Abstract
The performance of a new commercial PCR-enzyme-linked immunosorbent assay (ELISA) (Cryptodiag; Bio Advance, France) for the diagnosis of cryptosporidiosis and the identification of Cryptosporidium hominis and C. parvum from stool samples was examined. This test is based on PCR amplification of Cryptosporidium DNA extracted from stools, followed by an ELISA based on hybridization with Cryptosporidium sp.-, C. hominis-, or C. parvum-specific probes. In spiking experiments, approximately five oocysts were detected either in water or in stool suspensions while assessing for the efficient removal of stool PCR inhibitors. No cross-reactivity was observed in the detection of C. parvum and C. hominis using the respective specific probes. Thirty-three fecal samples from patients with microscopically proven cryptosporidiosis and 118 from patients with or without other digestive protozoan infections were tested by Cryptodiag, blinded to the results of microscopy. Compared to microscopy, the sensitivity of Cryptodiag was 97.0% (32/33) and 100% (33/33), including the gray zone, and specificity was 98.3% (116/118) and 96.6% (114/118), including the gray zone. Among 34 positive results, Cryptodiag identified 19 due to C. hominis, 8 due to C. parvum, and 7 due to Cryptosporidium spp. Genotyping by Cryptodiag agreed with reference typing methods in 85% of cases of C. parvum or C. hominis infections. Cryptodiag proved to be reliable and sensitive for the diagnosis of cryptosporidiosis. The use of specific probes allowed the identification of C. hominis and C. parvum, i.e., the two main species responsible for human cryptosporidiosis, and rapidly provided information on the possible source of infection.
Cryptosporidiosis occurs worldwide and is responsible for significant morbidity and mortality, especially in human immunodeficiency virus-infected patients (4). Most cases of cryptosporidiosis are due to sporadic rather than outbreak-associated infections (11). However, waterborne and food-borne outbreaks are reported frequently and represent around 10% of all cases of Cryptosporidium infection (11). There is still no curative treatment (1), making cryptosporidiosis a major public health issue and economic problem (7). Among the 16 validated species of Cryptosporidium, 2 are more frequently found in humans: Cryptosporidium parvum and C. hominis, and the latter is considered anthroponotic and more frequently responsible for extraintestinal sequelae than C. parvum (10).
Diagnosis of cryptosporidiosis is generally based on microscopic detection of oocysts in stools, but this offers no information on the infecting species and is unhelpful in epidemiological investigations (9). The development of genetic tools now has made possible the detection of Cryptosporidium oocysts by PCR and species identification by sequencing, restriction fragment length polymorphism (RFLP), or the use of species-specific probes (2, 3, 6, 8, 21). However, genotyping for specific identification remains restricted to reference or specialized laboratories.
The aim of this study was to assess the performance of a new method for the detection and identification (genotyping) of Cryptosporidium in biological samples. Based on PCR combined with genus- or species-specific probe hybridization and visualization by enzyme-linked immunosorbent assay (ELISA), this test provides a simple and sensitive tool for routine application in human diagnosis and genotyping surveillance.
MATERIALS AND METHODS
Parasites.
C. parvum oocysts obtained from the feces of a naturally infected calf were used for spiking experiments. The oocysts were concentrated from the feces by a one-step diethyl ether method. Briefly, 10 ml of dichromate fecal suspension was vigorously mixed with 5 ml of diethyl ether and then centrifuged at 1,000 × g for 10 min at 4°C. The pellet was washed twice in Tris-EDTA by centrifugation at 1,000 × g for 10 min at 4°C. The pellet was resuspended in 1 ml of Tris-EDTA before being applied to an ice-cold two-layer potassium bromide (KBr) gradient consisting of two solutions of 7 ml 16% KBr and 3 ml 6% KBr diluted in Tris-EDTA. The pellet, resuspended in Tris-EDTA, was carefully layered on the top of the gradient and centrifuged at 3,000 × g for 30 min at 4°C. Purified oocysts were collected from the interface between the KBr layers, diluted 1/3 with distilled water, and centrifuged at 3,000 × g for 10 min at 4°C. The pellet was resuspended, washed twice in phosphate-buffered saline (PBS), and kept at +4°C until it was used. The oocysts were enumerated in triplicate in a Kova-slide hemocytometer (Hycor).
Clinical samples.
We studied 151 human fecal samples collected in our laboratory hospital. Thirty-three samples were from patients with microscopically proven cryptosporidiosis. Cryptosporidiosis was diagnosed by microscopic demonstration of typical Cryptosporidium oocysts on a Ziehl-Neelsen-stained fecal smear after concentration by the one-step ethyl ether method. For one patient, who was initially found negative by microscopy but was found within the gray zone by Cryptodiag, the diagnosis of cryptosporidiosis was made retrospectively after careful reexamination of the slides. Five patients with cryptosporidiosis were coinfected with other intestinal protozoal parasites (Giardia duodenalis, two; Entamoeba coli, Endolimax nana, and Blastocysts hominis, one; Entamoeba histolytica, one; Pentatrichomonas hominis and Chilomastix mesnilii, one), two samples were from patients infected with Isospora belli, and 115 samples were from patients with no diagnosed intestinal parasitosis.
Molecular diagnosis by PCR-ELISA (Cryptodiag).
As recommended by the manufacturer, approximately 5 g of stools was diluted in 5 ml of distilled water and kept at 4°C before DNA extraction. DNA was extracted and purified using the extraction kit provided following the manufacturer's instructions. Briefly, the sample was subjected to a chemical and enzymatic lysis, followed by precipitation of proteins and resin filtration. Then, DNA collected from the column eluate was amplified and characterized in a two-step procedure. The first step, consisting of PCR amplification of a repeated Cryptosporidium target DNA, was performed in a 50-μl volume containing 5 μl of DNA template, 75 mM Tris-HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween 20, 4 mM MgCl2, 0.15 mM of each deoxyribonucleoside triphosphate, 1 μl of each primer provided with the kit, and 2 U of Taq DNA polymerase (Eurogentec, Angers, France). Amplifications were performed using a TC-412 thermal cycler (Techne; Bioblock, Illkirch, France), following the manufacturer's instructions, with initial denaturation for 5 min at 94°C, 39 cycles of amplification (denaturation for 30 s at 94°C, annealing for 45 s at 58°C, and extension for 60 s at 72°C), and a final extension step for 5 min at 72°C, and then cooling to 4°C. Carryover contamination can be prevented by the use of uracil-N-glycosylase with dUTP/deoxynucleoside triphosphate (Eurogentec, Angers, France).
In the second step, the amplified products were detected with an ELISA, as recommended by the manufacturer. Briefly, the amplified products were transferred into streptavidin-precoated microwells, incubated for 30 min at 50°C, and then subjected to chemical denaturation within the well with the provided denaturation solution for 10 min at room temperature. After four washing steps with the washing buffer, the Cryptosporidium-specific labeled probe diluted in a hybridization buffer was added to each microwell containing the denatured products and was incubated for 1 h at 44°C. After four washing steps with washing buffer, the diluted conjugate was added to each well and incubated for 1 h at 25°C. After four washing steps, a colorimetric substrate was added to each well and incubated for 15 min at 25°C in the dark. The reaction was stopped with the stop solution provided. Optical-density (OD) values were recorded spectrophotometrically at 450 nm, using 620 nm as a reference wavelength. Under these conditions, the OD values were <0.1 for the negative control (distilled water) and >2.0 with the positive control (provided with the kit), consisting of plasmid DNA containing the target sequence of Cryptosporidium.
Study design and analysis.
Spiking experiments were performed to determine the limit of detection of Cryptodiag for detection of Cryptosporidium oocysts in water and stools. Suspensions of purified oocysts (10,000, 1,000, 100, 10, and 0 oocysts/ml) were prepared in PBS and in stools, either undiluted or diluted at 1/2 in PBS. DNA extraction and purification were carried out as described above. Experiments were conducted using the Cryptosporidium sp. probe. Each dilution was tested in duplicate wells.
The specificity of Cryptodiag was examined experimentally and on clinical samples by testing DNA extracted from C. parvum, C. hominis, and C. felis and human DNA. Cross-experiments were performed to test the specificity of Cryptosporidium sp., C. hominis, and C. parvum probes. Each test was performed in triplicate wells.
Experiments on clinical samples allowed us to calculate the sensitivity and specificity of Cryptodiag for diagnosis of cryptosporidiosis, using as a reference the results of microscopic examination of stools (Ziehl-Neelsen stain). For genotyping of clinical isolates, we used a nested PCR-RFLP method as a reference (8), completed by sequence analysis of the 18S rRNA gene for some species not identified by PCR-RFLP. Cryptodiag tests, including C. hominis and C. parvum determinations, were performed blinded to the results of microscopic examination and PCR-RFLP genotyping.
RESULTS
Detection of Cryptosporidium DNA.
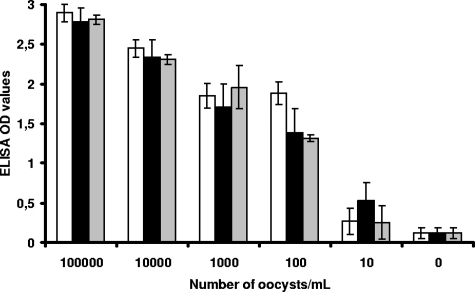
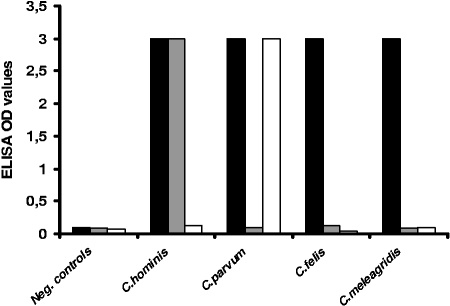
Spiking experiments performed on serial dilutions of C. parvum oocysts showed good correlation between oocyst counts and ELISA OD values (Fig. 1). The regression coefficients were 0.94, 0.91, and 0.96 for dilutions in PBS, stools, and diluted stools, respectively. The threshold of detection was estimated at 5 to 10 oocysts for all diluents, assessing for efficient removal of PCR inhibitors during the DNA extraction. The capacity of the Cryptosporidium sp. probe to differentiate Cryptosporidium DNA from that of other species was assessed by the strong positivity of the test performed with C. parvum and C. hominis plasmids (Fig. 2), as well as with the genomic DNA of C. felis, C. meleagridis, and C. canis (data not shown).
FIG. 1.
Spiking experiments performed on serial dilutions of purified C. parvum oocysts in PBS (white bars) and stools, either undiluted (black bars) or diluted at 1/10 in PBS (gray bars). Cryptosporidium DNA was detected by Cryptodiag using the Cryptosporidum sp. probe (R2 = 0.94 for dilution in PBS; R2 = 0.91 for dilution in stools; R2 = 0.96 for 1/10 dilution in diluted stool). Shown are the means of two OD values. The error bars indicate standard deviations.
FIG. 2.
Specificities of Cryptodiag probes for the detection of DNA from four species of Cryptosporidium using Cryptosporidium sp. (black bars), C. hominis (gray bars), or C. parvum (white bars) probes. Shown are the means of three OD values recorded for each point (maximum OD value = 3). Neg., negative.
Testing serial dilutions of C. hominis plasmids resulted in positive OD values for samples containing at least five target copies, corresponding approximately to less than 1 fg of the target parasite DNA.
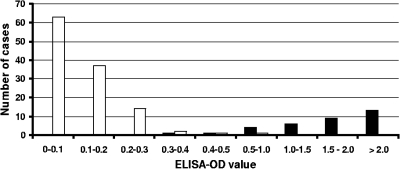
The sensitivity and specificity of Cryptodiag for diagnosis of cryptosporidiosis were assessed by testing 151 clinical samples. The results were expressed as the OD value obtained with the sample minus the OD value of the negative control. The distribution of OD values is presented in Fig. 3. The mean OD values were 0.113 ± 0.090 for samples that were negative by microscopy and 1.747 ± 0.701 for microscopy-positive samples (median value = 1.764). From this OD value distribution, a threshold of 0.4 was selected (the mean of the negative samples plus 3 standard deviations) for definitive diagnosis of cryptosporidiosis, with a gray zone (uncertain diagnosis) for OD values between 0.3 and 0.4. Following these criteria, the sensitivity and specificity were 100% and 96.6%, respectively, including the gray zone (Table 1). Comparison of the proportions of positive and negative results obtained by microscopy and by Cryptodiag showed no significant difference between the two techniques regardless of the cutoff used for Cryptodiag (McNemar's test; P = 0.125 and P = 1 for cutoffs of 0.3 and 0.4, respectively).
FIG. 3.
Distribution of ELISA OD values obtained by Cryptodiag for 151 stool samples, including 33 that were positive by microscopy (black bars) and 118 that were negative by microscopy (open bars) (maximum recorded OD value = 3).
TABLE 1.
Comparative performances of Cryptodiag and microscopy for diagnosis of cryptosporidiosis in 151 stool samples
| Microscopy, Ziehl-Neelsen result | No. with Cryptodiag resulta:
|
|||
|---|---|---|---|---|
| Negative | Gray zone | Positive | Total | |
| Negative | 114 | 2 | 2 | 118 |
| Positive | 0 | 1 | 32 | 33 |
| Total | 114 | 3 | 34 | 151 |
Positive, OD ≥ 0.4; gray zone, 0.3 ≤ OD < 0.4; negative, OD < 0.3. Cryptodiag sensitivity and specificity were as follows: for threshold of OD ≥ 0.4, sensitivity, 97.0% (32/33), and specificity, 98.3% (116/118); for threshold of OD ≥ 0.3 (including gray zone), sensitivity, 100% (33/33), and specificity, 96.6% (114/118).
The four discrepant results between microscopy and Cryptodiag were reviewed. For the two patients positive by Cryptodiag and the two patients within the gray zone, microscopy was confirmed to be negative, as was PCR-RFLP.
Genotypic identification by Cryptodiag.
Experiments performed with purified plasmid DNAs of C. parvum and C. hominis tested by Cryptodiag using the respective specific probes showed good specificity for the detection of the respective species and no cross-reactivity with other species. As expected, the genomic DNAs of C. felis, C. meleagridis, and C. canis were not detected by the C. parvum or C. hominis probes (Fig. 2).
Thirty-four clinical samples that were positive with the Cryptosporidium sp. probes were processed by Cryptodiag using the C. parvum- and C. hominis-specific probes and by PCR-RFLP as the reference genotyping method. In one case, genotyping by PCR-RFLP could not be achieved because of the presence of PCR inhibitors, whereas Cryptodiag identified C. parvum. Of the 33 remaining samples, all 6 samples identified as C. parvum by PCR-RFLP and 18 out of 20 samples identified as C. hominis by PCR-RFLP were correctly identified by Cryptodiag. In addition, PCR-RFLP identified C. felis in four cases (Cryptodiag was positive by the Cryptosporidium sp. probe). In only one case, the infecting species could not be determined by PCR-RFLP or by Cryptodiag.
Finally, combining PCR-RFLP and Cryptodiag results, C. hominis was found more often than C. parvum (19/34 versus 8/34), possibly due to the urban origin of most of our patients (Table 2).
TABLE 2.
Comparative performances of Cryptodiag and nested PCR-RFLP for identification of Cryptosporidium species
| Nested PCR-RFLP result | No. with Cryptodiag result:
|
|||
|---|---|---|---|---|
| C. parvum | C. hominis | Cryptosporidium spp. | Total | |
| C. parvum | 6 | 0 | 0 | 6 |
| C. hominis | 0 | 18 | 2 | 20 |
| C. felis | 0 | 0 | 4 | 4 |
| Cryptosporidium spp. | 0 | 0 | 1 | 1 |
| Not determined | 2 | 1 | 0 | 3 |
| Total | 8 | 19 | 7 | 34 |
DISCUSSION
Despite reports of an increasing number of cases of cryptosporidiosis in Europe and in the United States (18, 25) and the persistent occurrence of food-borne and waterborne outbreaks (13), cryptosporidiosis remains underreported in many countries, due to a lack of official reporting and/or difficulties in diagnosing cryptosporidiosis. In most public and private laboratories, testing for Cryptosporidium is usually not included in routine examination of stool for ova and parasites, while oocysts often cannot be detected by routine microscopic examination of stool without staining or immunolabeling.
The PCR-ELISA method Cryptodiag, developed for diagnosis of cryptosporidiosis, offers several advantages for diagnostic practice. First, it is commercialized as a kit in which the reagents needed for DNA extraction, amplification by PCR of Cryptosporidium DNA, and visualization and identification of hybridized products are provided in an easy-to-use format, and the equipment required, i.e., a conventional PCR thermal cycler and an ELISA spectophotometric reader, is routinely used in many laboratories.
Second, the improved DNA extraction procedure provided with the kit allows the elimination of most PCR inhibitors present in stools. This is a major advantage, since the presence of PCR inhibitors and their removal have been frequently found to be limitations in many PCR studies of stools or water samples (12, 17, 19).
Third, experimental tests on spiked samples showed that Cryptodiag has good sensitivity, with a threshold of detection of 5 to 10 oocysts/ml of water or stools. This represents a >100-fold increase in sensitivity compared to the sensitivity of microscopy as estimated in other studies performed on fecal samples seeded with various numbers of oocysts (23, 24). Tests performed on clinical samples confirmed this, showing 97 to 100% sensitivity compared to the reference microscopic examination following formol ether concentration and Ziehl-Neelsen staining and no cross-reactivity with other pathogens present in stools. In one case, the diagnosis of cryptosporidiosis was made only after careful reexamination of microscopic slides in view of a Cryptodiag result within the gray zone. In the cases where Cryptodiag was positive and microscopic examination was negative, the high sensitivity of Cryptodiag and its specificity support the hypothesis of mild infection not detectable microscopically. Discrepancy between PCR-positive and microscopy-negative results due to the higher sensitivity of detection of PCR has been highlighted by others using real-time PCR (22). In addition, Cryptodiag can be partially automated, so the test offers benefits of sensitivity, specificity, and practicability for its use in clinical laboratories intending to perform diagnosis of cryptosporidiosis in stools.
Besides the basic diagnosis of cryptosporidiosis, there is a growing interest in identifying the infecting species of Cryptosporidium. C. parvum and C. hominis account almost equally for 95 to 98% of infections in immunocompetent individuals (4, 16). Five other species have been reported to be human pathogens, mainly in immunocompromised individuals. There are marked differences in the epidemiologies of C. parvum and C. hominis. C. hominis primarily infects humans but has occasionally been reported to naturally infect animals. By contrast, C. parvum is mainly zoonotic, as it naturally infects several animal species, including livestock. Therefore, identification of the infecting species in patients with cryptosporidiosis, especially during outbreaks, is particularly relevant to a better understanding of the transmission dynamics and to the identification of risk factors and reservoir hosts. Large genotypic studies have already been performed, mainly in the United Kingdom, and have allowed a demonstration of the seasonality of transmission of C. parvum and C. hominis and the social factors influencing their distribution (14), as well as an assessment of the efficacy of the preventive measures adopted for the reduction of transmission (15, 20).
Differences in pathogenesis between the two species was also suggested in a study by Hunter et al. (10), who found a higher rate of extraintestinal sequelae in patients previously infected by C. hominis than in those infected by C. parvum. In human immunodeficiency virus-infected patients, more subtle relationships were found between C. hominis subtype family I, C. canis, and C. felis infections and the occurrence of diarrhea (5). Given the lack of effective antiparasitic treatment, such a finding has no direct therapeutic significance at present, but it should be taken into account for long-term patient management.
For the reasons mentioned above, species identification of at least C. parvum and C. hominis is essential for public health and patient management. Presently, species identification relies on genotyping, based on a time-consuming two-step procedure combining PCR, RFLP, or sequencing. Because of the lack of a ready-to-use commercial kit, genotyping remains restricted to reference or specialized laboratories. Based on its good performance for identification of C. hominis and C. parvum in a single test procedure, Cryptodiag could facilitate species identification in nonspecialized laboratories. This would restrict genotyping in specialized laboratories to other species detected by Cryptosporidium sp. probes but not hybridized by the C. hominis or C. parvum probes (i.e., less than 4% of cases).
In conclusion, this new commercial PCR-ELISA proved to be sensitive and specific for the diagnosis of cryptosporidiosis from stool samples with efficient removal of inhibitors during DNA extraction. Diagnosis and genotyping can be performed concurrently from the same DNA extract, with PCR amplification within 48 h. As it allows identification of the major species of Cryptosporidium infecting humans, this test will have relevant applications in routine clinical diagnosis and epidemiological investigations, with potential applications in human diagnosis and environmental surveillance.
Acknowledgments
We thank the clinicians who referred the patients and samples to the laboratory, the technicians for processing the samples, and Annie Sulahian for reviewing the manuscript.
This study was supported by a grant from Bio-Advance (Bussy-Saint-Martin, France).
We have no conflicts of interest.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Abubakar, I., S. H. Aliyu, C. Arumugam, N. K. Usman, and P. R. Hunter. 2007. Treatment of cryptosporidiosis in immunocompromised individuals: systematic review and meta-analysis. Br. J. Clin. Pharmacol. 63387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, K., K. L. Kellar, I. Moura, M. C. Casaqui Carollo, T. K. Graczyk, S. Slemenda, S. P. Johnston, and A. J. da Silva. 2007. Rapid microsphere assay for identification of Cryptosporidium hominis and Cryptosporidium parvum in stool and environmental samples. J. Clin. Microbiol. 452835-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccio, S. M. 2005. Molecular epidemiology of human cryptosporidiosis. Parassitologia 47185-192. [PubMed] [Google Scholar]
- 4.Caccio, S. M., and E. Pozio. 2006. Advances in the epidemiology, diagnosis and treatment of cryptosporidiosis. Expert Rev. Anti Infect Ther. 4429-443. [DOI] [PubMed] [Google Scholar]
- 5.Cama, V. A., J. M. Ross, S. Crawford, V. Kawai, R. Chavez-Valdez, D. Vargas, A. Vivar, E. Ticona, M. Navincopa, J. Williamson, Y. Ortega, R. H. Gilman, C. Bern, and L. Xiao. 2007. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196684-691. [DOI] [PubMed] [Google Scholar]
- 6.Chalmers, R. M., C. Ferguson, S. Caccio, R. B. Gasser, Y. G. Abs EL-Osta, L. Heijnen, L. Xiao, K. Elwin, S. Hadfield, M. Sinclair, and M. Stevens. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35397-410. [DOI] [PubMed] [Google Scholar]
- 7.Corso, P. S., M. H. Kramer, K. A. Blair, D. G. Addiss, J. P. Davis, and A. C. Haddix. 2003. Cost of illness in the 1993 waterborne Cryptosporidium outbreak, Milwaukee, Wisconsin. Emerg. Infect. Dis. 9426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coupe, S., C. Sarfati, S. Hamane, and F. Derouin. 2005. Detection of cryptosporidium and identification to the species level by nested PCR and restriction fragment length polymorphism. J. Clin. Microbiol. 431017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fall, A., R. C. Thompson, R. P. Hobbs, and U. Morgan-Ryan. 2003. Morphology is not a reliable tool for delineating species within Cryptosporidium. J. Parasitol. 89399-402. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., S. Hughes, S. Woodhouse, N. Raj, Q. Syed, R. M. Chalmers, N. Q. Verlander, and J. Goodacre. 2004. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin. Infect. Dis. 39504-510. [DOI] [PubMed] [Google Scholar]
- 11.Hunter, P. R., S. Hughes, S. Woodhouse, Q. Syed, N. Q. Verlander, R. M. Chalmers, K. Morgan, G. Nichols, N. Beeching, and K. Osborn. 2004. Sporadic cryptosporidiosis case-control study with genotyping. Emerg. Infect. Dis. 101241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 711135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karanis, P., C. Kourenti, and H. Smith. 2007. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J. Water Health 51-38. [DOI] [PubMed] [Google Scholar]
- 14.Lake, I. R., F. C. Harrison, R. M. Chalmers, G. Bentham, G. Nichols, P. R. Hunter, R. S. Kovats, and C. Grundy. 2007. Case-control study of environmental and social factors influencing cryptosporidiosis. Eur. J. Epidemiol. 22805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake, I. R., G. Nichols, G. Bentham, F. C. Harrison, P. R. Hunter, and S. R. Kovats. 2007. Cryptosporidiosis decline after regulation, England and Wales, 1989-2005. Emerg. Infect. Dis. 13623-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leoni, F., C. Amar, G. Nichols, S. Pedraza-Diaz, and J. McLauchlin. 2006. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 55703-707. [DOI] [PubMed] [Google Scholar]
- 17.Nichols, R. A., J. E. Moore, and H. V. Smith. 2006. A rapid method for extracting oocyst DNA from Cryptosporidium-positive human faeces for outbreak investigations. J. Microbiol. Methods 65512-524. [DOI] [PubMed] [Google Scholar]
- 18.Semenza, J. C., and G. Nichols. 2007. Cryptosporidiosis surveillance and water-borne outbreaks in Europe. Euro Surveill. 12E13-E14. [DOI] [PubMed] [Google Scholar]
- 19.Sluter, S. D., S. Tzipori, and G. Widmer. 1997. Parameters affecting polymerase chain reaction detection of waterborne Cryptosporidium parvum oocysts. Appl. Microbiol. Biotechnol. 48325-330. [DOI] [PubMed] [Google Scholar]
- 20.Sopwith, W., K. Osborn, R. Chalmers, and M. Regan. 2005. The changing epidemiology of cryptosporidiosis in North West England. Epidemiol. Infect. 133785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stroup, S. E., S. Roy, J. McHele, V. Maro, S. Ntabaguzi, A. Siddique, G. Kang, R. L. Guerrant, B. D. Kirkpatrick, R. Fayer, J. Herbein, H. Ward, R. Haque, and E. R. Houpt. 2006. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J. Med. Microbiol. 551217-1222. [DOI] [PubMed] [Google Scholar]
- 22.ten Hove, R., T. Schuurman, M. Kooistra, L. Möller, L. van Lieshout, and J. J. Verweij. 2007. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin. Microbiol. Infect. 131001-1007. [DOI] [PubMed] [Google Scholar]
- 23.Weber, R., R. T. Bryan, and D. D. Juranek. 1992. Improved stool concentration procedure for detection of Cryptosporidium oocysts in fecal specimens. J. Clin. Microbiol. 302869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao, L., R. P. Herd, and D. M. Rings. 1993. Diagnosis of Cryptosporidium on a sheep farm with neonatal diarrhea by immunofluorescence assays. Vet. Parasitol. 4717-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoder, J. S., and M. J. Beach. 2007. Cryptosporidiosis surveillance—United States, 2003-2005. MMWR Surveill. Summ. 561-10. [PubMed] [Google Scholar]