Abstract
Candida orthopsilosis and Candida metapsilosis are recently described species, having previously been grouped with the more prevalent species Candida parapsilosis. Current literature contains very little data pertaining to the distributions and antifungal susceptibilities of these Candida species. We determined the species and antifungal susceptibilities of 1,929 invasive clinical isolates from the ARTEMIS antifungal surveillance program collected between 2001 and 2006 and identified as C. parapsilosis using Vitek and conventional methods. Of the 1,929 isolates of presumed C. parapsilosis tested, 117 (6.1%) were identified as C. orthopsilosis and 34 (1.8%) as C. metapsilosis. The percentage of presumed C. parapsilosis isolates found to be C. orthopsilosis varied greatly by region, with the highest percentage (10.9%) from South America and the lowest (0.7%) from Africa. The MIC distributions of the C. orthopsilosis and C. metapsilosis isolates were statistically significantly lower than those of C. parapsilosis for all drugs except fluconazole, for which they were significantly higher (P < 0.001 for all). No C. orthopsilosis or C. metapsilosis isolates were fluconazole resistant, and all were susceptible to caspofungin, anidulafungin, and micafungin.
Candida parapsilosis is a cause of serious nosocomial infections and is the second most common Candida species isolated from bloodstream infections in many regions of the world (1, 21, 25, 29), particularly in Latin America (2, 4). Of further concern are recent reports of reduced azole and echinocandin susceptibilities in this species (1, 2, 3, 14, 19, 20, 21, 26).
C. parapsilosis has long been considered a three-species complex. Since the early 1990s, it has been shown by randomly amplified polymorphic DNA analysis (10), karyotyping (13), multilocus enzyme electrophoresis and ribosomal internal transcribed spacer sequencing (11), DNA reassociation analysis (24), complex DNA probe pattern by Southern blot analysis (6), and multigenic sequence analysis (5) that the phenotypic and genotypic patterns fall into three distinct groupings. Tavanti and coworkers (27) used multigenic sequence analysis and internal transcribed spacer sequencing to define the C. parapsilosis complex as the three separate species, C. parapsilosis, Candida orthopsilosis, and Candida metapsilosis.
Despite the fact that the C. parapsilosis complex has been recognized for a number of years, very little is known about the epidemiology of the two rare species within the complex, C. orthopsilosis (formerly C. parapsilosis group II) and C. metapsilosis (formerly C. parapsilosis group III). There have been at least two outbreaks reported in which C. orthopsilosis (C. parapsilosis type II) was reported to be part of the outbreak. The first was an outbreak in a hospital in San Antonio reported by Lin and coworkers (11). In 2000, Zancope-Oliviera and coworkers (31) described a small nosocomial outbreak of C. parapsilosis group II in a hospital in Brazil. There are other outbreaks that were reported as being caused by C. parapsilosis in which a distinction was not made between the types, and thus they could have been caused by either C. orthopsilosis or C. metapsilosis. No other epidemiological data appeared until Tavanti and coworkers (28) described a number of C. orthopsilosis strains from 13 patients in Pisa, Italy.
In this study, we used a large worldwide collection of presumed C. parapsilosis isolates, collected as part of the ARTEMIS Global Surveillance study, to screen for C. orthopsilosis and C. metapsilosis. This is the first global description of the prevalence, distribution, and antifungal susceptibility of C. orthopsilosis and C. metapsilosis.
MATERIALS AND METHODS
Study collection and test sites.
The isolates were those previously analyzed as part of the ARTEMIS Global Surveillance study (19). One hundred thirty-four medical centers submitted isolates over the course of the study, and at least 101 of these hospitals submitted both adult and pediatric isolates. Yeasts from all body sites and tissues that were considered pathogens responsible for invasive disease were submitted. Yeasts considered by the collecting physicians to be colonizers were excluded, as were duplicate isolates from a single patient as determined locally. Identification was performed locally and confirmed at the University of Iowa using the Vitek Yeast Biochemical Card (bioMérieux Vitek, Durham, NC).
DNA isolation and amplification.
Isolates were stored as water-based stocks, passaged on potato dextrose agar, and then plated fresh on potato dextrose agar and incubated at 30°C for 24 h. A 3-μl equivalent of yeast was scraped from the plate and resuspended in 20 μl of sterile water. The yeast was “shocked” by heating it to 95°C for 8 min and was then placed in a −70°C freezer for >1 h.
Taq DNA polymerase reaction mixtures were prepared as suggested by the manufacturer (New England Biolabs, Ipswich, MA). Each 50-μl reaction mixture contained 4 μl of the prepared yeast supernatant. Primers S1F and S1R, which amplify the secondary alcohol dehydrogenase gene (27), were used for the PCR. The reaction consisted of one 5-min cycle at 95°C, followed by 40 cycles as follows: 1 min at 92°C, 1 min at 45°C, and 1 min at 68°C. The reaction was terminated by one 7-min cycle at 68°C. Following completion of the cycle, 1 μl of BanI (New England Biolabs, Ipswich, MA) was added directly to the reaction mixture, and the mixture was incubated for an additional 2 hours at 37°C. Isolates of C. parapsilosis, C. orthopsilosis, C. metapsilosis, and Lodderomyces elongisporus were identified by the size of the major restriction fragment on an agarose gel following digestion. All isolates identified as C. orthopsilosis, C. metapsilosis, and L. elongisporus were amplified and digested a second time for confirmation. ATCC isolates 96139, 96144, and 22688 were used as positive controls for C. orthopsilosis, C. metapsilosis, and L. elongisporus, respectively.
Antifungal agent susceptibility testing.
Standard antifungal susceptibility testing was performed as described previously (22) and in the Clinical and Laboratory Standards Institute (CLSI) document M27-A2 (15). The final concentration ranges were 0.007 to 8.0 μg/ml for anidulafungin, caspofungin, and micafungin and 0.12 to 128.0 μg/ml for fluconazole. Amphotericin B Etest concentrations ranged from 0.002 to 32 μg/ml. The MIC was defined as the lowest concentration at which there was a visually prominent reduction in growth (approximately 50%) relative to the drug-free growth control after 24 h of incubation (anidulafungin, caspofungin, and micafungin) (17) or after 48 h of incubation (fluconazole). The interpretive criteria for fluconazole were those published by the CLSI (15) and by Rex et al. (23). Isolates for which the fluconazole MICs were ≤8.0 μg/ml were susceptible, those for which the MICs were 16 to 32 μg/ml were susceptible dose dependent, and those for which the MICs were ≥64 μg/ml were resistant. Interpretive criteria for the echinocandins (anidulafungin, caspofungin, and micafungin) were those recently established by the CLSI (isolates with an MIC of ≤2.0 μg/ml to the echinocandins were considered susceptible [minutes of the June 2007 meeting of the CLSI Antifungal Subcommittee {unpublished}]). Interpretive criteria for amphotericin B have not yet been established by the CLSI. However, for purposes of comparison, isolates with amphotericin B MICs of ≤1.0 μg/ml were considered susceptible. Quality control was performed using Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 for each batch of isolates tested.
Statistical analyses.
Differences in antifungal MIC distributions were examined using the Wilcoxon rank sum test. Differences in species distribution by year were examined using a chi-square test. Alpha was set at 0.05, and all P values were two tailed. SPSS version 15.0 (Chicago, IL) was used for all statistical analyses.
RESULTS
Demographic distribution of C. orthopsilosis isolates.
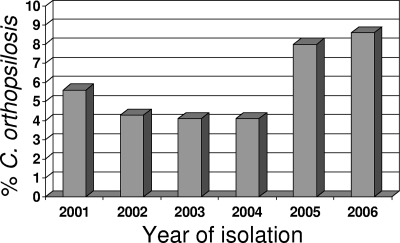
Between 2001 and 2006, 1,976 isolates of C. parapsilosis were banked at the University of Iowa as part of the ARTEMIS Global Surveillance study. Of those isolates, 1,929 could be recovered and species identification could be performed. These isolates came from 89 study centers in 29 countries on six continents. C. parapsilosis accounted for 1,762 (91.3%) of the isolates, while 117 (6.1%) were C. orthopsilosis, 34 (1.8%) were C. metapsilosis, and 16 (0.8%) were L. elongisporus (described previously [12]). The percentage of C. parapsilosis complex isolates that were C. orthopsilosis increased longitudinally during this surveillance study (Fig. 1). While in the first 4 years of the study the average percentage of the isolates that were C. orthopsilosis was 4.5%, the average percentage over the last 2 years was 8.3% (P = 0.01).
FIG. 1.
Percentages of C. parapsilosis isolates that were C. orthopsilosis by year (P = 0.01 for an increasing trend over time in the proportion of all isolates that were C. orthopsilosis).
Of the isolates for which the site of isolation was provided, 77% of C. orthopsilosis isolates and 60% of C. metapsilosis isolates were from bloodstream infections compared to 79% of C. parapsilosis isolates. C. orthopsilosis was also isolated from ascites fluid, abscesses, catheters, cerebrospinal fluid, and pulmonary sources (e.g., pleural and bronchial alveolar lavage fluids). C. metapsilosis was also isolated from abscesses, ascites fluid, bronchial alveolar lavage fluid, and joint fluid.
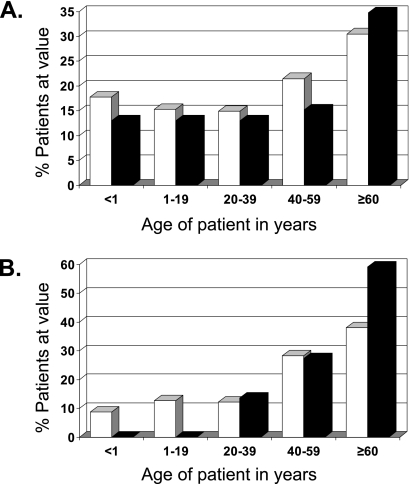
C. parapsilosis is often associated with infections of neonates and infants (1, 8). In this study, the median age range for patients with C. parapsilosis isolates was 40 to 49 years old, with the highest percentage of isolates (30.5%) falling into the ≥60-year-old age group (Fig. 2A). Likewise, the median age range for patients with C. orthopsilosis isolates was 40 to 49 years old, and the highest percentage of isolates (34.8%) fell into the ≥60-year-old age group. Interestingly, when only the C. parapsilosis and C. orthopsilosis isolates from the United States were analyzed, there was a difference between the two groups (Fig. 2B). While 33.6% of the patients with C. parapsilosis isolates were in the ≤39-year-old age group, only 13.6% of the patients with C. orthopsilosis isolates fell into this age group, and while only 38.1% of the patients with C. parapsilosis isolates were in the ≥60-year-old age group, 59.1% of the patients with C. orthopsilosis isolates were in this age group.
FIG. 2.
Distribution by age of patients with C. parapsilosis (white) and C. orthopsilosis (black) globally (A) and in the United States (B).
Regional distribution of C. orthopsilosis and C. metapsilosis.
C. orthopsilosis isolates were identified from all six of the continents from which isolates were received, but the isolates were not evenly distributed by continent or by country (Table 1). Asia had the highest percentage of isolates that were C. orthopsilosis (11%). However, if a series of 10 isolates from a single pediatric ward in one hospital, a possible outbreak, was removed, the percentage in Asia dropped to 6.1%. Of the isolates from South America, 10.9% were C. orthopsilosis. The distribution on this continent varied from 16.5% and 16.0% C. orthopsilosis isolates from Venezuela and Brazil, respectively, to 4.3% and 3.1% C. orthopsilosis isolates from Ecuador and Argentina, respectively. In North America, 5.0% of the C. parapsilosis complex isolates were C. orthopsilosis. In the United States, 4.9% of the isolates were C. orthopsilosis, while in Mexico, 10.7% of the isolates were C. orthopsilosis. None of the 52 isolates from Canada were C. orthopsilosis. Overall, Europe and the Middle East had 17 (3.5%) C. orthopsilosis isolates, with almost half of them coming from Italy, where 8.0% of the isolates were C. orthopsilosis. No C. orthopsilosis isolates were recovered from Finland, the United Kingdom, Poland, Portugal, Russia, Hungary, and Israel, despite their combined total of 177 isolates of C. parapsilosis. Interestingly, although there were 146 C. parapsilosis complex isolates from multiple centers in South Africa, only a single isolate of C. orthopsilosis was identified from that country.
TABLE 1.
Regional distribution of C. parapsilosis, C. orthopsilosis, and C. metapsilosis
| Country or region | No. of isolates by species (%)c
|
|||
|---|---|---|---|---|
| C. parapsilosis | C. orthopsilosis | C. metapsilosis | L. elongisporus | |
| United States | 506 | 26 | 2 | 2 |
| Canada | 52 | 2 | ||
| Mexico | 42 | 6 | 8 | |
| North America | 600 | 32 (5.0) | 4 (0.6) | 10 |
| Venezuela | 94 | 19 | 2 | |
| Chile | 7 | |||
| Ecuador | 45 | 2 | ||
| Brazil | 80 | 16 | 4 | |
| Argentina | 62 | 2 | 1 | |
| Columbia | 49 | 3 | ||
| South America | 337 | 42 (10.9) | 5 (1.3) | |
| Finland | 12 | |||
| Netherlands | 2 | 1 | ||
| Hungary | 16 | |||
| Czech Republic | 27 | 2 | ||
| Slovak Republic | 44 | 3 | ||
| United Kingdom | 25 | |||
| Spain | 46 | 2 | 1 | |
| Turkey | 72 | 1 | 2 | |
| Poland | 25 | 4 | ||
| Portugal | 66 | 1 | ||
| Italy | 86 | 8 | 5 | 1 |
| Russia | 18 | 1 | ||
| Israel | 15 | 1 | ||
| Europe and Middle East | 454 | 17 (3.5) | 14 (2.9) | 2 |
| Thailand | 2 | 1 | ||
| Malaysia | 38 | 15b | 2 | |
| South Korea | 72 | 2 | ||
| Taiwan | 29 | 2 | ||
| China | 22 | 3 | 3 | |
| Asia | 163 | 21 (11.0) | 5 (2.6) | 2 |
| Australia | 64 | 4 (5.5) | 5 (6.8) | |
| Africaa | 144 | 1 (0.7) | 1 (0.7) | |
All African isolates in this study came from the country of South Africa.
Five isolates were not included in the perceived outbreak.
The percentages of C. orthopsilosis and C. metapsilosis in each geographic region are indicated in parentheses.
Like C. orthopsilosis, C. metapsilosis isolates could be found on all six continents. Only 4 of 646 (0.6%) North American isolates and only 1 of 146 (0.7%) South African isolates were C. metapsilosis. Although no C. orthopsilosis isolates came from Canada, two C. metapsilosis isolates did. Europe had almost as many C. metapsilosis isolates, 14 (2.9%), as C. orthopsilosis isolates. Poland had the highest percentage with 13.8% of all C. parapsilosis complex isolates being C. metapsilosis. C. metapsilosis isolates also came from Spain, Portugal, Turkey, and Italy. Australia had more C. metapsilosis isolates than C. orthopsilosis isolates, and 6.8% of all C. parapsilosis complex isolates from Australia were C. metapsilosis. Both China and Taiwan had C. metapsilosis isolates, and 2.6% of all Asian isolates were C. metapsilosis. Despite the very high number of C. orthopsilosis isolates from South America, only 1.7% of South American C. parapsilosis complex isolates were C. metapsilosis: four from Brazil and one from Argentina.
Antifungal susceptibility testing.
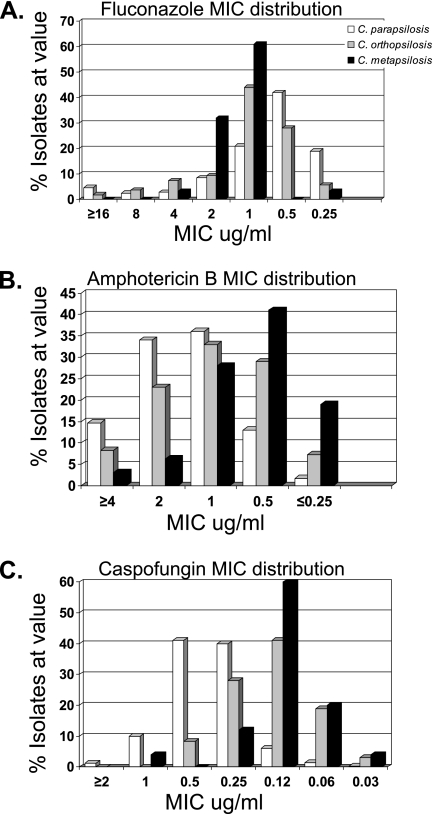
Susceptibility test results are summarized in Fig. 3 and Table 2. All of the C. orthopsilosis isolates were fluconazole susceptible. There were no resistant or susceptible dose-dependent C. metapsilosis isolates, and the highest fluconazole MIC was 4 μg/ml (Table 2 and Fig. 3A). By comparison, 0.6% of C. parapsilosis isolates were resistant and 4.0% were susceptible dose dependent. The mean MIC of fluconazole against C. parapsilosis was higher than the mean MICs of fluconazole against C. orthopsilosis and C. metapsilosis. However, both the MIC90s and the overall MIC distributions were higher for C. orthopsilosis and C. metapsilosis than for C. parapsilosis (Table 2 and Fig. 3A).
FIG. 3.
MIC distributions of C. parapsilosis (white), C. orthopsilosis (gray), and C. metapsilosis (black) isolates against fluconazole (A), amphotericin B (B), and caspofungin (C).
TABLE 2.
MIC50/MIC90 and mean MIC of C. parapsilosis, C. orthopsilosis, and C. metapsilosis isolates
| Drug |
C. parapsilosis
|
C. orthopsilosisa
|
C. metapsilosisa
|
|||
|---|---|---|---|---|---|---|
| MIC50/MIC90 | Mean MIC | MIC50/MIC90 | Mean MIC | MIC50/MIC90 | Mean MIC | |
| Fluconazole | 0.5/2 | 2.3 | 1/4 | 2.1 | 1/2 | 1.4 |
| Amphotericin B | 1/4 | 1.5 | 1/2 | 1.1 | 0.5/1 | 0.7 |
| Caspofungin | 0.5/1 | 0.4 | 0.12/0.25 | 0.2 | 0.12/0.25 | 0.2 |
| Anidulafungin | 2/2 | 1.9 | 1/2 | 1.0 | 0.5/1 | 0.6 |
| Micafungin | 1/2 | 1.1 | 0.25/0.5 | 0.4 | 0.25/0.5 | 0.4 |
P < 0.001 for the difference in MIC distribution compared with C. parapsilosis for all drugs tested. For fluconazole, C. orthopsilsosis and C. metapsilosis MIC distributions were higher than those of C. parapsilosis, while for amphotericin B and all the echinocandins, C. orthopsilosis and C. metapsilosis MIC distributions were lower than those of C. parapsilosis.
The MICs of C. orthopsilosis and C. metapsilosis to amphotericin B were lower than those of C. parapsilosis (Table 2 and Fig. 2B). While 14.7% of C. parapsilosis isolates had an amphotericin B MIC of ≥4 μg/ml, only 8.3% of C. orthopsilosis isolates and 3.1% of C. metapsilosis isolates had MICs of ≥4 μg/ml. While only 15% of C. parapsilosis isolates had MICs of ≤0.5 μg/ml, 36.3% of C. orthopsilosis isolates and 60% of C. metapsilosis isolates had MICs of ≤0.5 μg/ml.
Recent reports have indicated that the MICs of the echinocandins for C. parapsilosis isolates are elevated compared to those for C. albicans (18, 20, 21). As is demonstrated in Fig. 3 and Table 2, the echinocandin MICs of C. parapsilosis were higher than those of C. orthopsilosis or C. metapsilosis. In this study, the MIC90s for C. parapsilosis against caspofungin, anidulafungin, and micafungin were 1 μg/ml, 2 μg/ml, and 2 μg/ml, respectively. By contrast, the MIC90s for C. orthopsilosis against caspofungin, anidulafungin and micafungin were 0.25 μg/ml, 2 μg/ml, and 0.5 μg/ml, respectively. Similar results were seen for C. metapsilosis, with MIC90 values of 0.25 μg/ml, 1 μg/ml, and 0.5 μg/ml against caspofungin, anidulafungin, and micafungin, respectively.
DISCUSSION
This is the most comprehensive study of the antifungal susceptibilities of C. orthopsilosis and C. metapsilosis to date. There have been reports of smaller numbers of isolates, some dating to before they were formally separated into species. In 1995, Lin and coworkers (11) reported that two isolates of C. metapsilosis had MICs to amphotericin B that were lower on average than those of C. parapsilosis in the same study. Similar results were reported for two isolates of C. metapsilosis from Hungary (9). In a study of 27 isolates of C. orthopsilosis from Pisa, Italy, by Tavanti and coworkers (28), the mean MICs for fluconazole (5.89 μg/ml) were higher than we report here (2.1 μg/ml). The same study reported mean MICs of 0.045 μg/ml for amphotericin B and 0.18 μg/ml for caspofungin. The caspofungin MIC is similar to what we report here (0.17 μg/ml), but the mean MIC for amphotericin B is more than fourfold lower that the global average of 1.1 μg/ml reported here.
To date, there have been very few studies looking at the prevalence of C. orthopsilosis and C. metapsilosis among C. parapsilosis complex isolates. The most recent data, a population-based study from Spain (7), found that C. metapsilosis comprised 6.9% (6/87) and C. orthopsilosis comprised 5.7% (5/87) of all C. parapsilosis complex isolates collected between 2002 and 2003. A survey of Candida bloodstream infections in Scotland during 2005 and 2006 reported 35 isolates of C. parapsilosis but none of C. orthopsilosis or C. metapsilosis (16). We likewise report no C. orthopsilosis or C. metapsilosis isolates among 25 C. parapsilosis complex isolates from the United Kingdom. A similar survey in Hungary of isolates collected in 2004 and 2005 found that out of 22 C. parapsilosis complex isolates, 2 were C. metapsilosis and none were C. orthopsilosis (9). We found no C. orthopsilosis or C. metapsilosis isolates among 16 C. parapsilosis complex isolates from Hungary. Tavanti and coworkers (28) reported that 4.5% of all C. parapsilosis complex isolates from Pisa, Italy, were C. orthopsilosis, and none were C. metapsilosis. The actual incidence in that study may have been somewhat lower, as all 33 C. orthopsilosis isolates came from only 13 patients. Although our results were pooled from eight different sites in Italy, we found 8% C. orthopsilosis and 5% C. metapsilosis, similar to the numbers reported for Spain (7). There is also a very recent report of C. orthopsilosis causing a bloodstream infection in Malaysia (30), another country from which we have isolated a number of C. orthopsilosis isolates.
Although we do not have the clinical data from the hospital or the fingerprint patterns of the isolates, there were 10 isolates of C. orthopsilosis from a pediatrics unit isolated over the course of a year in a single hospital in Malaysia. If this was an outbreak, it would bring the number of known nosocomial outbreaks of C. orthopsilosis to three.
In conclusion, we report the species distribution and antifungal susceptibilities of a large global collection of C. parapsilosis complex isolates. We found that C. orthopsilosis and C. metapsilosis together account for fewer than 10% of C. parapsilosis complex infections overall, with substantial regional variation noted. However, the proportion of isolates represented by C. orthopsilosis may have been increasing in recent years. Both C. orthopsilosis and C. metapsilosis have amphotericin B and echinocandin MICs lower than those of C. parapsilosis. Although they have slightly higher fluconazole MICs overall than C. parapsilosis, all of the C. orthopsilosis and all of the C. metapsilosis isolates were susceptible to fluconazole. The differences in antifungal susceptibility that we describe are therefore not categorical differences, suggesting that routine discrimination among these C. parapsilosis complex species is not necessary for the clinical laboratory. However, ongoing surveillance is needed to monitor changes in the species distribution and antifungal susceptibility within this species complex. In addition, discrimination of species within the complex may be important for future clinical trials of candidemia treatment, in order to determine if the MIC differences noted by our group and others are associated with differences in clinical outcomes.
Acknowledgments
We thank Shailesh Tendolkar and Richard Hollis for their contributions to the manuscript.
This study was funded by grants from Merck & Co., Inc., and Pfizer, Inc.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Almirante, B., D. Rodriguez, M. Cuenca-Estrella, M. Almela, F. Sanchez, J. Ayata, C. Alonso-Tarres, J. L. Rodriguez-Tudela, A. Pabissa, et al. 2006. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 441681-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito, L. R., T. Guimaraes, M. Nucci, R. C. Rosas, L. P. Almeida, D. A. DaMatta, and A. L. Colombo. 2006. Clinical and microbiological aspects of candidemia due to Candida parapsilosis in Brazilian tertiary care hospitals. Med. Mycol. 44261-266. [DOI] [PubMed] [Google Scholar]
- 3.Clark, T. A., S. A. Slavinski, J. Morgan, T. Lott, B. A. Arthington-Skaggs, M. E. Brandt, R. M. Webb, M. Currier, R. H. Flowers, S. K. Fridkin, and R. A. Hajjeh. 2004. Epidemiologic and molecular characterization of an outbreak of Candida parapsilosis bloodstream infections in a community hospital. J. Clin. Microbiol. 424468-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo, A. L., M. Nucci, R. Salomao, M. L. M. Branchini, R. Richtmann, A. Derossi, and S. B. Wey. 1999. High rate of non-albicans candidemia in Brazilian tertiary care hospitals. Diagn. Microbiol. Infect. Dis. 34281-286. [DOI] [PubMed] [Google Scholar]
- 5.Diezmann, S., C. J. Cox, G. Schonian, R. J. Vilgalys, and T. G. Mitchell. 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 425624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enger, L., S. Joly, C. Pujol, P. Simonson, M. A. Pfaller, and D. R. Soll. 2001. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J. Clin. Microbiol. 39658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Lopez, A., A. Alastruey-Izquierdo, D. Rodriguez, B. Almirante, A. Pahissa, J. L. Rodriguez-Tudela, M. Cuenca-Estrella, and the Barcelona Candidemia Project Study Group. 2008. Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis. Results from population-based surveillance of candidemia in Spain. Antimicrob. Agents. Chemother 521506-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao, A. S., M. E. Brandt, W. R. Pruitt, L. A. Conn, B. A. Perkins, D. S. Stephens, W. S. Baughman, A. L. Reingold, G. A. Rothrock, M. A. Pfaller, R. W. Pinner, and R. A. Hajjeh. 1999. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin. Infect. Dis. 291164-1170. [DOI] [PubMed] [Google Scholar]
- 9.Kocsubé, S., M. Tóth, C. Vágvölgyi, I. Dóczi, M. Pesti, I. Pócsi, J. Szabó, and J. Varga. 2007. Occurrence and genetic variability of Candida parapsilosis sensu lato in Hungary. J. Med. Microbiol. 56190-195. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann, P. F., D. Lin, and B. A. Lasker. 1992. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J. Clin. Microbiol. 303249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, D., L. C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 331815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockhart, S. R., S. A. Messer, M. A. Pfaller, and D. J. Diekema. 2008. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of blood stream infections. J. Clin. Microbiol. 46374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lott, T. J., R. J. Kuykendall, S. F. Welbel, A. Pramanik, and B. A. Lasker. 1993. Genomic heterogeneity in the yeast Candida parapsilosis. Curr. Genet. 23463-467. [DOI] [PubMed] [Google Scholar]
- 14.Moudgal, V., T. Little, D. Boikov, and J. A. Vazquez. 2005. Multiechinocandin- and multiazole-resistant Candida parapsilosis isolates serially obtained during therapy for prosthetic valve endocarditis. Antimicrob. Agents Chemother. 49767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth microdilution antifungal susceptibility testing of yeast, 2nd ed. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 16.Odds, F. C., M. F. Hanson, A. D. Davidson, M. D. Jacobsen, P. Wright, J. A. Whyte, N. A. Gow, and B. L. Jones. 2007. One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 561066-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odds, F. C., M. Motyl, R. Andrade, J. Bille, E. Cantón, M. Cuenca-Estrella, A. Davidson, C. Durussel, D. Ellis, E. Foraker, A. W. Fothergill, M. A. Ghannoum, R. A. Giacobbe, M. Gobernado, R. Handke, M. Laverdière, W. Lee-Yang, W. G. Merz, L. Ostrosky-Zeichner, J. Pemán, S. Perea, J. R. Perfect, M. A. Pfaller, L. Proia, J. H. Rex, M. G. Rinaldi, J. L. Rodriguez-Tudela, W. A. Schell, C. Shields, D. A. Sutton, P. E. Verweij, and D. W. Warnock. 2004. Interlaboratory comparison of results of susceptibility testing with caspofungin against Candida and Aspergillus species. J. Clin. Microbiol. 423475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrosky-Zeichner, L., J. H. Rex, P. G. Pappas, R. J. Hamill, R. A. Larsen, H. W. Horowitz, W. G. Powderly, N. Hyslop, C. A. Kauffman, J. Cleary, J. E. Mangino, and J. Lee. 2003. Antifungal susceptibility survey of 2,000 bloodstream Candida isolates in the United States. Antimicrob. Agents Chemother. 473149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J. Clin. Microbiol. 46150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., L. Boyken, R. J. Hollis, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2006. Global surveillance of in vitro activity of micafungin against Candida: a comparison with caspofungin by CLSI-recommended methods. J. Clin. Microbiol. 443533-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller, M. A., D. J. Diekema, D. L. Gibbs, V. A. Newell, K. P. Ng, A. Colombo, J. Finquelievich, R. Barnes, J. Wadula, and the Global Antifungal Surveillance Group. 2008. Geographic and temporal trends in isolation and antifungal susceptibility of Candida parapsilosis: a global assessment from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J. Clin. Microbiol. 46842-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, and R. J. Hollis. 2003. Activity of fluconazole and voriconazole determined by broth microdilution, disk diffusion, and Etest methods against 1,586 recent clinical isolates of Candida species: report from the ARTEMIS global antifungal susceptibility program, 2001. J. Clin. Microbiol. 411440-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rex, J., M. Pfaller, J. Galgiani, M. Bartlett, A. Espinel-Ingroff, M. Ghannoum, M. Lancaster, F. Odds, M. Rinaldi, T. Walsh, and A. Barry. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24235-247. [DOI] [PubMed] [Google Scholar]
- 24.Roy, B., and S. A. Meyer. 1998. Confirmation of the distinct genotype groups within the form species Candida parapsilosis. J. Clin. Microbiol. 36216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Miguel, L. G., J. Cobo, E. Otheo, A. Sánchez-Sousa, V. Abraira, and S. Moreno. 2005. Secular trends of candidemia in a large tertiary-care hospital from 1988 to 2000: emergence of Candida parapsilosis. Infect. Control Hosp. Epidemiol. 26548-552. [DOI] [PubMed] [Google Scholar]
- 26.Sarvikivi, E., O. Lyyttikainen, D. R. Soll, C. Pujol, M. A. Pfaller, M. Richardson, P. Koukila-Kahkola, P. Luukkainen, and H. Saxen. 2005. Emergence of fluconazole resistance in a Candida parapsilosis strain that caused infections in a neonatal intensive care unit. J. Clin. Microbiol. 432729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavanti, A., A. D. Davidson, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavanti, A., L. A. M. Hensgens, E. Ghelardi, M. Campo, and S. Senesi. 2007. Genotyping of Candida orthopsilosis clinical isolates by amplification fragment length polymorphism reveals genetic diversity among independent isolates and strain maintenance within patients. J. Clin. Microbiol. 451455-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viscoli, C., C. Girmenia, A. Marinus, L. Collette, P. Martino, B. Vandercam, C. Doyen, B. Lebeau, D. Spence, V. Krcmery, B. De Pauw, F. Meunier, and the Invasive Fungal Infection Group of the EORTC. 1999. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin. Infect. Dis. 281071-1079. [DOI] [PubMed] [Google Scholar]
- 30.Yong, P. V., P. P. Chong, L. Y. Lau, R. S. Yeoh, and F. Jamal. 2008. Molecular identification of C. orthopsilosis isolated from blood culture. Mycopathologia 16581-87. [DOI] [PubMed] [Google Scholar]
- 31.Zancope-Oliveira, R. M., M. J. James, A. P. Derossi, J. L. Sampaio, M. M. Muniz, R. K. Li, A. S. Nascimento, J. M. Peralta, and E. Reiss. 2000. Strain characterization of Candida parapsilosis fungemia by molecular typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 19514-520. [DOI] [PubMed] [Google Scholar]