Abstract
Hepatocellular carcinoma (HCC) and cirrhosis are important causes of mortality worldwide. Persistent hepatitis B virus (HBV) infection is a major cause of these diseases. Double mutations in the basal core promoter (BCP) (A1762T and G1764A) and precore (pre-C) (G1896A) regions of the virus are associated with progression to HCC. The current study is aimed at developing a simple method for screening and detecting BCP and pre-C mutations in HBV carriers. We have developed and validated an oligonucleotide ligation assay (OLA) to detect point mutations in the HBV core gene. We have applied OLA methods to samples from HBV-infected carriers recruited from the Gambia Liver Cancer Study (GLCS) comprising asymptomatic HBsAg carriers, patients with cirrhosis, and patients with HCC. We observed an 89.3% and 95.8% concordance between the OLA and DNA sequencing for BCP and pre-C mutations, respectively. OLA detected the mutations in single-strain infections and in infections with mixtures of wild-type and mutant viruses under conditions where sequencing detected only the single dominant strains. BCP mutations were detected in 75.7% of patients with advanced liver disease (cirrhosis/HCC) compared to 47.6% of asymptomatic carriers, while pre-C mutations were detected in 34.5% of advanced liver disease patients and in 47.6% of asymptomatic HBsAg carriers. There was a significant association between the presence of BCP mutations and advanced liver disease. In conclusion, OLA is a simple, economical, and reliable assay for detection of pre-C and BCP mutations. Its application can lead to improvement in diagnosis and clinical care in regions where HBV is endemic.
Chronic hepatitis B virus (HBV) infection is a major health problem worldwide, particularly in sub-Saharan Africa (22). The prevalence of the HBV carrier state in unvaccinated Gambians is 15 to 20% (35). Chronic HBV carriers are at high risk of developing hepatocellular carcinoma (HCC), the most important cause of cancer morbidity and mortality in many parts of the world including Asia and sub-Saharan Africa. Fifty-three percent of HCC cases worldwide are related to HBV (31). HCC is the most common cancer in Gambian men and the second most common in Gambian women, with HCC incidence rates of 61.0 and 55.7 per 100,000, respectively (1). Although the development of primary liver cancer frequently is associated with persistent HBV infection (4), the contribution of HBV to the pathogenesis of HCC is multifactorial and is complicated by the involvement of HBV mutants (MTs) that are capable of modulating the carcinogenesis process (6).
HBV is a small DNA virus with a 3.2-kb, partially double-stranded, relaxed circular genome and contains four overlapping open reading frames (pre-S/S, precore [pre-C]/core, polymerase, and X gene) (25). The pre-C region encodes the HBV e antigen (HBeAg), which has been used clinically as an indicator of active viral replication and proposed to have an immunoregulatory role in natural infection (38). The enhancer II (EnhII) and basal core promoter (BCP) reside in the overlapping X gene. The HBx protein is capable of transactivating HBV promoters (37). EnhII/BCP play an important role in the viral replicative cycle by regulating the formation of the pregenomic RNA which is important for the production of the viral core and polymerase proteins and HBeAg (25). Several nucleotide alterations that frequently occur in the BCP/EnhII/pre-C regions have been identified, but their effects on the development of HCC are unclear.
The predominant mutation in the pre-C region involves a G-to-A alteration at nucleotide 1896, changing a tryptophan codon (TGG) into a premature stop codon (TAG) at codon 28 (10). Such pre-C variants were first described in Mediterranean patients with significant liver disease in the absence of HBeAg (7).
Mutations in the BCP at nucleotide 1762 (A1762T) and nucleotide 1764 (G1764A) have been described elsewhere (8). These mutations result in coding changes at codons 130 and 131 in the HBX protein changing lysine to methionine and valine to isoleucine, respectively. The relationship between these pre-C/BCP MTs, serum HBV DNA levels, and the severity of liver disease is unclear (15, 23, 27). Although the pre-C and BCP MTs have been found in asymptomatic carriers in Africa (11), there are limited data on the association of these MTs with the development of HCC in sub-Saharan Africa, mainly because currently available detection methods are expensive and require laboratory facilities and equipment not commonly available in this region. A study in South Africa conducted on 50 HCC patients and 47 asymptomatic HBsAg carriers reported BCP prevalence rates of 66% and 11%, respectively (3). In another study, the pre-C region was sequenced in sera from 27 HCC patients from The Gambia and 33 asymptomatic HBsAg carriers. Fourteen (52%) patients compared to 12 (36%) asymptomatic carriers were found to harbor the pre-C MT (20).
Three methods are currently available for detecting HBV genome variants: the point mutation assay, the line probe assay, and direct DNA sequencing. The point mutation assay is either a radioactivity-based technique (16) or a colorimetric assay (2). The line probe assay will reliably identify pre-C mutation but has a low sensitivity in detecting BCP mutation (9). The use of the oligonucleotide ligation assay (OLA) was demonstrated by Tobe et al. as a typing assay for two alleles in a single microtiter well (33). Later, Edelstein et al. developed a high-throughput OLA method for the detection of mutations in patients with human immunodeficiency virus type 1 (HIV-1) (12). More recently OLA has been used to detect mutations causing reverse transcriptase inhibitor resistance (Q151M and M184V) in HIV-2-infected Gambian patients (17). The overall sensitivity of OLA in detecting mutations in HIV among Gambian patients was 98.8% with 99.2% concordance compared to sequencing. Minor variants, when present in mixtures of wild-type (WT) and MT viruses, were detected in cases where sequencing detected only the major population.
We have developed a novel point mutation assay using the OLA methodology for the detection of single base changes in the pre-C and BCP region of HBV. The HBV OLA was validated against consensus sequencing of the serum HBV DNA derived from asymptomatic carriers and from patients with HBV-related liver disease. We assessed the potential of OLA as a screening tool in carriers at risk of developing HCC.
MATERIALS AND METHODS
Subjects.
The study used stored serum samples from the Gambia Liver Cancer Study conducted in The Gambia between 1997 and 2001 (18). The inclusion criterion for the OLA study was positivity for HBsAg. The study used samples from 138 HCC patients, 49 cirrhosis patients, and 40 HBV-infected asymptomatic HBsAg carriers. HCC and cirrhosis cases were confirmed by clinical examination, ultrasound, and histology or α-fetoprotein (AFP) level. Asymptomatic HBsAg carriers were HBV carriers who had no evidence of liver disease, and they served as controls in the study.
AFP was detected and quantified by a standard radiometric assay (DiaSorin SA, Sallugia, Italy), as previously described (18). HBsAg was determined as a marker of chronic HBV carriage by reverse passive hemagglutination assay (Murex Diagnostics Limited, Dartford, United Kingdom) with radioimmunoassay testing of negative samples (Sorin Biomedica Diagnostics, Vercelli, Italy). Participants positive for HBsAg were tested for HBV “e” antigenemia (HBeAg) as a surrogate marker of active replication using a radioimmunoassay kit (DiaSorin).
PCR amplification of the HBV core region.
HBV DNA was extracted from 200 μl of serum using the QIAamp DNA minikit (Qiagen, United Kingdom). Extracted DNA was amplified in nested PCRs to generate a 274-bp fragment encoding HBV nucleotides 1674 to 1947 encompassing the basal core, pre-C, and core regions of the viral genome. The first-round PCR mixture contained 20 picomoles of primers HBV1 and HBVR1 (Table 1), 5 μl of DNA extract equivalent to 10 μl serum, and 12.5 μl of HotStar PCR Master Mix (Qiagen, United Kingdom) in a final PCR volume of 25 μl. Cycling conditions were 95°C for 15 min (1 cycle); 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 min (35 cycles); and a final extension at 72°C for 7 min. The second round of PCR used primers CPREF1 and CPRER1, the same reaction mix, and the same cycling conditions except for an annealing temperature of 55°C. Samples that were negative after the first nested PCR using the outer and inner primers were repeated using only the inner primers (CPRF1 and CPRER1) to obtain shorter fragments, and two rounds of 35 cycles were performed as described above with an annealing temperature of 55°C.
TABLE 1.
Primers and probes used in PCR amplification and OLAs
| Primer or probe | Positions in HBV genome | Sequenceb |
|---|---|---|
| Nested PCR primers for HBV core amplificationa | ||
| HBV1 (OF) | 1246-1266 | 5′-TTTGTGGCTCCTCTGCCGATC-3′ |
| HBVR1 (OR) | 2322-2300 | 5′-GATAAG ATAGGGGCATTTGGTGG-3′ |
| CPRF1 (IF) | 1674-1694 | 5′-TCTGCAATGTCAACGACCGAC-3′ |
| CPRER1 (IR) | 1947-1927 | 5′-AGTAACTCCACAGTAGCTCCA-3′ |
| M3 (sense) | 1730-1747 | 5′-CTGGGAGGAGTTGGGGGA-3′ |
| 5C (antisense) | 2466-2486 | 5′-CCCACCTTATGAGTCCAAGG-3′ |
| Probes for mutation type | ||
| Pre-C | ||
| 1896G | 1876-1896 | Dig-5′-GCTGTGCCTTGGGTGGCTTTG-3′ |
| 1896A | 1876-1896 | Fluo-5′-GCTGTGCCTTGGGTGGCTTTA-3′ |
| 1896/Com | 1897-1917 | 2-Phos-5′-GGGCATGGACATTGACCCTTA-3′ biotin |
| BCP | ||
| 1762AE | 1741-1762 | Dig-5′-CGGGGGAGGAGATTAGATTAAA-3′ |
| 1762TE | 1741-1762 | Fluo-5′-CGGGGGAGGAGATTAGATTAAT-3′ |
| 1762 ComE | 1763-1784 | Phos-5′-GRTCTTTGTACTAGGAGGCTGTA-3′ biotin |
| 1764GE | 1741-1764 | Dig-5′-CGGGGGAGGAGATTAGATTAAWGG-3′ |
| 1764AE | 1741-1764 | Fluo-5′-CGGGGGAGGAGATTAGATTAAWGA-3′ |
| 1764 ComE | 1765-1785 | 3-Phos-5′-TCTTTGTACTAGGAGGCTGTA-3′ biotin |
OF, outer forward; OR, outer reverse; IF, inner forward; IR, inner reverse.
Bases comprising the codons of interest are in boldface. Dig, digoxigenin; Phos, phosphorylated; Fluo, fluorescein.
OLA probes.
OLA probes were designed against the core gene sequence of genotype E (ayw4) (GenBank accession number X75657) (28). Ligation primers specific for WT sequences were labeled at the 5′ end with digoxigenin; MT-specific primers were labeled with fluorescein. The common oligonucleotide which hybridizes downstream of the WT and MT was labeled at the 3′ end with biotin and phosphorylated at the 5′ end (Table 1). This primer anneals to nucleotides in the specific regions of the HBV pre-C/BCP gene immediately adjacent to the 3′ end of both the MT and WT genomes.
OLA.
An equivalent of 10 μl of serum was used in PCR amplification (total reaction volume of 25 μl); 2 μl of the PCR mixture was then used in the OLA reaction. Thus, the amount of patient serum used in the OLA reaction is approximately 12.5 times less than that used in the PCR amplification. All tests were performed in duplicate, and ligation assays were performed according to published protocols (5, 12) utilizing a reaction mixture containing 1× ligase buffer (100 mM MgCl2, 10 mM dithiothreitol, 10 mM NAD [Sigma, United Kingdom], 80 mM KCl [Sigma], 0.1% Triton X-100), 3 U of thermostable ligase (Epicentre Technologies, United Kingdom), and 3 pmol of each ligation primer.
Detection of MT and WT genotypes was achieved by using a streptavidin-coated microtiter plate and the simultaneous addition of anti-digoxigenin-peroxidase (Boehringer Mannheim) and anti-fluorescein-alkaline phosphatase (Boehringer Mannheim) followed by sequential washing and addition of Gibco enzyme-linked immunosorbent assay (ELISA) amplification system (Invitrogen) and tetramethylbenzidine substrate (Promega) to detect MT and WT sequences, respectively. The optical densities (ODs) were measured using a Multiscan Ascent ELISA plate reader (Thermo Labsystems, Finland) at wavelengths of 490 nm and 450 nm for MT and WT, respectively.
The cutoff values for the OLAs were obtained by testing 30 replicates of MT-positive samples. The standard deviations of the ODs for the positive WT samples in the MT assay (OD at 490 nm [OD490]) and the MT samples in the WT assay (OD450) were multiplied by 3. For example, 3 × standard deviation for OLA 1896 is 0.1 and that for the OLA 1762/1764 is 0.25. Thus, following this experiment, 0.1 (OLA 1896) and 0.25 (OLA 1762) were added to the mean ODs of the negative samples to obtain the cutoff values. It was also possible to read the plates visually, as this may be important for resource-poor settings where ELISA plate readers are not easily available (data not shown). Samples were considered MT (magenta and positive OD490), WT (yellow and positive OD450), or mixed WT and MT if magenta and yellow (plus positive OD490 and OD450) were obtained in the MT and WT assays, respectively.
DNA sequencing.
Following a DNA extraction method similar to that indicated for the OLA, HBV basal core, pre-C, and core gene regions were amplified prior to DNA sequencing using primers M3 and 5C (Table 1) (11). To avoid contamination, samples that tested negative for markers of HBV (anti-HBc and HBsAg) were used as negative controls and were included in each experiment. The same controls were included in every step: DNA extraction, PCR amplification, and OLA reaction.
PCRs were carried out using HotStar Taq (Qiagen, United Kingdom) in a mixture containing 2.5 μl of 10× buffer, 5 μl of 5× Q solution, 1 μl of deoxynucleoside triphosphate (5 mM), 1 μl each of primers (20 picomoles/μl) M3 and 5C, 0.2 μl of HotStar Taq (5 μg/μl), and 5 μl of the DNA (100 to 300 μg/μl). The thermocycling conditions were 95°C for 15 min followed by 45 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 7.0 min. The PCR products (5 μl) were purified with 1.5 μl of ExoSAP-IT (Amersham) by incubation for 15.0 min at 37°C and 15.0 min at 80°C.
DNA sequencing was performed using the Big Dye Terminator v1.1 cycle sequencing kit reaction mix containing 1.5 μl PCR product, 1.25 μl of 5× buffer, 1 μl of 5 μM primer, and 30 cycles of 95°C for 10 s, 50°C for 5 s, and 60°C for 4.0 min. Samples were sequenced in both directions, and consensus sequences were obtained from alignment of the forward and reverse sequences.
Analysis of DNA sequences.
Seventy-six DNA sequences obtained from nine asymptomatic HBV carriers, 15 cirrhosis patients, and 52 HCC cases were aligned with genotype E (X75664 and X75657) reference sequences obtained from GenBank (28) using the DNAstar Lasergene program. Nucleotide variations at positions 1762, 1764, and 1896 were compared with the reference sequences.
HBV viral load measurement.
Serum HBV DNA load was measured using a real-time PCR technique as previously described (26).
Statistical analysis.
Statistical analysis was performed using STATA 8.0 (College Station, TX) and SAS 9.1 (SAS Institute, North Carolina). The sequence data were transferred into Excel spreadsheets for analysis of the selected codons. Comparisons between OLA and sequencing were made using two-tailed χ2 analysis, Fisher's exact test, and the Wilcoxon-Mann-Whitney test. Statistical test differences were considered significant if P values were <0.05. Comparisons between viral DNA quantity and dichotomous groups (the three study groups, HBsAg, HBeAg, BCP, and pre-C mutations) were made using the Wilcoxon-Mann-Whitney test.
RESULTS
Performance of OLA.
There was no overlap in the ranges of OD readings for the positive and negative controls. In general, samples that tested negative (<260 DNA copies/ml) in the PCR amplification assay were also negative in the OLA. The lower detection limit of the OLA was derived as follows: DNA was extracted from 100 μl of serum containing 2.4 × 108 copies/ml and eluted into 50 μl buffer, 5.0 μl (equivalent to 10 μl serum) was added into the pre-OLA mixture, and the PCR amplification reaction (total volume of mixture, 25 μl) was performed. A 2.0-μl amount of the PCR mixture was used in the OLA (equivalent to 0.8 μl serum). Six serial dilutions containing between 2.4 × 108 and 2.4 × 103 DNA copies/ml were tested and resulted in a detection limit of 2.4 × 104 copies/ml or 19.2 copies/OLA reaction.
The lower (i.e., more sensitive) detection limit of DNA observed in the quantitative PCR (qPCR) assay compared to OLA could be due to limitations on OLA template preparation, i.e., probe mismatch and nonspecific inhibitors.
Assay validation.
The sensitivity of OLA for detecting minority populations of MT virus was determined by assaying serial dilutions of MT DNA amid a constant amount of WT DNA. The DNA samples used in the mixtures tested positive for MT alone or WT alone in both OLA and DNA sequencing assays and were considered as pure MT or WT. A positive MT sample with a DNA quantity of 4.8 × 108 copies/ml was serially diluted into a solution containing 3.7 × 106 copies/ml WT DNA. By mixing equal volumes of 2.4 × 108 copies/ml of MT DNA and 3.7 × 106 copies/ml of WT DNA, we obtained a mixture of approximately 98% molecules of MT DNA. Additional dilutions were prepared containing 88% (2.4 × 107 copies), 39% (2.4 × 106 copies), 6.1% (2.4 × 105 copies), 0.6% (2.4 × 104 copies), and 0.06% (2.4 × 103 copies) MT DNA in a constant amount of WT DNA (Table 2). The lower detection limit for MT sequence was 2.4 × 104 copies/ml amid 3.7 × 106 copies WT DNA. We also found that WT sequence was detectable in the mixture containing 98% MT sequences as well as in those with lower amounts of MT (Table 2).
TABLE 2.
MT (G1896) and WT (A1896) detection in mixtures of the WT and MT strainsa
| IND sample | Concn of MT in mixture (no. of copies/ml) | % of MT in mixture | Expt no. | OD reading
|
|
|---|---|---|---|---|---|
| MT | WT | ||||
| 1 | MT alone (2.4 × 108) | 100 | 1 | 1.29 | 0.26 |
| 2 | 1.44 | 0.21 | |||
| 2 | WT alone (3.7 × 106) | 0 | 1 | 0.49 | 3.43 |
| 2 | 0.44 | 3.61 | |||
| 3 | 2.4 × 108 | 98 | 1 | 0.98 | 2.02 |
| 2 | 1.01 | 2.10 | |||
| 4 | 2.4 × 107 | 90 | 1 | 1.45 | 3.01 |
| 2 | 1.31 | 3.11 | |||
| 5 | 2.4 × 106 | 40 | 1 | 1.33 | 3.13 |
| 2 | 1.24 | 3.36 | |||
| 6 | 2.4 × 105 | 6.1 | 1 | 1.21 | 3.40 |
| 2 | 1.10 | 3.10 | |||
| 7 | 2.4 × 104 | 0.6 | 1 | 1.02 | 3.45 |
| 2 | 0.98 | 3.34 | |||
| 8 | 2.4 × 103 | 0.06 | 1 | 0.48 | 3.40 |
| 2 | 0.46 | 3.20 | |||
Samples 3 to 8 contain various concentrations of MT sequences diluted in a constant amount of WT (3.7 × 106 copies/ml). Mixtures were tested with both the MT and WT probes. The cutoff values obtained in this assay were 0.53 for MT and 0.56 for WT.
The performance of the assay in detecting MT in samples with a single strain was determined by testing samples of MT DNA diluted in water and compared with detection of MT diluted in WT DNA. The sample was OLA and sequencing positive for MT alone. The limit of the assay in detecting the target MT was 2.4 × 104 copies/ml in the MT/WT mixture and 2.4 × 105 in the MT/water mixture. The reduced sensitivity of the assay when MT was diluted in water compared to MT dilution in WT DNA is possibly due to loss of DNA in very dilute solutions, or perhaps the presence of WT contributes to the lower limit of detection and in the absence of WT the limit of detection increases by a log.
Comparison of OLA with direct DNA sequencing.
One hundred one HBsAg-positive subjects were selected for DNA sequencing; 25 had undetectable virus (<260 DNA copies/ml) in the qPCR assay; they failed to generate PCR products and were therefore not tested for OLA. DNA samples from the remaining 76 subjects (52 HCC patients, 15 cirrhosis patients, and nine asymptomatic HBsAg carriers) were tested for BCP mutations by both OLA and direct DNA sequencing of the PCR fragment. The sensitivity of OLA in detecting the A1896, T1762, and A1764 MTs was 94.2%, 89.7%, and 89.5%, respectively, and the specificity of the assay was 97.3%, 88.2%, and 90.4% in detecting the A1896, T1762, and A1764 MTs, respectively (Table 3).
TABLE 3.
Comparison of OLA and sequencing methods for detecting MTs
| OLA (nucleotide position) and result type | No. of results found by sequencing
|
|||
|---|---|---|---|---|
| WT | MT | IND | Total | |
| 1896 | ||||
| WT | 37 | 1 | 1a | 39 |
| MT | 2 | 33 | 35 | |
| IND | 1b | 1 | ||
| Total | 76 | |||
| 1762 | ||||
| WT | 15 | 2 | 5a | 22 |
| MT | 5 | 44 | 49 | |
| IND | 5b | 5 | ||
| Total | 76 | |||
| 1764 | ||||
| WT | 19 | 2 | 1a | 22 |
| MT | 5 | 43 | 48 | |
| IND | 6b | 6 | ||
| Total | 76 | |||
Sequencing assay.
OLA.
Samples that had mixed strains and tested positive for both WT and MT in the OLA were considered indeterminate (IND), while in the DNA sequencing assay IND was defined as the presence of a nucleotide other than WT or MT in the target region.
Three (3.9%) out of the 76 samples were IND by sequencing, and seven (9.2%) were IND in the OLA (Table 4). Thus, the OLA detected a significantly higher proportion of IND samples than did sequencing (P < 0.05). Similarly there was a higher proportion of BCP IND than of pre-C IND samples. This was true for both DNA sequencing and OLA, i.e., five (6.5%) BCP IND samples compared to no pre-C IND samples in the DNA sequencing assay and 6.5% (5/76) BCP IND samples compared to 2.6% (2/76) pre-C IND samples in the OLA. Three OLA IND samples (samples 1, 2, and 10), although they had viral loads above the limit of detection, tested positive for MT alone or WT alone in the DNA sequencing assay. The discrepancy between DNA sequencing and OLA in these samples could be due to infection with mixed virus strains and suggests reduction in the sensitivity of the DNA sequencing assay in detecting minor virus sequences in mixed infections. However, another two OLA IND samples (T1762/G1764 and T1762/T1764) were confirmed by DNA sequencing (Table 4).
TABLE 4.
Samples with IND or negative OLA results, their corresponding viral loads, nucleotide sequences in the target regions, and sequence changes in one, two, or three bases from the target regionsa
| OLA and expt no. | Result type | Viral load (no. of copies per ml) | DNA sequence | Change(s) in one, two, or three bases from target |
|---|---|---|---|---|
| Pre-C | ||||
| 1 | IND | 1.5 × 107 | MT | None |
| 2 | IND | 3.6 × 104 | MT | None |
| 3 | Negativeb | 2.8 × 102 | WT | G1899A |
| 4 | Negative | 3.3 × 102 | MT | G1899A |
| 5 | Negative | 5.9 × 102 | WT | |
| 6 | Negative | 3.7 × 102 | MT | |
| BCP | ||||
| 7 | IND | 1.2 × 105 | T1762T/G1764 | None |
| 8 | IND | 6.5 × 103 | WT | None |
| 9 | IND | 4.3 × 105 | MT | T1768A |
| 10 | IND | 3.6 × 104 | WT | None |
| 11 | IND | 1.0 × 105 | T1762/T1764 | None |
| 12 | Negative | 1.2 × 107 | A1762/T1764 | None |
| 13 | Negative | 2.5 × 105 | WT | G1763A, C1766G |
| 14 | Negative | 1.7 × 107 | WT | T1765G, C1766T |
| 15 | Negative | 7.0 × 102 | WT | None |
Samples 3 to 6 and 15 had viral loads of <103 DNA copies/ml, which is lower than the detectability limit of the OLA. The OLA detected mixtures of WT and MT in samples 1, 2, 8, 9, and 10, while consensus sequencing detected the dominant strain. Sequences in positions 1762 and 1764 are discordant in sample 7, with the presence of MT in position 1762 and WT in 1764. Samples 13 and 14 are negative in the OLA because of the nucleotide changes in one, two, or three bases from the target region, although consensus sequencing revealed WT sequences. Sample 11 was OLA positive for MT 1762 and WT 1764; the discordant result was due to a mixture of MT T1762 and T1764. The two polymorphisms are different from the WT or MT sequences. Nucleotide 1762 or 1764 in sample 7 or 12 is neither WT nor MT, hence the IND/negative result in the OLA.
“Negative” represents samples with no visible color change in either the MT or the WT assay.
Eight of the 76 sequences are OLA negative but PCR positive (>260 DNA copies/ml). Three of them had base changes located at one to four bases from the ligation sites, which are expected to prevent binding and ligation. An additional OLA-negative sample had a DNA concentration of 1.2 × 107 copies but had a change other than the specific mutation (1764T) in the target region which would disrupt OLA primer binding (Table 4).
Application of OLA.
Blood samples collected from 227 HBsAg carriers (138 HCC patients, 49 cirrhosis patients, and 40 asymptomatic HBsAg carriers) were tested for the pre-C and BCP mutations by OLA. Details of the subject recruitment are described above, with additional details being previously published (18). Forty-four samples (15.8%) had undetectable DNA (≤260 copies/ml) in the qPCR assay, while 183 and 178 samples gave valid results for pre-C or BCP mutation, respectively (Table 5). Five samples were not tested for BCP.
TABLE 5.
Viral polymorphisms in liver disease patients and asymptomatic HBsAg carriersa
| Subject group (n) | Result for OLA
|
|||||
|---|---|---|---|---|---|---|
| 1896
|
1762/1764
|
|||||
| No. (%) of subjects
|
P | No. (%) of subjects
|
P | |||
| WT | Pre-C MT | WT | BCP MT | |||
| Asymptomatic HBsAg carriers (21) | 11 (52.3) | 10 (47.6) | >0.5 | 11 (52.3) | 10 (47.6) | >0.5 |
| HCC patients (119 for OLA 1896, 114 for OLA 1762/1764) | 75 (63.0) | 44 (36.9) | 0.356 | 28 (24.6) | 86 (75.4) | 0.01 |
| Cirrhosis patients (43) | 31 (72.1) | 12 (27.9) | 0.119 | 10 (23.2) | 33 (76.7) | 0.02 |
| Total (183 for OLA 1896, 178 for OLA 1762/1764) | 117 | 66 | 49 | 129 | ||
Two hundred twenty-seven subjects were originally included in the study; 44 had undetectable viral DNA (<260 DNA copies/ml) and tested negative in the OLA. Data from the remaining 183 are presented here. The prevalences of pre-C and BCP mutations in patients with liver diseases, i.e., HCC and cirrhosis, were 34.5% and 75.7%, respectively (P < 0.01).
In the OLA pre-C mutation assay, 36.5.1% (66/183) and 63.9.4% (117/183) tested positive for MT and WT, respectively. Thirteen (7.1%) individuals had IND results, while 31 (16.9%) failed genotyping; they were OLA negative (i.e., the color representing neither WT nor MT failed to appear). Twenty-six of these 31 (14.2%) had DNA values lower than the OLA detection limit (>260 to ≤2.4 × 104 DNA copies/ml) but higher than the PCR detection limit. Another five (2.7%) OLA-negative samples were positive by PCR with HBV DNA levels of >2.4 × 104 to 1.7 × 107 copies/ml.
The pre-C mutation was not associated with disease; the variant is detected in 36.9% (44/119) of HCC patients, 27.9% (12/43) of cirrhosis patients, and 47.68% (10/21) of asymptomatic HBsAg carriers (P = 0.241, Table 5).
In the OLA BCP assay, 72.4% (129/178) and 27.5% (49/178) of the subjects tested positive for MT and WT, respectively (Table 5). A significantly (P = 0.01 for HCC and P = 0.02 for cirrhosis) higher proportion of individuals with HBV-related liver disease than of asymptomatic carriers had the BCP mutation. BCP mutations were detected in 75.4% (86/114) of HCC subjects, 76.7% (33/43) of cirrhotic subjects, and 47.6% (10/21) of asymptomatic HBsAg carriers (Table 5).
DISCUSSION
We have developed a simple OLA-based point mutation assay to detect pre-C and BCP mutations in chronic HBV carriers. Having optimized and validated the OLA assay, we used it to study sera from a well-characterized HCC case control study. The assay, which included DNA extraction from serum PCR amplification of a region of the HBV core gene, ligation of oligonucleotides, and color development, can be readily performed within 1 working day. The OLA allows large numbers of samples to be handled with ease and speed and thus has the potential to be used for rapid screening.
Apart from rapidity and economy, another advantage of the OLA method in genotyping point mutations compared to other genotypic approaches and DNA sequencing is the high ease of interpretation of ligation assays; the assay yields clearly positive or negative outcomes that are easy to interpret visually or that can be interpreted by a spectrophotometer and computer program with samples on microtiter plates.
The reproducibility of the assay is very good as demonstrated by the consistent results obtained from 30 WT, MT, or IND samples that were tested on two (25 samples) or three (5 samples) separate occasions (data not shown). The assay can detect MT DNA with a lower detection limit of 19.2 copies per reaction. OLA has the important feature of being able to detect a low amount of MT DNA (2.4 × 104 copies/ml) amid a high amount of WT (3.7 × 106 copies/ml). Thus, MT sequences can be detected when they constitute only 0.6% of the total HBV sequence concentration, improving the screening potential of the assay, as it will facilitate early detection of MT HBV genomes that may presage HCC development.
As expected, due to the highly specific nature of OLA, the assay produced inconclusive or negative results when tested against nucleotide sequences other than the WT or the specific MT form. The specificity of the assay was reduced when it was used with samples bearing additional changes in nucleotide positions located one to four bases from the target site. For example, DNA sequencing of two OLA-negative samples revealed 1762T/1764T and 1762T/1762G sequences. This is potentially a disadvantage; however, once a study population has been characterized, a panel of oligonucleotides covering all possible mutations can be readily prepared.
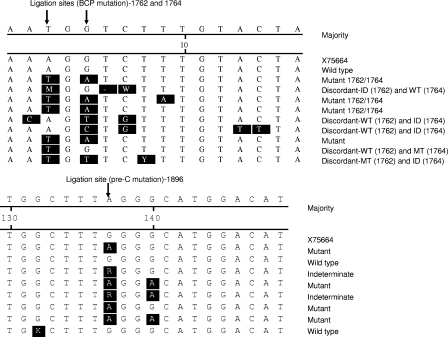
The discordant results between DNA sequencing and OLA in the samples that had mismatches of nucleotides 1762 and 1764 but were clearly OLA positive for MT alone or WT alone likely reflect the higher sensitivity of OLA in detecting specific point mutations (Fig. 1). Although sequencing is the gold standard that was used in this study, it has limitations in detection of minority sequences. We found that where a sequence was clearly pure WT or MT, OLA did not disagree with the gold standard sequencing but in the case of minor MT or WT populations OLA was able to detect these when sequencing was unable. Although the testing of artificial mixtures of WT and MT (Table 2) shows some evidence that OLA is superior to sequencing in detecting minor populations, we plan to perform sequencing and OLA on multiple clones from samples with mixed populations that are detectable by OLA but not by sequencing to further develop this assay.
FIG. 1.
Alignment of samples with WT, MT, discordant, and IND results. Nucleotide differences from X75664 (reference genotype E sequence obtained from GenBank) are shaded in black.
It has been previously shown that although the identification of novel mutations is more readily obtained by consensus sequencing, OLA is more sensitive for the detection of specific variants within a mixed virus population in drug resistance mutation studies of HIV-1 infection (13). Another important advantage of the OLA method is that it can detect both WT and MT sequences in a population of mixed strains (Table 2) (17). We detected 0.6% of MT amid WT sequences, unlike consensus sequencing, which will detect only the predominant virus in a mixed infection.
OLA samples reactive in both WT and MT OLAs are classified as IND, and further investigation including sequencing of clones will be needed to confirm mixed populations and to fully correctly characterize all viruses present.
Some samples failed to provide a clear OLA signal (i.e., no WT or MT signal resulting in negative results). Predominantly, this occurred in cases of insufficient target DNA arising from low copy numbers in samples, suggesting that samples with very low input copy numbers can affect the accuracy of OLA. We previously observed that a DNA quantity of >261 copies per ml can be measured in the samples by qPCR (26). However, the OLA was unable to genotype 16.9% (31/183) of PCR-positive samples despite having target DNA concentrations within the detection range. These false-negative results are most likely to be caused by probe-target mismatch in samples with low copy numbers of HBV DNA. This may be a limitation to our assay compared to DNA sequencing and alternative methods using specific primers in real-time PCR quantification (30). False-negative results could also be due to insertions, deletions, or duplications in the target region (19, 21).
The lower sensitivity could not be explained by lower sample viral load since a number of untypeable samples had viral loads of up to 107 copies/ml.
As an important demonstration of the utility of this new method, the clinical significance of the presence of HBV pre-C and BCP mutations was assessed in samples for our previous case control study.
We found that patients with liver diseases (cirrhosis or HCC) were significantly more likely to have the BCP mutation than were asymptomatic carriers.
The association of BCP mutations with HCC obtained in our study compared well with results from a study in Taiwan conducted on 200 HCC patients and 160 infected chronic HBV asymptomatic HBsAg carriers (24). The frequency of the BCP mutation in the Taiwanese study was 76.9% in patients with HCC and 14.4% in chronic carriers with a statistically significant odds ratio for the BCP mutation (P < 0.001). The conclusion from that study was that of the factors examined (age, sex, HBV genotype, viral load, and pre-C mutations) the BCP mutation is the strongest independent predictor for the development of HCC. Our data also demonstrate a strong association of the BCP mutation with HBV-associated liver disease. The higher overall proportion of asymptomatic carriers with BCP mutations in the Gambian population assessed herein than in the Taiwanese population could be related to different HBV genotypes circulating in the two countries. In The Gambia genotype E is predominant at 87.3% with genotype A occurring at 12.7% (27). The importance of genotypic differences is highlighted in the Taiwanese study, where genotype C was associated with a significantly higher incidence of BCP mutations than was genotype B. It is still unclear whether the pre-C mutation has any pathogenic role for more severe disease, although it was initially thought to cause more severe liver disease because it was found in patients with active liver diseases or fulminant hepatitis (29). However, in our study, the pre-C mutation was not associated with advanced HBV-related liver disease, consistent with results from several prior studies (14).
AFP, which is a widely used marker for HCC detection because of its ease of use and low cost, suffers from low specificity and sensitivity, being elevated in diseases other than liver cancer and absent in a proportion of confirmed HCC cases (32, 34, 36). Further studies are needed to determine the role of molecular biomarkers such as BCP mutations and HBV genotypes. These markers can be used in combination with other biomarkers as a reliable screening tool with HBV chronic carriers at risk of developing HCC.
In conclusion, we have developed a simple, economical, and reliable assay for detecting HBV core mutations. This point mutation assay may be extremely useful as a tool for screening blood samples from large numbers of HBV carriers for the presence of HBV pre-C and BCP mutations. Based on our and others' findings demonstrating an association between BCP mutations and liver cancer or cirrhosis, this assay may have important clinical applications as a screening tool in assessing an individual's risk for HCC and also could prove an invaluable tool for conducting epidemiology studies in areas of HBV endemicity with the highest HBV burden of liver disease but with limited resources to carry out expensive and sophisticated laboratory tests.
Acknowledgments
Sincere thanks go to Fumi Lesi for clinical support in recruiting HCC patients and asymptomatic HBsAg carriers, Alasana Bah for laboratory assistance, and Yusupha Bah and Ebrima Bah for invaluable assistance in the collection of field and cancer registry data.
This work was supported by the Medical Research Council (United Kingdom); the International Agency for Research on Cancer, Lyon, France; and the National Cancer Institute, NIH, DHHS, Bethesda, MD.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Bah, E., D. M. Parkin, A. J. Hall, et al. 2001. Cancer in the Gambia: 1988-97. Br. J. Cancer 841207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballard, A. L., and E. H. Boxall. 1997. Colourimetric point mutation assay: for detection of precore mutants of hepatitis B. J. Virol. Methods 67143-152. [DOI] [PubMed] [Google Scholar]
- 3.Baptista, M., A. Kramvis, and M. C. Kew. 1999. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29946-953. [DOI] [PubMed] [Google Scholar]
- 4.Beasley, R. P. 1988. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 611942-1956. [DOI] [PubMed] [Google Scholar]
- 5.Beck, I. A., M. Mahalanabis, G. Pepper, et al. 2002. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J. Clin. Microbiol. 401413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Block, T. M., A. S. Mehta, C. J. Fimmel, et al. 2003. Molecular viral oncology of hepatocellular carcinoma. Oncogene 225093-5107. [DOI] [PubMed] [Google Scholar]
- 7.Brunetto, M. R., U. A. Rodriguez, and F. Bonino. 1999. Hepatitis B virus mutants. Intervirology 4269-80. [DOI] [PubMed] [Google Scholar]
- 8.Buckwold, V. E., Z. Xu, M. Chen, et al. 1996. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 705845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron-Wilson, C. L., P. Muir, A. L. Ballard, et al. 2003. Evaluation of a line probe assay for identification of hepatitis B virus precore variants in serum from chronic hepatitis B carriers. J. Virol. Methods 11497-103. [DOI] [PubMed] [Google Scholar]
- 10.Carman, W. F., M. R. Jacyna, S. Hadziyannis, et al. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii588-591. [DOI] [PubMed] [Google Scholar]
- 11.Dumpis, U., M. Mendy, A. Hill, et al. 2001. Prevalence of HBV core promoter/precore/core mutations in Gambian chronic carriers. J. Med. Virol. 65664-670. [DOI] [PubMed] [Google Scholar]
- 12.Edelstein, R. E., D. A. Nickerson, V. O. Tobe, et al. 1998. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J. Clin. Microbiol. 36569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, G. M., M. Mahalanabis, I. A. Beck, et al. 2004. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J. Clin. Microbiol. 423670-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang, Z. L., J. Yang, X. Ge, et al. 2002. Core promoter mutations (A(1762)T and G(1764)A) and viral genotype in chronic hepatitis B and hepatocellular carcinoma in Guangxi, China. J. Med. Virol. 6833-40. [DOI] [PubMed] [Google Scholar]
- 15.Grandjacques, C., P. Pradat, L. Stuyver, et al. 2000. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J. Hepatol. 33430-439. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins, A. E., R. J. Gilson, S. V. Beath, et al. 1994. Novel application of a point mutation assay: evidence for transmission of hepatitis B viruses with precore mutations and their detection in infants with fulminant hepatitis B. J. Med. Virol. 4413-21. [DOI] [PubMed] [Google Scholar]
- 17.Jallow, S., S. Kaye, M. Schutten, et al. 2007. Development and evaluation of an oligonucleotide ligation assay for detection of drug resistance-associated mutations in the human immunodeficiency virus type 2 pol gene. J. Clin. Microbiol. 451565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirk, G. D., O. A. Lesi, M. Mendy, et al. 2004. The Gambia liver cancer study: infection with hepatitis B and C and the risk of hepatocellular carcinoma in West Africa. Hepatology 39211-219. [DOI] [PubMed] [Google Scholar]
- 19.Kramvis, A., and M. C. Kew. 1999. The core promoter of hepatitis B virus. J. Viral Hepat. 6415-427. [DOI] [PubMed] [Google Scholar]
- 20.Laskus, T., M. Radkowski, M. Nowicki, et al. 1998. Association between hepatitis B virus core promoter rearrangements and hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 244812-814. [DOI] [PubMed] [Google Scholar]
- 21.Laskus, T., J. Rakela, M. J. Nowicki, et al. 1995. Hepatitis B virus core promoter sequence analysis in fulminant and chronic hepatitis B. Gastroenterology 1091618-1623. [DOI] [PubMed] [Google Scholar]
- 22.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 3371733-1745. [DOI] [PubMed] [Google Scholar]
- 23.Lin, D. Y., I. S. Sheen, C. T. Chiu, et al. 1993. Ultrasonographic changes of early liver cirrhosis in chronic hepatitis B: a longitudinal study. J. Clin. Ultrasound 21303-308. [DOI] [PubMed] [Google Scholar]
- 24.Liu, C. J., B. F. Chen, P. J. Chen, et al. 2006. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J. Infect. Dis. 1931258-1265. [DOI] [PubMed] [Google Scholar]
- 25.Locarnini, S. 2004. Molecular virology of hepatitis B virus. Semin. Liver Dis. 24(Suppl. 1)3-10. [DOI] [PubMed] [Google Scholar]
- 26.Mendy, M. E., S. Kaye, M. van der Sande, et al. 2006. Application of real-time PCR to quantify hepatitis B virus DNA in chronic carriers in The Gambia. Virol. J. 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriyama, K., H. Okamoto, F. Tsuda, et al. 1996. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology 226269-280. [DOI] [PubMed] [Google Scholar]
- 28.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198489-503. [DOI] [PubMed] [Google Scholar]
- 29.Omata, M., T. Ehata, O. Yokosuka, et al. 1991. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N. Engl. J. Med. 3241699-1704. [DOI] [PubMed] [Google Scholar]
- 30.Pang, A., M. F. Yuen, H. J. Yuan, et al. 2004. Real-time quantification of hepatitis B virus core-promoter and pre-core mutants during hepatitis E antigen seroconversion. J. Hepatol. 401008-1017. [DOI] [PubMed] [Google Scholar]
- 31.Parkin, D. M., F. Bray, J. Ferlay, et al. 2001. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer 94153-156. [DOI] [PubMed] [Google Scholar]
- 32.Soresi, M., C. Magliarisi, P. Campagna, et al. 2003. Usefulness of alpha-fetoprotein in the diagnosis of hepatocellular carcinoma. Anticancer Res. 231747-1753. [PubMed] [Google Scholar]
- 33.Tobe, V. O., S. L. Taylor, and D. A. Nickerson. 1996. Single-well genotyping of diallelic sequence variations by a two-color ELISA-based oligonucleotide ligation assay. Nucleic Acids Res. 243728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong, M. J., L. M. Blatt, and V. W. Kao. 2001. Surveillance for hepatocellular carcinoma in patients with chronic viral hepatitis in the United States of America. J. Gastroenterol. Hepatol. 16553-559. [DOI] [PubMed] [Google Scholar]
- 35.Whittle, H. C., H. Inskip, A. K. Bradley, et al. 1990. The pattern of childhood hepatitis B infection in two Gambian villages. J. Infect. Dis. 1611112-1115. [DOI] [PubMed] [Google Scholar]
- 36.Wong, I. H., W. Y. Lau, T. Leung, et al. 2000. Quantitative comparison of alpha-fetoprotein and albumin mRNA levels in hepatocellular carcinoma/adenoma, non-tumor liver and blood: implications in cancer detection and monitoring. Cancer Lett. 156141-149. [DOI] [PubMed] [Google Scholar]
- 37.Yen, T. S. 1996. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 320-30. [DOI] [PubMed] [Google Scholar]
- 38.Yim, H. J., and A. S. Lok. 2006. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology 43S173-S181. [DOI] [PubMed] [Google Scholar]