymus, spleen, lung, heart, kidney, and pancreas tissue specimens. The tissues of all negative control chickens were seronegative throughout the study, and thus, they were not tested by RT-PCR in this study.
TABLE 1.
Detection of positive-strand HEV RNA as well as replicative negative-strand HEV RNA in nonliver tissues from chickens experimentally infected with avian HEV via a natural route of exposure
| Tissue | No. of chickens positive for positive-strand HEV RNA (no. positive for replicative negative-strand HEV RNA)/total no. of chickens tested on the following dpi:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 7 | 10 | 16 | 20 | 24 | 28 | 35 | 42 | 56 | |
| Colorectum | 1 (0)/2 | 1 (0)/2 | 2 (1)/2 | 1 (0)/2 | 2 (0)/2 | 1 (1)/2 | 2 (2)/2 | 1 (0)/2 | 0 (na)/2 | 2 (1)/2 | 0 (n)/2 | 2 (0)/4 |
| Jejunum | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 2 (0)/2 | 1 (0)/2 | 2 (2)/2 | 0 (n)/2 | 0 (n)/2 | 2 (1)/2 | 0 (n)/2 | 0 (n)/4 |
| Ileum | 2 (0)/2 | 0 (n)/2 | 0 (n)/2 | 1 (1)/2 | 2 (2)/2 | 0 (n)/2 | 2 (1)/2 | 0 (n)/2 | 1 (0)/2 | 1 (1)/2 | 0 (n)/2 | 0 (n)/4 |
| Duodenum | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 2 (1)/2 | 0 (n)/2 | 1 (0)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Cecum | 1 (0)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 2 (0)/2 | 2 (0)/2 | 2 (2)/2 | 1 (0)/2 | 2 (0)/2 | 2 (1)/2 | 1 (0)/2 | 0 (n)/4 |
| Cecal tonsils | 1 (0)/2 | 0 (n)/2 | 2 (0)/2 | 2 (0)/2 | 0 (n)/2 | 0 (n)/2 | 2 (0)/2 | 0 (n)/2 | 0 (n)/2 | 1 (1)/2 | 1 (0)/2 | 1 (1)/4 |
| Thymus | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Spleen | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Lung | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 2 (0)/4 |
| Heart | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/2 | 2 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Kidney | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Pancreas | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 0 (n)/2 | 1 (0)/2 | 0 (n)/2 | 0 (n)/4 |
| Liverb | (0)/2 | (0)/2 | (0)/2 | (0)/2 | (0)/2 | (1)/2 | (2)/2 | (1)/2 | (0)/2 | (1)/2 | (0)/2 | (0)/4 |
n, not tested by the negative-strand-specific RT-PCR since they were negative for the positive-strand viral RNA by the regular RT-PCR.
The results for positive-strand HEV RNA detection in liver samples were reported previously (2).
Detection of replicative negative-strand viral RNA in extrahepatic tissues in chickens experimentally infected with avian HEV via the natural route.
Since HEV is a positive-strand RNA virus, the detection of positive-strand viral RNA in tissues does not necessarily mean that the virus replicates in the tissues. Therefore, all the tissues that were positive for the positive-strand viral RNA were retested by the standardized negative-strand-specific RT-PCR assay. Replicative negative-strand viral RNA was detected in the liver as well as in the GI tissues but not in thymus, spleen, lung, heart, kidney, or pancreas tissue (Table 1). Replicative negative-strand viral RNA was detected from 5 to 56 dpi in the GI tissues (Table 1). Liver tissues were positive from 16 dpi. The earliest appearance of replicative negative-strand viral RNA was found in colorectal tissues at 5 dpi and in ileal tissues at 7 dpi. Most of the GI tissues and sera were positive for replicative viral RNA at both 20 and 35 dpi, the times when viremia was detected (2). Cecal tonsil tissues were was positive for replicative viral RNA even at 56 dpi (Table 1).
DISCUSSION
Knowledge of the mechanisms of HEV pathogenesis and replication is very limited due to the lack of an efficient cell culture system and a practical animal model. Most of the scant knowledge about the pathogenesis of HEV was obtained from human HEV infection in nonhuman primates (25, 32). With the discovery of swine HEV (21), a pig model that can be used to study HEV has been developed (16-18, 22, 34). Unlike nonhuman primates, swine are the natural hosts of swine HEV. However, swine HEV causes a subclinical infection in pigs and only microscopic hepatitis lesions are visible (11, 18, 34); thus, HEV-infected pigs are not suitable for the study of all aspects of HEV replication and pathogenesis.
Recently, another animal strain of HEV, avian HEV, was identified from chickens with HS syndrome in the United States (13). Avian HEV is antigenically and genetically similar to human HEV (12, 14-15). Although HEV is thought to be transmitted by the fecal-oral route (7, 25), the experimental reproduction of HEV infections in nonhuman primates and pigs by the oral route was difficult (18), and the intravenous route is still the preferred route for inducing of experimental HEV infections (11, 22, 32). However, chickens can easily be infected with the chicken strain of HEV, avian HEV, by the natural oral route (2).
The mechanism by which HEV causes hepatic damage is not known. It has been suggested that liver damage is caused by the immune response to the invading virus and not by the direct replication of the virus in the liver (25). It is unclear how the virus reaches the liver, as HEV is transmitted by the fecal-oral route. It is believed that the virus first replicates in the GI tract following oral ingestion and then reaches the target organ, the liver. Extrahepatic sites of HEV replication have been demonstrated in pigs intravenously inoculated with swine HEV and human HEV (34). However, the intravenous route of inoculation limited the ability to study the initial sites of virus replication following exposure by the natural oral route.
In the present study, by successfully developing and employing a negative-strand-specific RT-PCR assay, we demonstrated the sites of extrahepatic HEV replication in chickens experimentally infected with a strain of HEV by the natural route of infection. The tissues collected from chickens oronasally infected with avian HEV were first tested by a regular RT-PCR to detect positive-strand viral RNA. All the tissues were positive for viral RNA at some point during the course of the study. Positive-strand HEV RNA was prominently distributed in the liver and GI tissues; but it was rarely detected in non-GI tissues, like thymus, spleen, kidney, lung, and heart tissues, suggesting that the liver and GI tissues may be the sites of virus replication. The detection of positive-strand viral RNA in tissues does not necessarily mean that the virus actually replicates in the tissues, since the virus is circulating in the blood as a result of viremia. However, positive-strand viral RNA was also detected in some tissues in the absence of viremia, suggesting that the positive-strand RNA detected in these tissues in the absence of viremia is likely the result of viral replication and is not due to circulating virus.
Because HEV is a single-strand positive-sense RNA virus, it produces an intermediate negative-strand viral RNA during virus replication. Therefore, detection of negative-strand viral RNA in tissues would be indicative of virus replication. A negative-strand-specific RT-PCR was performed with all the tissues containing positive-strand viral RNA to detect the replicative negative-strand viral RNA. The results showed that replicative viral RNA was detected only in the liver and GI tissues, including colorectal, jejunal, ileal, duodenal, cecal, and cecal tonsil tissues, but not in non-GI tissues, thus strongly indicating that the GI tissues are the extrahepatic sites of HEV replication. It is interesting to note that the lymphoid tissues associated with the GI tract (but not the thymus) are also positive for replicative viral RNA. From the fact that the colorectal tissue was positive for replicative viral RNA from as early as 5 dpi and the ileal tissue was positive from 7 dpi, while the liver tissue was positive from 16 dpi, it can be concluded that HEV initially replicates in the GI tissues following fecal-oral transmission. However, the cecal tonsil tissues were positive for replicative RNA at 35 and 56 dpi but not early in infection, suggesting that the cecal tonsils may be the sites of secondary viral replication.
Serum was also tested by the negative-strand-specific RT-PCR, and interestingly, replicative viral RNA was detected at 20 dpi and 35 dpi in some chickens, corresponding to the times of detection of viremia and the presence of replicative viral RNA in most GI tissues, suggesting that the replicative viral RNA in serum may be due to an overflow from the peak level of replication in many GI tissues at the same time. The detection of replicative viral RNA in serum could also be due to the circulating cell-associated viruses during the period of viremia.
In summary, this represents the first report on the identification of the extrahepatic sites of HEV replication after experimental HEV infection by the natural route. We showed that replicative viral RNA is detected in liver tissue as well as in GI tissues and that GI tissues are the initial sites of HEV replication before HEV reaches its target organ, the liver. The biological and pathological significance of these extrahepatic replication sites remains unknown, but the findings from this study may aid in the eventual development of an efficient in vitro cell culture system for HEV propagation, which is currently lacking. The negative-strand-specific RT-PCR assay developed in this study should be of value for the future study of HEV replication and pathogenesis.
Acknowledgments
This study was supported in part by grants from the National Institutes of Health (grants AI074667 and AI050611).
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Arankalle, V. A., M. S. Chadha, S. A. Tsarev, S. U. Emerson, A. R. Risbud, K. Banerjee, and R. H. Purcell. 1994. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc. Natl. Acad. Sci. USA 913428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billam, P., F. F. Huang, Z. F. Sun, F. W. Pierson, R. B. Duncan, F. Elvinger, D. K. Guenette, T. E. Toth, and X. J. Meng. 2005. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J. Virol. 793429-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billam, P., Z. F. Sun, and X. J. Meng. 2007. Analysis of the complete genomic sequence of an apparently avirulent strain of avian hepatitis E virus identified major genetic differences compared to the prototype avian HEV. J. Gen. Virol. 881538-1544. [DOI] [PubMed] [Google Scholar]
- 4.Clemente-Casares, P., S. Pina, M. Buti, R. Jardi, M. MartIn, S. Bofill-Mas, and R. Girones. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9448-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerson, S. U., D. Anderson, V. A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 851-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 6.Emerson, S. U., H. Nguyen, U. Torian, and R. H. Purcell. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 8010457-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson, S. U., and R. H. Purcell. 2003. Hepatitis E virus. Rev. Med. Virol. 13145-154. [DOI] [PubMed] [Google Scholar]
- 8.Feagins, A. R., T. Opriessnig, D. K. Guenette, P. G. Halbur, and X. J. Meng. 2007. Detection and characterization of infectious hepatitis E virus from commercial pig livers sold in local grocery stores in the USA J. Gen. Virol. 88912-917. [DOI] [PubMed] [Google Scholar]
- 9.Guo, H., E. M. Zhou, Z. F. Sun, X. J. Meng, and P. G. Halbur. 2006. Identification of B-cell epitopes in the capsid protein of avian hepatitis E virus (avian HEV) that are common to human and swine HEVs or unique to avian HEV. J. Gen. Virol. 87217-223. [DOI] [PubMed] [Google Scholar]
- 10.Guo, H., E. M. Zhou, Z. F. Sun, and X. J. Meng. 2007. Egg whites from eggs of chickens infected experimentally with avian hepatitis E virus contain infectious virus, but evidence of complete vertical transmission is lacking. J. Gen. Virol. 881532-1537. [DOI] [PubMed] [Google Scholar]
- 11.Halbur, P. G., C. Kasorndorkbua, C. Gilbert, D. K. Guenette, M. B. Potters, R. H. Purcell, S. U. Emerson, T. E. Toth, and X. J. Meng. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haqshenas, G., F. F. Huang, M. Fenaux, D. K. Guenette, F. W. Pierson, C. T. Larsen, H. L. Shivaprasad, T. E. Toth, and X. J. Meng. 2002. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J. Gen. Virol. 832201-2209. [DOI] [PubMed] [Google Scholar]
- 13.Haqshenas, G., H. L. Shivaprasad, P. R. Woolcock, D. H. Read, and X. J. Meng. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 822449-2462. [DOI] [PubMed] [Google Scholar]
- 14.Huang, F. F., G. Haqshenas, H. L. Shivaprasad, D. K. Guenette, P. R. Woolcock, C. T. Larsen, F. W. Pierson, F. Elvinger, T. E. Toth, and X. J. Meng. 2002. Heterogeneity and seroprevalence of a newly identified avian hepatitis E virus from chickens in the United States. J. Clin. Microbiol. 404197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, F. F., Z. F. Sun, S. U. Emerson, R. H. Purcell, H. L. Shivaprasad, F. W. Pierson, T. E. Toth, and X. J. Meng. 2004. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 851609-1618. [DOI] [PubMed] [Google Scholar]
- 16.Huang, Y. W., G. Haqshenas, C. Kasorndorkbua, P. G. Halbur, S. U. Emerson, and X. J. Meng. 2005. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 791552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, Y. W., T. Opriessnig, P. G. Halbur, and X. J. Meng. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 813018-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasorndorkbua, C., P. G. Halbur, P. J. Thomas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2002. Use of a swine bioassay and a RT-PCR assay to assess the risk of transmission of swine hepatitis E virus in pigs. J. Virol. Methods 10171-78. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, A., M. Beniwal, P. Kar, J. B. Sharma, and N. S. Murthy. 2004. Hepatitis E in pregnancy. Int. J. Gynaecol. Obstet. 85240-244. [DOI] [PubMed] [Google Scholar]
- 20.Meng, X. J. 2005. Hepatitis E as a zoonosis, p. 611-623. In H. Thomas, A. Zuckermann, and S. Lemon (ed.), Viral hepatitis, 3rd ed. Blackwell Publishing Ltd., Oxford, United Kingdom.
- 21.Meng, X. J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 949860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng, X. J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 729714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Nishizawa, T., M. Takahashi, H. Mizuo, H. Miyajima, Y. Gotanda, and H. Okamoto. 2003. Characterization of Japanese swine and human hepatitis E virus isolates of genotype IV with 99% identity over the entire genome. J. Gen. Virol. 841245-1251. [DOI] [PubMed] [Google Scholar]
- 25.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 26.Schlauder, G. G., G. J. Dawson, J. C. Erker, P. Y. Kwo, M. F. Knigge, D. L. Smalley, J. E. Rosenblatt, S. M. Desai, and I. K. Mushahwar. 1998. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J. Gen. Virol. 79447-456. [DOI] [PubMed] [Google Scholar]
- 27.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65282-292. [DOI] [PubMed] [Google Scholar]
- 28.Sun, Z. F., C. T. Larsen, A. Dunlop, F. F. Huang, F. W. Pierson, T. E. Toth, and X. J. Meng. 2004. Genetic identification of avian hepatitis E virus (HEV) from healthy chicken flocks and characterization of the capsid gene of 14 avian HEV isolates from chickens with hepatitis-splenomegaly syndrome in different geographical regions of the United States. J. Gen. Virol. 85693-700. [DOI] [PubMed] [Google Scholar]
- 29.Sun, Z. F., C. T. Larsen, F. F. Huang, P. Billam, F. W. Pierson, T. E. Toth, and X. J. Meng. 2004. Generation and infectivity titration of an infectious stock of avian hepatitis E virus (HEV) in chickens and cross-species infection of turkeys with avian HEV. J. Clin. Microbiol. 422658-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, M., T. Nishizawa, and H. Okamoto. 2003. Identification of a genotype III swine hepatitis E virus that was isolated from a Japanese pig born in 1990 and that is most closely related to Japanese isolates of human hepatitis E virus. J. Clin. Microbiol. 411342-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas, D. L., P. O. Yarbough, D. Vlahov, S. A. Tsarev, K. E. Nelson, A. J. Saah, and R. H. Purcell. 1997. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J. Clin. Microbiol. 351244-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsarev, S. A., S. U. Emerson, T. S. Tsareva, P. O. Yarbough, M. Lewis, S. Govindarajan, G. R. Reyes, M. Shapiro, and R. H. Purcell. 1993. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J. Infect. Dis. 1671302-1306. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Y. C., H. Y. Zhang, N. S. Xia, G. Peng, H. Y. Lan, H. Zhuang, Y. H. Zhu, S. W. Li, K. G. Tian, W. J. Gu, J. X. Lin, X. Wu, H. M. Li, and T. J. Harrison. 2002. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 67516-521. [DOI] [PubMed] [Google Scholar]
- 34.Williams, T. P., C. Kasorndorkbua, P. G. Halbur, G. Haqshenas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 393040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yazaki, Y., H. Mizuo, M. Takahashi, T. Nishizawa, N. Sasaki, Y. Gotanda, and H. Okamoto. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 842351-2357. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, E. M., H. Guo, F. F. Huang, Z. F. Sun, and X. J. Meng. 2008. Identification of two neutralization epitopes on the capsid protein of avian hepatitis E virus. J. Gen. Virol. 89500-508. [DOI] [PubMed] [Google Scholar]