Abstract
Human herpesvirus 6 (HHV-6) is a ubiquitous virus with which infections have been associated with pathologies ranging from delayed bone marrow engraftment to a variety of neurological diseases. The lack of a standardized assay that can be used to detect and estimate HHV-6 DNA contents in various clinical specimens can lead and has led to discordant results among investigators and on the potential association of HHV-6 to diseases. To identify the most reliable and sensitive assays, an identical set of 11 coded serum samples spiked with various quantities of the HHV-6A variant (range, 4 to 400,000 genome copies/ml) was sent to eight independent laboratories around the world. Each laboratory was asked to estimate the HHV-6 DNA content by use of its own protocols and assays. Among the various assays, three TaqMan-based real-time PCR assays yielded quantities that were closest to the quantity of HHV-6 that had been spiked. To provide better homogeneity between the results from the different laboratories working on HHV-6, we propose that investigators interested in quantifying HHV-6 in clinical samples adopt one of these assays.
Human herpesvirus 6 (HHV-6) was isolated more than 20 years ago from patients with AIDS and various lymphoproliferative disorders (41). HHV-6 has a worldwide distribution, and by the age of 3 years, most individuals have contracted HHV-6 (37, 56). Two distinct HHV-6 variants exist, HHV-6A and HHV-6B (1). With the exception of a few genes or regions, the coding sequences of HHV-6A and HHV-6B are >90% identical (14, 27). Despite this high degree of similitude in DNA content, these two variants have distinct biological properties, such as their ability to be propagated on different cell lines and their reactivity with monoclonal antibodies, and their association with specific pathological conditions is such that these viruses are considered distinct pathogens (2). The most common clinically defined disease associated with HHV-6 is the common childhood illness roseola, also referred to as exanthem subitum (51). Nearly all (99%) episodes of roseola are linked to HHV-6B infection. Other complications of primary HHV-6B infection include hepatitis, meningitis, fatal hemophagocytic syndrome, and fatal disseminated infection (7, 22, 23, 39). Furthermore, HHV-6 reactivation occurs in 40 to 50% of bone marrow and solid-organ transplant recipients, making this virus a growing medical concern (6, 11, 19, 36, 53-55). HHV-6B reactivation is most common after allogeneic hematopoietic stem cell transplantation and is associated with subsequent delayed monocyte and platelet engraftment, increased platelet transfusion requirements, all-cause mortality, graft-versus-host disease, and central nervous system dysfunction (58). The reactivation of HHV-6 (A and B variants) is also observed in various other neurological diseases. such as multiple sclerosis and epilepsy (9, 15, 17, 43, 48, 52). Although the detection of HHV-6A is less frequent than that of HHV-6B in patients, the former variant is believed to act as a pathogen in the context of severe central nervous system and systemic infections (8, 40). In addition, concomitant infections with both HHV-6A and HHV-6B have been described in lung and lymphoid tissue samples (12, 16).
The difficulty in linking HHV-6 infection in the case of either primary infection or reactivation to disease resides partly in the lack of a standardized assay that can be used to detect and quantify HHV-6 DNA in biological samples. The use of different assays to monitor HHV-6 DNA is one potential source for discordant results. At present, the detection and the quantitation of HHV-6 DNA in biological fluids, such as serum, plasma, and cerebrospinal fluid, are considered markers of active HHV-6 infection (an exception is for chromosomally integrated HHV-6; see Discussion). Several types of PCR assays are currently used to detect HHV-6 DNA in biological fluids, but a comparison of the various assays used has never been performed. The sensitivity of the assay is a very important issue, considering that HHV-6 generally remains cell associated and the detection of cell-free HHV-6 in biological samples can be clinically meaningful (17, 35, 47). The purpose of this study was to identify the most reliable and most sensitive PCR assays for the quantitative detection of HHV-6 in human serum. To do so, identical and coded sets of HHV-6A-spiked serum samples were sent to various independent investigators throughout the world for testing by each laboratory's specific methodology.
MATERIALS AND METHODS
Study design.
Sucrose gradient-purified HHV-6A (strain GS) (1010 viral particles/ml, as determined by electron microscopy) was purchased from Advanced Biotechnologies Inc. (Columbia, MD). Human serum negative for HHV-6 DNA (as determined by TaqMan PCR) was spiked with various quantities of infectious HHV-6A virions (0 to 400,000 genome equivalents) per ml. After the serum and HHV-6A were mixed, 250-μl aliquots were prepared, frozen at −80°C, and shipped on dry ice to eight laboratories for testing. Each laboratory received 11 coded samples. All samples were assayed in a blinded fashion, and the participating laboratories were informed of their performance once all laboratories had turned in their results. Of the nine assays tested, three (assays 3, 5, and 7) yielded qualitative results and six yielded quantitative results. Brief methodologies for each of these assays are provided below.
Assay 1.
To remove cells and debris, the clinical specimens (0.2 to 1 ml) were centrifuged twice (first at 900 × g for 15 min at 4°C and then at 1,500 × g for 15 min at 4°C). Precleared samples were then subjected to high-speed centrifugation (26,000 × g, 120 min, 4°C) in order to concentrate the viral particles. Synthetic calibrator plasmid DNA (104 copies of calibrator per sample) was added prior to DNA extraction to control the efficiency of each step of the analytical procedure. Extraction of DNA from biological fluids was performed with 0.5 ml of a lysis buffer containing 10 mM Tris-HCl (pH 8), 5 mM EDTA, 0.5% (vol/vol) sodium dodecyl sulfate, and 0.1 μg of proteinase K per ml. Lysis was followed by phenol-chloroform extraction and high-salt-concentration isopropanol precipitation; the purified DNA was resuspended in 100 μl of a 5 mM Tris-HCl-0.5 mM EDTA buffer and was stored at 20°C until use. Ten microliters of the purified material was tested in each PCR in triplicate to measure both the calibrator and the HHV-6 copy numbers. Primers that amplify a fragment of the highly conserved U67 open reading frame of HHV-6 are listed in Table 1. The TaqMan reaction was performed in a final volume of 25 μl containing 100 μM each dATP, dCTP, and dGTP; 200 μM dUTP; 5.5 mM magnesium chloride; TaqMan buffer A (1×); the primers at 300 nM; the probe at 200 nM; 0.625 U of AmpliTaq Gold; 0.25 U of uracil-N-glycosylase; and 10 μl of DNA template. The TaqMan PCR cycling conditions were 2 min at 50°C, followed by 15 min of denaturation at 95°C and 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 60 s. A standard curve was drawn by using serial dilutions of known copy numbers of plasmid DNA containing a portion of the U67 gene.
TABLE 1.
Primers and probes used in the various assaysa
| Assay | Target | Primer | Primer | Probe (variant specific) | Reference |
|---|---|---|---|---|---|
| 1 | orf67 | 5′-CGAAACGCCTACACAGAAT-3′ | 5′-CAAAGCCAAATTATCCAGAGCG-3′ | 5′-CGTCACACCCGAAGGAAT-3′ | 29 |
| 2 | orf67 | 5′-CAAAGCCAAATTATCCAGAGCG-3′ | 5′-CGCTAGGTTGAGGATGATCGA-3′ | 5′-CCCGAAGGAATAACGCTC-3′ | 25 |
| 3 | orf57 | External: 5′-GCGTTTTCAGTGTGTAGTTCGGCAG-3′ | External: 5′-GGCCGCATTCGTACAGATACGGAGG-3′ | NA | 42 |
| Internal: 5′-GCTAGAACGTATTTGCTGCAGAACG-3′ | Internal: 5′-ATCCGAAACAACTGTCTGACTGGCA-3′ | NA | |||
| 4 | orf31 | 5′-TTTGCAGTCATCACGATCGG-3′ | 5′-AGAGCGACAAATTGGAGGTTTC-3′ | 5′-AGCCACAGCAGCCATCTACATCTGTCAA-3′ | 44 |
| 5 | orf31 | BIP-5′-ACGAAGAACGCATCACAGAGCT ACTGCGTATTCTTTGGAGAGC-3′ | FIP-5′-TCCGTAGACGGAGTGGCTGATAGTATCACGTTCCTCACGAAAA-3′ | NA | 26 |
| B3-5′-GCATCTTGTGACGAGAGAAGA-3′ | F3-5′-CAGAGATCGACCAAATGCAAAA-3′ | NA | |||
| LPB-5′-GAGACGAACAAAATAAAAGAAC-3′ | LPF-5′-TATATTTATTAGTGGAGGTAGC-3′ | NA | |||
| 6 | orf65/66 | 5′-GACAATCACATGCCTGGATAATG-3′ | 5′-TGTAAGCGTGTGGTAATGGACTAA-3′ | 5′-AGCAGCTGGCGAAAAGTGCTGTGC-3′ | 18 |
| 7 | orf89 | 5′-ATAAATTTGATGGGTTAGTGAAAAAG-3′ | 5′-GTCAGGATTGGACATCTCTTTGT-3′ | 5′-Biotin-CATGTTGATGATGATGGCACAC-3′ | 46 |
| 5′-Biotin-CTCATTGTTGTTGATGGCACAC-3′ |
Abbreviations: BIP, backward inner primer; FIP, forward inner primer; LPB, loop primer backward; LPF, loop primer forward; NA, not applicable.
Assay 2.
DNA was extracted from serum samples by using Qiagen columns (QIAamp DNA blood mini kit), according to the manufacturer's guidelines. After elution with Tris-EDTA buffer (10 mM Tris-HCl, 0.5 mM EDTA, pH 9.0), the DNA was aliquoted to avoid repeated freezing and thawing and was stored at −80°C. Two negative controls, which consisted of reagents only, were processed with each set of 10 samples in order to evaluate the samples for a possible cross-reaction. Each sample was confirmed to be negative for beta-globin by PCR amplification with specific primers to ensure no cellular contamination. The quantitative real-time PCR was performed in a Rotor-Gene 3000 real-time cycler (Corbett Research). In this study, primers and probes that amplify and detect a 115-bp fragment located in the U67 gene were used (Table 1). The reaction mixture for each PCR test contained 12.5 μl of 2× SuperHot PCR master mix (Bioron GmbH, Germany), 0.4 μM of each HHV-6-specific primer, 0.2 μM of the HHV-6-specific probe, and 10 μl of previously extracted DNA in a final volume of 25 μl. Each sample was analyzed in duplicate. The cycling conditions were as follows: after a preincubation at 95°C for 10 min to activate the DNA polymerase, two-step thermocycling was performed for 50 cycles at 95°C for 15 s and 58°C for 60 s. To exclude the possibility of contamination during the PCR, one negative control consisting of all reagents except the sample DNA was amplified for every five samples in each experiment. For the final calculation of the DNA copy number, we generated a standard curve of known amounts (5, 50, 500, and 5,000 copies) of DNA isolated and quantified from HHV-6B (Z-29) virions (Advanced Biotechnologies Inc.). Each point was obtained in triplicate.
Assay 3.
The QIAamp blood kit was used to extract DNA from plasma samples according to the manufacturer's guidelines (Qiagen). DNA amplification was performed with nested primers specific for a highly conserved sequence corresponding to the major capsid protein gene of HHV-6 (Table 1) (4, 42). The external primers amplified a 520-bp sequence, and the internal primers amplified a 258-bp sequence. PCR was performed with the Taq PCR master mix kit (Qiagen), according to the manufacturer's instructions. DNA was amplified with a 0.5 μM final primer concentration for 35 cycles by using the following conditions: denaturation at 92°C for 0.3 min, annealing at 55°C for 0.3 min, and extension at 72°C for 0.32 min. A total of 5 μl of the primary PCR product was amplified with the internal primers under the same PCR conditions. A total of 10 μl of PCR product was subjected to electrophoresis on a 1.5% agarose gel and was visualized by ethidium bromide staining.
Assay 4.
Viral DNA from plasma was extracted with a QIAamp blood kit, according to the manufacturer's recommendations (Qiagen, Chatsworth, CA). The PCR was performed with a TaqMan PCR kit (PE Applied Biosystems, Foster City, CA), as described previously (28). Briefly, the DNA extracted from plasma was added to a PCR mixture containing 10 mM Tris (pH 8.3); 50 mM KCl; 10 mM EDTA; 5 mM MgCl2; 100 μM each dATP, dCTP, dGTP, and dTTP; 0.2 μM each primer; 0.1 μM fluorogenic probe; and 1.25 U of AmpliTaq Gold (PE Applied Biosystems). Following activation of the AmpliTaq Gold for 10 min at 95°C, 45 cycles of 15 s at 95°C and 1 min at 62°C were carried out on a model 7700 sequence detector (PE Applied Biosystems). A standard graph was constructed by using the threshold cycle (CT) values obtained from serially diluted HHV-6 PSTY05 plasmid, which contained the U31 gene (50). The copy number was expressed per ml.
Assay 5.
The loop-mediated isothermal amplification (LAMP) reaction was carried out with a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd., Tokyo, Japan), according to the manufacturer's instructions. The primers used for the HHV-6 LAMP reaction are listed in Table 1. The LAMP reaction was carried out with 25 μl of a mixture containing 2.4 μM primers H6U31FIP and H6U31BIP, 0.4 μM each outer primer (primers H6U31F3 and H6U31B3), 1.2 μM each loop primer (primers H6U31LPF and H6U31LPB), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2SO4, 0.8 M betaine, 1.4 mM each deoxynucleoside triphosphate, and 0.1% Tween 20. The mixture was heated to 96°C for 30 s, after which 1 μl (8 U) of Bst DNA polymerase was added. The mixture was incubated at 63°C for 60 min, and increased turbidity was monitored by using an LA-200 apparatus (Teramecs Co., Ltd., Kyoto, Japan) (33). The cutoff value for discrimination between positive and negative samples was 0.1, which was determined by analyzing five virus-negative samples.
Assay 6.
DNA extraction from samples (200 μl) was carried out with a QIAamp DNA mini kit (Qiagen, Tokyo, Japan), according to the manufacturer's instructions. Twenty micrograms of glycogen was added to each sample as a carrier DNA. The elution was performed in 50 μl of elution buffer. The HHV-6 DNA load was measured by a real-time PCR assay, as reported previously (18). Briefly, real-time PCR was carried out in an automated Sequence Detector 7700 system (PE Applied Biosystems). The 50-μl reaction mixture contained 25 μl of TaqMan PCR master mix, 200 nM each primer, and 100 nM probe specific for HHV-6 (Table 1). After 2 min at 50°C and 10 min at 95°C, samples were submitted to 45 cycles. Each cycle consisted of a step at 95°C for 15 s, followed by a step at 60°C for 1 min. The CT value was defined by the first cycle number at which the fluorescence was greater than the threshold. The threshold of quantitation was 25 copies/ml of serum. The HHV-6 variant identity was confirmed by using a variant-specific real-time PCR assay, as described previously (8).
Assay 7.
Nucleic acids were extracted from plasma by using an Easy MAG platform (bioMérieux, Durham, NC), according to the instructions of the manufacturer. Total extracted nucleic acids were eluted into 55 μl of water. A PCR-enzyme immunoassay (EIA) was performed to detect HHV-6 DNA by a procedure published previously (46). In brief, a 50-μl PCR mixture contained the following: 1× buffer; 1.5 mM MgCl2; 10% glycerol; 200 μM each dATP, dCTP, and dGTP; 100 μM dTTP; 90 μM dUTP; 10 μM digoxigenin-11-dUTP (Roche Biochemicals, Indianapolis, IN); 1 μM each primer; 0.01 units/μl uracil-N-glycosylase (Epicenter Technologies, Madison, WI); 0.025 U/μl AmpliTaq gold DNA polymerase (Applied Biosystems, Foster City, CA); and 10 μl of specimen DNA extract. The reaction mixtures were placed in an ABI 9700 thermal cycler (Applied Biosystems) that was programmed for 50 cycles with a three-step PCR procedure (94°C for 1 min, 60°C for 1 min, 72°C for 1 min). This was preceded by an initial uracil-N-glycosylase activation (2 min at 50°C). The primer set (Table 1) was designed to amplify the HHV-6-specific immediate-early gene. A sequence-specific, biotinylated capture probe (Table 1) was hybridized to the denatured amplicon, and the complexes were captured in avidin-coated wells. Detection was completed with enzyme-linked antidigoxigenin antibodies (46). The output signal was measured at an optical density at 450 nm (OD450). A positive result was defined as an OD450 − OD490 value greater than or equal to 0.1.
Assay 8.
DNA was extracted from 200 μl of plasma by the MagNA Pure automated extraction method (Roche Diagnostics, Indianapolis, IN). Carrier RNA [RNA-homopolymer poly(A)] was added to the lysis buffer at a concentration of 1 μg carrier RNA/100 μl lysis buffer. Nucleic acids were eluted into 50 μl of elution buffer. For PCR, the RealArt HHV-6 assay (Qiagen GmbH, Hamburg, Germany), a yet-to-be-commercialized ready-to-use system containing reagents and enzymes for the amplification of a 129-bp region of the HHV-6 genome, was used. The PCR mixture was prepared with 15 μl of master mixture and 5 μl of target DNA. Quantitative standards included with the kit were used to generate a standard curve to determine the viral load. A second heterologous amplification system was used to identify possible PCR inhibition. Amplification was performed with a LightCycler apparatus (Roche Diagnostics) and was initiated by a 10-min incubation at 95°C, followed by 10 cycles at 95°C for 5 s, 65°C for 20 s, and 72°C for 15 s. Forty additional cycles consisting of 95°C for 5 s, 55°C for 20 s, and 72°C for 15 s were then executed. The CT value was defined by the first cycle number at which the fluorescence was greater than the threshold, and the specificity of amplification was assessed by melting curve analysis.
Assay 9.
DNA from plasma was extracted with a MagNA Pure LC DNA extraction system with a total nucleic acid large volume kit (Roche Diagnostics, Mannheim, Germany) with elution into 50 μl. A negative extraction control was included with each group of samples. A real-time PCR assay targeted to a portion of the green fluorescent protein (GFP) gene contained in plasmid pEGFP-C2 (BD Biosciences, Palo Alto, CA) was performed as an internal control. The plasmid containing GFP was added to each specimen aliquot prior to extraction to serve both as a positive extraction control and as a control for possible PCR inhibitors. The primers and probes used in this assay (undisclosed by the company) are specific for known strains of HHV-6. All oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA), and all dual-labeled (TaqMan) probes were synthesized by Applied Biosystems. Real-time PCR assays were performed with an ABI Prism 7500 real-time thermocycler (Applied Biosystems). The HHV-6 target and the internal amplification control reactions were multiplexed. The PCR was performed in a 30-μl reaction volume with 20 μl TaqMan universal PCR master mixture (Applied Biosystems). For the viral gene target, each forward and reverse primer was used at a final concentration of 2.3 μM, and the dual-labeled probe was used at a final concentration of 0.2 μM. For the internal control gene target, the forward and reverse primers and the dual-labeled probe were all used at a final concentration of 0.2 μM. All DNA samples were analyzed in duplicate, and 10 μl sample DNA was added to each reaction mixture. The thermocycling profile was as follows: stage 1 (1 repetition), 50°C for 2 min; stage 2 (1 repetition), 95°C for 10 min; and stage 3 (45 repetitions), 95°C for 15 s and 60°C for 1 min, with fluorescence data collection performed at the 60°C step for each cycle. HHV-6 plasmid DNA (50 to 5 × 107 copies per reaction) was used to construct a seven-point standard curve. The software of the ABI 7500 thermocycler performed the calculations to convert the sample CT values to copy numbers, which were normalized to 1 ml of sample.
RESULTS
Eight different laboratories using nine different assays were asked to participate in this study. Assays varied from nested PCR, PCR linked to EIA, LAMP, and real-time PCR. Each laboratory received an identical set of 11 coded samples. Each sample contained 0.25 ml of serum spiked with various quantities (0 to 100,000 genome equivalents) of HHV-6A virus, and each laboratory proceeded to extract the viral DNA and perform the analysis by the protocol used in each laboratory. Upon completion, each laboratory was asked to send its results to the project coordinator. Once all results were turned in, the code was broken and the performance results were sent to the participating laboratories. The results obtained by all laboratories are presented in Table 2.
TABLE 2.
Estimation of HHV-6 DNA copy number in spiked serum samples by various PCR assays
| No. of spiked HHV-6A genome copies | Resulta by the following assay (assay type):
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 (real-time PCR) | 2 (real-time PCR) | 3 (nested PCR) | 4 (real-time PCR) | 5 (LAMP) | 6 (real-time PCR) | 7 (PCR-EIA) | 8 (real-time PCR) | 9 (real-time PCR) | |
| 100,000 | 122,071 | 16,707 | + | 39,090 | + | 56,406 | + | 245,250 | 172,000 |
| 10,000 | 10,326 | 1,736 | + | 4,575 | + | 8,631 | + | 10,690 | 21,800 |
| 1,000 | 425b | 223 | + | 8 | + | 903 | + | 1,265 | 2,500 |
| 100 | 111 | 28 | + | 13 | − | 113 | + | 51 | 300 |
| 100 | 31 | 19 | + | 0 | + | 30 | + | 0 | 200 |
| 10 | +c | 1 | − | 46 | − | 27 | + | 0 | 16 |
| 10 | 16 | 7 | + | 0 | + | 29 | + | 0 | 11 |
| 1 | 0 | 19 | − | 0 | + | 0 | + | 0 | 0 |
| 1 | 0 | 0 | + | 0 | − | 0 | + | 0 | 0 |
| 0 | 0 | 0 | − | 0 | − | 0 | − | 0 | 0 |
| 0 | 0 | 0 | − | 0 | − | 0 | − | 0 | 0 |
Results are presented quantitatively (in numbers of copies) or qualitatively (a positive [+] or a negative [−] result).
This sample contained PCR inhibitors.
One of three PCR replicates gave a positive amplification.
Nonquantitative assays.
Three assays (assays 3, 5, and 7) yielded nonquantitative results. Of these three assays, assay 7 proved to be the most sensitive, with the detection of virions in all samples containing HHV-6, including those spiked with one virion (Table 2). This assay combines magnetic bead-based nucleic acid extraction, PCR amplification, and colorimetric microtiter plate identification, based on the detection of HHV-6 digoxigenin-labeled amplicons with a biotinylated probe complementary to the region of the amplified PCR products. The fact that the detection was based on an enzymatic reaction likely contributes to the sensitivity of the assay. In comparison, the nested PCR assay (assay 3; 35 cycles of amplification performed twice) detected virions in only one of two samples spiked with 10 and 1 HHV-6 particles. For detection, this assay relies on gel electrophoresis of the PCR products and visualization of the products by ethidium bromide staining. The LAMP assay, based on isothermal amplification of target DNA, failed to detect half (one of two) of the samples containing 100, 10, and 1 copies of HHV-6. During amplification, a large amount of a by-product, pyrophosphate ion, is produced, yielding a white precipitate of magnesium pyrophosphate in the reaction mixture. Measurement of the turbidity of the reaction mixture provides an estimate of the amount of DNA synthesized. No false-positive results were reported for any of these three assays.
Quantitative assays.
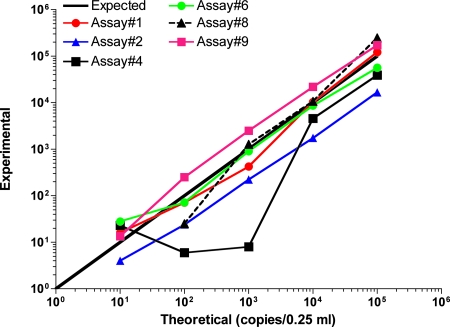
A total of six quantitative assays, all based on the TaqMan technology, were tested. The data presented (Table 2) correspond to those for a single assay run, with the exception of assay 1, by which the analysis was performed in triplicate and whose results correspond to the means of the triplicate values obtained. The quantitative assays were compared on the basis of sensitivity (how many copies could be detected) and the accuracy of the copy number estimates relative to the spiked copy numbers. Among the various quantitative PCR assays, three of them (assays 1, 6, and 9) yielded results that were the closest to the expected spiked HHV-6 DNA copy numbers (Fig. 1 and Table 2). Assays 1, 6, and 9 yielded comparable results, with a correlation coefficient (r2) of >0.99 (Fig. 1). All three assays detected HHV-6 DNA in samples containing 10 copies of HHV-6 (equivalent to 40 copies/ml of serum), while none of these three assays detected HHV-6 in samples containing 1 HHV-6 copy (4 copies/ml). No false-positive results were reported.
FIG. 1.
Relationship between experimentally detected and expected (theoretical) number of HHV-6 DNA copies in spiked serum samples. Series of coded samples were analyzed in a blinded fashion by different real-time PCR assays according to the protocols described in the Materials and Methods section. The estimated numbers of HHV-6 DNA copies obtained experimentally were plotted against the expected values. For samples containing 10 and 100 copies of spiked HHV-6, the mean copy number from two samples is presented.
DISCUSSION
HHV-6 is an understudied pathogen with which infections are linked to diverse pathologies, such as roseola (primary infection) (51), complications in organ transplant recipients (34, 35, 48, 57, 58), and neurological disorders (5, 9, 15, 17, 21, 32, 43, 52). Considering that nearly 100% of individuals past the age of 3 years have been infected with HHV-6 (37, 56, 59), qualitative diagnostic tests designed to report on the presence or the absence of this virus in whole blood (plasma plus leukocytes) can be of limited value. However, detection of HHV-6 DNA in cell-free plasma can be of biological significance. However, as recently reported by Achour et al. (3), it is recommended that plasma samples be treated with DNase prior to any extraction procedure to avoid potential false-positive results emanating from the release of cellular and viral DNA (possibly due to the lysis of infected cells) in the absence of active viral infection.
Among the nonquantitative assays that were compared, PCR-EIA (assay 7) demonstrated the greatest sensitivity and detected virions in all spiked samples, including those containing one copy of HHV-6. This assay offers two relative advantages over assays 3 and 5. First, detection of the amplified product occurs through the use of a probe that is complementary to an internal region of the amplicon, ensuring that a positive signal originates from the detection of the HHV-6 target sequence. The nested PCR, which relies on the visualization of ethidium bromide-stained DNA fragments following electrophoresis of the PCR, and the LAMP assay, which measures the turbidity of the mixture, do not offer the same level of confidence since no validation of the presence of the target sequence is made. Second, because it uses enzyme-labeled antibodies, which amplify the signal and lower the detection level, the detection of the amplified fragments in the PCR-EIA assay is enhanced. Nested PCR also suffers from the increased possibility of false-positive signals due to the presence of carryover products from the first amplification round into the second amplification round. Thus, for routine screening for the presence or the absence of HHV-6 in biological samples, the PCR-EIA is recommendable.
In the absence of reliable serological assays that can discriminate between latent and active HHV-6 infection, molecular assays that can precisely quantitate the HHV-6 viral loads in various biological samples would offer a benefit over nonquantitative assays to establish potential links between HHV-6 and diseases. The use of reverse transcriptase quantitative PCR to detect and quantitate viral transcripts associated with active viral infection is certainly one valid method that could be used to monitor infection. However, this assay is complicated by the fact that detailed knowledge of the sites where HHV-6 replicates is not yet available, making it hard to know which tissue should be biopsied. For example, HHV-6 can be found in the cerebrospinal fluid after it has disappeared from the peripheral blood (20). In addition, several tissues or organs cannot be biopsied simply.
Over the years, several reports describing the quantitation of HHV-6 DNA in biological samples have been reported. Diversity in the methodology used as well as the heterogeneity between the various biological samples analyzed has made it difficult to compare the results obtained by independent laboratories, sometimes leading to conflicting results. None of these assays were ever compared systematically. One of the goals of the present study was to identify the most reliable assays available for the quantitation of HHV-6 in serum (and other biological samples) and to determine the assay(s) that could serve as a reference standard for the quantitation of HHV-6 in biological samples. To avoid variations between the samples assayed, laboratories around the world were sent a series of identically coded serum samples spiked with various quantities of HHV-6A and were asked to analyze the HHV-6 content by using their own methodologies and assays. The extraction procedures varied from standard phenol-chloroform extraction to the use of commercial DNA affinity spin columns. Three assays, based on the TaqMan PCR technology, provided results that were the closest to the expected values (Table 2 and Fig. 1). All three assays can detect both HHV-6 variants. The sensitivities of these assays were below 40 HHV-6 copies/ml. Perhaps the only limiting factor for the general use of this methodology is the access to a real-time PCR apparatus. However, because the cost of such equipment has drastically decreased in recent years, the constantly increasing ability of scientists and hospitals to access a real-time PCR machine is making this limitation not a real concern.
Although the detection of HHV-6 in serum or cerebrospinal fluid is suggestive of active infection, it does not constitute absolute proof. In fact, data suggest that HHV-6 can, on rare occasions (∼1%), integrate into the host's chromosome (CIHHV-6) and be transmitted through the germ line (10, 13, 30, 31, 45). Individuals carrying CIHHV-6 constantly display abnormally high HHV-6 DNA contents in plasma (≥3.5 log10 copies/ml) and whole blood (>6 log10 copies/ml) in the absence of documented active infection (49). Such individuals are sometimes misdiagnosed as having active HHV-6 infection and are put on antiviral therapy, without any effect on the plasmatic HHV-6 DNA load. Thus, the results of a high viral load (>3.5 log10 copies/ml) should be interpreted with caution, and the conclusion that a patient has an active infection should be made once the presence of CIHHV-6 has been excluded, as suggested previously (49). Active infection could be confirmed by analyzing leukocytes for the presence of HHV-6 transcripts associated with active infection (38). Alternatively, the presence of CIHHV-6 could be confirmed by the detection of HHV-6 DNA in hair follicles (24, 49).
In our opinion, the use of a validated and standardized assay for the quantitation of HHV-6 DNA in clinical samples is essential to better evaluate the numerous associations of HHV-6 with diseases. We propose that TaqMan-based PCR assays identical or similar to those used in assay 1, 6, or 9 be used as reference assays by investigators interested in setting up their own HHV-6 assay or in monitoring various biological samples for the presence of HHV-6 DNA.
Acknowledgments
This work was sponsored by the HHV-6 Foundation.
Special thanks go to Kristin Loomis, president and executive director of the HHV-6 Foundation, for her constant devotion in bringing closer together investigators working on HHV-6 and her help organizing this study.
No author has declared a conflict of interest.
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Ablashi, D. V., H. Agut, Z. Berneman, G. Campadilli-Fiume, D. Carrigan, L. Ceccerini-Nelli, B. Chandran, S. Chou, H. Collandre, R. Cone, T. Dambaugh, S. Dewhurst, D. DiLuca, L. Foà-Tomasi., B. Fleckeinstein, N. Frenkel, Gallo, R. U. Gomples, C. Hall, M. Jones, G. Lawrence, M. Martin, L. Montagnier, F. Neipel, J. Nicholas, P. Pellett, A. Razzaque, G. Torrelli, B. Thomson, S. Salahuddin, L. Wyatt, and K. Yamanishi. 1993. Human herpesvirus-6 strain groups: a nomenclature. Arch. Virol. 129363-366. [DOI] [PubMed] [Google Scholar]
- 2.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184545-552. [DOI] [PubMed] [Google Scholar]
- 3.Achour, A., D. Boutolleau, A. Slim, H. Agut, and A. Gautheret-Dejean. 2007. Human herpesvirus-6 (HHV-6) DNA in plasma reflects the presence of infected blood cells rather than circulating viral particles. J. Clin. Virol. 38280-285. [DOI] [PubMed] [Google Scholar]
- 4.Akhyani, N., R. Berti, M. B. Brennan, S. S. Soldan, J. M. Eaton, H. F. McFarland, and S. Jacobson. 2000. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: increased prevalence of HHV-6A in patients with multiple sclerosis. J. Infect. Dis. 1821321-1325. [DOI] [PubMed] [Google Scholar]
- 5.Asano, Y., T. Yoshikawa, S. Suga, I. Kobayashi, T. Nakashima, T. Yazaki, Y. Kajita, and T. Ozaki. 1994. Clinical features of infants with primary human herpesvirus 6 infection (exanthem subitum, roseola infantum). Pediatrics 93104-108. [PubMed] [Google Scholar]
- 6.Asano, Y., T. Yoshikawa, S. Suga, T. Nakashima, T. Yazaki, M. Fukuda, S. Kojima, and T. Matsuyama. 1991. Reactivation of herpesvirus type 6 in children receiving bone marrow transplants for leukemia. N. Engl. J. Med. 324634-635. [DOI] [PubMed] [Google Scholar]
- 7.Asano, Y., T. Yoshikawa, S. Suga, T. Yazaki, K. Kondo, and K. Yamanishi. 1990. Fatal fulminant hepatitis in an infant with human herpesvirus-6 infection. Lancet 335862-863. [DOI] [PubMed] [Google Scholar]
- 8.Boutolleau, D., C. Duros, P. Bonnafous, D. Caiola, A. Karras, N. D. Castro, M. Ouachee, P. Narcy, M. Gueudin, H. Agut, and A. Gautheret-Dejean. 2006. Identification of human herpesvirus 6 variants A and B by primer-specific real-time PCR may help to revisit their respective role in pathology. J. Clin. Virol. 35257-263. [DOI] [PubMed] [Google Scholar]
- 9.Challoner, P. B., K. T. Smith, J. D. Parker, D. L. MacLeod, S. N. Coulter, T. M. Rose, E. R. Schultz, J. L. Bennett, R. L. Garber, M. Chang, P. A. Schad, P. M. Stewart, R. C. Nowinski, J. P. Brown, and G. C. Burmer. 1995. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc. Natl. Acad. Sci. USA 927440-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, D. A., E. P. Nacheva, H. N. Leong, D. Brazma, Y. T. Li, E. H. Tsao, H. C. Buyck, C. E. Atkinson, H. M. Lawson, M. N. Potter, and P. D. Griffiths. 2006. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J. Infect. Dis. 193912-916. [DOI] [PubMed] [Google Scholar]
- 11.Cone, R. W., M. L. Huang, L. Corey, J. Zeh, R. Ashley, and R. Bowden. 1999. Human herpesvirus 6 infections after bone marrow transplantation: clinical and virologic manifestations. J. Infect. Dis. 179311-318. [DOI] [PubMed] [Google Scholar]
- 12.Cone, R. W., M. L. Huang, R. C. Hackman, and L. Corey. 1996. Coinfection with human herpesvirus 6 variants A and B in lung tissue. J. Clin. Microbiol. 34877-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daibata, M., T. Taguchi, Y. Nemoto, H. Taguchi, and I. Miyoshi. 1999. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 941545-1549. [PubMed] [Google Scholar]
- 14.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 738040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donati, D., N. Akhyani, A. Fogdell-Hahn, C. Cermelli, R. Cassiani-Ingoni, A. Vortmeyer, J. D. Heiss, P. Cogen, W. D. Gaillard, S. Sato, W. H. Theodore, and S. Jacobson. 2003. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology 611405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fillet, A. M., M. Raphael, B. Visse, J. Audouin, L. Poirel, H. Agut, et al. 1995. Controlled study of human herpesvirus 6 detection in acquired immunodeficiency syndrome-associated non-Hodgkin's lymphoma. J. Med. Virol. 45106-112. [DOI] [PubMed] [Google Scholar]
- 17.Fotheringham, J., D. Donati, N. Akhyani, A. Fogdell-Hahn, A. Vortmeyer, J. D. Heiss, E. Williams, S. Weinstein, D. A. Bruce, W. D. Gaillard, S. Sato, W. H. Theodore, and S. Jacobson. 2007. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLoS Med. 4e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautheret-Dejean, A., C. Manichanh, F. Thien-Ah-Koon, A. M. Fillet, N. Mangeney, M. Vidaud, N. Dhedin, J. P. Vernant, and H. Agut. 2002. Development of a real-time polymerase chain reaction assay for the diagnosis of human herpesvirus-6 infection and application to bone marrow transplant patients. J. Virol. Methods 10027-35. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths, P. D., M. Ait-Khaled, C. P. Bearcroft, D. A. Clark, A. Quaglia, S. E. Davies, A. K. Burroughs, K. Rolles, I. M. Kidd, S. N. Knight, S. M. Noibi, A. V. Cope, A. N. Phillips, and V. C. Emery. 1999. Human herpesviruses 6 and 7 as potential pathogens after liver transplant: prospective comparison with the effect of cytomegalovirus. J. Med. Virol. 59496-501. [DOI] [PubMed] [Google Scholar]
- 20.Hall, C. B., M. T. Caserta, K. C. Schnabel, C. Long, L. G. Epstein, R. A. Insel, and S. Dewhurst. 1998. Persistence of human herpesvirus 6 according to site and variant: possible greater neurotropism of variant A. Clin. Infect. Dis. 26132-137. [DOI] [PubMed] [Google Scholar]
- 21.Hall, C. B., C. E. Long, K. C. Schnabel, M. T. Caserta, K. M. McIntyre, M. A. Costanzo, A. Knott, S. Dewhurst, R. A. Insel, and L. G. Epstein. 1994. Human herpesvirus-6 infection in children. A prospective study of complications and reactivation. N. Engl. J. Med. 331432-438. [DOI] [PubMed] [Google Scholar]
- 22.Huang, L. M., C. Y. Lee, P. I. Lee, J. M. Chen, and P. J. Wang. 1991. Meningitis caused by human herpesvirus-6. Arch. Dis. Child. 661443-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, L. M., C. Y. Lee, K. H. Lin, W. M. Chuu, P. I. Lee, R. L. Chen, J. M. Chen, and D. T. Lin. 1990. Human herpesvirus-6 associated with fatal haemophagocytic syndrome. Lancet 33660-61. [DOI] [PubMed] [Google Scholar]
- 24.Hubacek, P., J. Maalouf, M. Zajickova, M. Kouba, O. Cinek, K. Hyncicova, I. Fales, and P. Cetkovsky. 2007. Failure of multiple antivirals to affect high HHV-6 DNAaemia resulting from viral chromosomal integration in case of severe aplastic anaemia. Haematologica 92e98-e100. [DOI] [PubMed] [Google Scholar]
- 25.Hymas, W., J. Stevenson, E. W. Taggart, and D. Hillyard. 2005. Use of lyophilized standards for the calibration of a newly developed real time PCR assay for human herpes type six (HHV6) variants A and B. J. Virol. Methods 128143-150. [DOI] [PubMed] [Google Scholar]
- 26.Ihira, M., T. Yoshikawa, Y. Enomoto, S. Akimoto, M. Ohashi, S. Suga, N. Nishimura, T. Ozaki, Y. Nishiyama, T. Notomi, Y. Ohta, and Y. Asano. 2004. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J. Clin. Microbiol. 42140-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 738053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, H., M. Morita, Y. Yabuta, K. Kuzushima, K. Kato, S. Kojima, T. Matsuyama, and T. Morishima. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locatelli, G., F. Santoro, F. Veglia, A. Gobbi, P. Lusso, and M. S. Malnati. 2000. Real-time quantitative PCR for human herpesvirus 6 DNA. J. Clin. Microbiol. 384042-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luppi, M., P. Barozzi, R. Marasca, and G. Torelli. 1994. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia 8(Suppl. 1)S41-S45. [PubMed] [Google Scholar]
- 31.Luppi, M., P. Barozzi, C. M. Morris, E. Merelli, and G. Torelli. 1998. Integration of human herpesvirus 6 genome in human chromosomes. Lancet 3521707-1708. [DOI] [PubMed] [Google Scholar]
- 32.Mock, D. J., J. M. Powers, A. D. Goodman, S. R. Blumenthal, N. Ergin, J. V. Baker, D. H. Mattson, J. G. Assouline, E. J. Bergey, B. Chen, L. G. Epstein, and B. M. Blumberg. 1999. Association of human herpesvirus 6 with the demyelinative lesions of progressive multifocal leukoencephalopathy. J. Neurovirol. 5363-373. [DOI] [PubMed] [Google Scholar]
- 33.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289150-154. [DOI] [PubMed] [Google Scholar]
- 34.Ogata, M., H. Kikuchi, T. Satou, R. Kawano, J. Ikewaki, K. Kohno, K. Kashima, E. Ohtsuka, and J. Kadota. 2006. Human herpesvirus 6 DNA in plasma after allogeneic stem cell transplantation: incidence and clinical significance. J. Infect. Dis. 19368-79. [DOI] [PubMed] [Google Scholar]
- 35.Ogata, M., T. Satou, R. Kawano, K. Goto, J. Ikewaki, K. Kohno, T. Ando, Y. Miyazaki, E. Ohtsuka, Y. Saburi, T. Saikawa, and J. I. Kadota. 2008. Plasma HHV-6 viral load-guided preemptive therapy against HHV-6 encephalopathy after allogeneic stem cell transplantation: a prospective evaluation. Bone Marrow Transplant. 41279-285. [DOI] [PubMed] [Google Scholar]
- 36.Okuno, T., K. Higashi, K. Shiraki, K. Yamanishi, M. Takahashi, Y. Kokado, M. Ishibashi, S. Takahara, T. Sonoda, K. Tanaka, et al. 1990. Human herpesvirus 6 infection in renal transplantation. Transplantation 49519-522. [DOI] [PubMed] [Google Scholar]
- 37.Okuno, T., K. Takahashi, K. Balachandra, K. Shiraki, K. Yamanishi, M. Takahashi, and K. Baba. 1989. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 27651-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oster, B., and P. Hollsberg. 2002. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J. Virol. 767578-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prezioso, P. J., J. Cangiarella, M. Lee, G. J. Nuovo, W. Borkowsky, S. J. Orlow, and M. A. Greco. 1992. Fatal disseminated infection with human herpesvirus-6. J. Pediatr. 120921-923. [DOI] [PubMed] [Google Scholar]
- 40.Razonable, R. R., C. Fanning, R. A. Brown, M. J. Espy, A. Rivero, J. Wilson, W. Kremers, T. F. Smith, and C. V. Paya. 2002. Selective reactivation of human herpesvirus 6 variant a occurs in critically ill immunocompetent hosts. J. Infect. Dis. 185110-113. [DOI] [PubMed] [Google Scholar]
- 41.Salahuddin, S. Z., D. V. Ablashi, P. D. Markham, S. F. Josephs, S. Sturzenegger, M. Kaplan, G. Halligan, P. Biberfeld, F. Wong-Staal, B. Kramarsky, et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234596-601. [DOI] [PubMed] [Google Scholar]
- 42.Secchiero, P., D. R. Carrigan, Y. Asano, L. Benedetti, R. W. Crowley, A. L. Komaroff, R. C. Gallo, and P. Lusso. 1995. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J. Infect. Dis. 171273-280. [DOI] [PubMed] [Google Scholar]
- 43.Soldan, S. S., R. Berti, N. Salem, P. Secchiero, L. Flamand, P. A. Calabresi, M. B. Brennan, H. W. Maloni, H. F. McFarland, H. C. Lin, M. Patnaik, and S. Jacobson. 1997. Association of human herpes virus 6 (HHV-6) with multiple sclerosis: increased IgM response to HHV-6 early antigen and detection of serum HHV-6 DNA. Nat. Med. 31394-1397. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, N., H. Kimura, Y. Hoshino, K. Kato, T. Yoshikawa, Y. Asano, K. Horibe, S. Kojima, and T. Morishima. 2000. Monitoring four herpesviruses in unrelated cord blood transplantation. Bone Marrow Transplant. 261193-1197. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka-Taya, K., J. Sashihara, H. Kurahashi, K. Amo, H. Miyagawa, K. Kondo, S. Okada, and K. Yamanishi. 2004. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J. Med. Virol. 73465-473. [DOI] [PubMed] [Google Scholar]
- 46.Tang, Y. W., P. N. Rys, B. J. Rutledge, P. S. Mitchell, T. F. Smith, and D. H. Persing. 1998. Comparative evaluation of colorimetric microtiter plate systems for detection of herpes simplex virus in cerebrospinal fluid. J. Clin. Microbiol. 362714-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vu, T., G. Carrum, G. Hutton, H. E. Heslop, M. K. Brenner, and R. Kamble. 2007. Human herpesvirus-6 encephalitis following allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 39705-709. [DOI] [PubMed] [Google Scholar]
- 48.Wainwright, M. S., P. L. Martin, R. P. Morse, M. Lacaze, J. M. Provenzale, R. E. Coleman, M. A. Morgan, C. Hulette, J. Kurtzberg, C. Bushnell, L. Epstein, and D. V. Lewis. 2001. Human herpesvirus 6 limbic encephalitis after stem cell transplantation. Ann. Neurol. 50612-619. [DOI] [PubMed] [Google Scholar]
- 49.Ward, K. N., H. N. Leong, A. D. Thiruchelvam, C. E. Atkinson, and D. A. Clark. 2007. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J. Clin. Microbiol. 451298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yalcin, S., T. Karpuzoglu, G. Suleymanlar, G. Mutlu, T. Mukai, T. Yamamoto, Y. Isegawa, and K. Yamanishi. 1994. Human herpesvirus 6 and human herpesvirus 7 infections in renal transplant recipients and healthy adults in Turkey. Arch. Virol. 136183-190. [DOI] [PubMed] [Google Scholar]
- 51.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i1065-1067. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa, T., and Y. Asano. 2000. Central nervous system complications in human herpesvirus-6 infection. Brain Dev. 22307-314. [DOI] [PubMed] [Google Scholar]
- 53.Yoshikawa, T., S. Kojima, and Y. Asano. 1992. Human herpesvirus-6 infection and bone marrow transplantation. Leuk. Lymphoma 865-73. [DOI] [PubMed] [Google Scholar]
- 54.Yoshikawa, T., S. Suga, Y. Asano, T. Nakashima, T. Yazaki, Y. Ono, T. Fujita, K. Tsuzuki, S. Sugiyama, and S. Oshima. 1992. A prospective study of human herpesvirus-6 infection in renal transplantation. Transplantation 54879-883. [DOI] [PubMed] [Google Scholar]
- 55.Yoshikawa, T., S. Suga, Y. Asano, T. Nakashima, T. Yazaki, R. Sobue, M. Hirano, M. Fukuda, S. Kojima, and T. Matsuyama. 1991. Human herpesvirus-6 infection in bone marrow transplantation. Blood 781381-1384. [PubMed] [Google Scholar]
- 56.Yoshikawa, T., S. Suga, Y. Asano, T. Yazaki, H. Kodama, and T. Ozaki. 1989. Distribution of antibodies to a causative agent of exanthem subitum (human herpesvirus-6) in healthy individuals. Pediatrics 84675-677. [PubMed] [Google Scholar]
- 57.Zerr, D. M. 2006. Human herpesvirus 6 and central nervous system disease in hematopoietic cell transplantation. J. Clin. Virol. 37(Suppl. 1)S52-S56. [DOI] [PubMed] [Google Scholar]
- 58.Zerr, D. M., L. Corey, H. W. Kim, M. L. Huang, L. Nguy, and M. Boeckh. 2005. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin. Infect. Dis. 40932-940. [DOI] [PubMed] [Google Scholar]
- 59.Zerr, D. M., A. S. Meier, S. S. Selke, L. M. Frenkel, M. L. Huang, A. Wald, M. P. Rhoads, L. Nguy, R. Bornemann, R. A. Morrow, and L. Corey. 2005. A population-based study of primary human herpesvirus 6 infection. N. Engl. J. Med. 352768-776. [DOI] [PubMed] [Google Scholar]