Abstract
We have developed a set of reverse transcription-PCR assays for the detection and identification of known and novel paramyxoviruses in clinical specimens. Primers were designed from the conserved motifs of the polymerase pol gene sequences to detect members of the Paramyxovirinae or Pneumovirinae subfamily or groups of genera within the Paramyxovirinae subfamily. The consensus-degenerate hybrid oligonucleotide primer design and seminested or nested PCR assay design were used to enhance the breadth of reactivity and sensitivity of the respective assays. Using expressed RNA and 10-fold dilution series of virus-infected tissue culture isolates from different members of the family or genera, these assays were able to detect on average between 100 and 500 copies of template RNA. The assays were specific to the respective group of genera or subfamily viruses. This set of primers enhances our ability to look for novel viruses in outbreaks and diseases of unknown etiology.
The paramyxoviruses are ubiquitous pathogens and have been identified in a variety of hosts, including birds (chickens and turkeys), aquatic animals (salmon, whale, seal, dolphin, and porpoise), rodents (mice and rats), dogs, cats, sheep, reptiles (snake and lizards), horses, cattle, bats, pigs, simians, and humans. In humans, paramyxoviruses have been associated with a wide range of diseases, including croup, bronchiolitis, pneumonia, encephalitis, meningitis, parotitis, orchitis, spontaneous abortion, rash illnesses, and persistent infections (7, 10, 12, 16). Recently a variety of new members of the Paramyxoviridae family have been identified in animals, and some, most notably Nipah and Hendra viruses, have caused serious and sometimes fatal human infections (1, 2, 4). It is likely that there are additional, as yet unidentified members of this family and some will likely infect and cause disease in humans. Therefore, we believe the Paramyxoviridae constitute an important target for improved methods to detect novel viruses.
Detection of most paramyxoviruses has been routinely carried out by cell culture isolation, electron microscopy, antigen detection assays (immunofluorescence assays or enzyme immunoassays [EIA]), serologic assays, and genome-based assays, such as PCR assays. Each system has limitations. Traditional genome-based, antigen-based, and antibody-based assays usually are too specific to detect novel viruses. Cell culture isolation will allow detection only of viruses that grow and replicate in the culture system used and will require further characterization, usually by antigen- or genome-based assays. Electron microscopy requires a fairly high titer of virus for visualization and also requires further characterization.
To increase our ability to detect novel viruses, we chose to develop broadly reactive PCR assays. This strategy has been used very successfully to identify and characterize a number of novel human viruses, including severe acute respiratory syndrome coronavirus (5), hepatitis G virus (17), Sin Nombre virus (8), human retrovirus 5 (3), and novel animal viruses, such as the macaque gammaherpesvirus (15) and pig endogenous retrovirus (9). The present article describes the development of a set of PCR assays that should detect all known and novel paramyxoviruses. The primers for these assays were developed from highly conserved regions of the genome. We applied the consensus-degenerate hybrid oligonucleotide primer methodology (13, 14) in primer design and seminested PCRs to optimize the specificity and sensitivity of these broadly reactive assays.
MATERIALS AND METHODS
Virus and isolation of viral RNA.
The reference viruses or viral RNA used in this study are listed in Table 1 and include three strains from the genus Avulavirus, two strains from the genus Henipavirus, five strains from the genus Morbillivirus, seven strains from the genus Respirovirus, five strains from the genus Rubulavirus, four strains from the genus Pneumovirus, three strains from the genus Metapneumovirus and three newly isolated unclassified paramyxoviruses. RNAs were extracted from 100 μl of supernatant fluid of virus-infected cells with the QIAamp viral RNA kit (Qiagen, Santa Clarita, CA) according to the manufacturer's instructions. The RNA was eluted from the column in 50 μl of RNase-free water. RNAs for the Hendra virus, Nipah virus, Menangle virus, and Fer-de-Lance virus (FDLV) strains were obtained from Paul Rota (Centers for Disease Control and Prevention) as extracts in Trizol, which were prepared according to the instructions provided by the commercial source (Invitrogen, Carlsbad, CA).
TABLE 1.
Reference viruses used in evaluating PCR in this work
| Virus and classification | Abbreviation | Source |
|---|---|---|
| Paramyxovirinae | ||
| Avulavirus | ||
| Newcastle disease virus | NDV | Jack King, Qingzhong Yu |
| Avian paramyxovirus-2 | APMV-2 | Jack King, Qingzhong Yu |
| Avian paramyxovirus-3 | APMV-3 | Jack King, Qingzhong Yu |
| Henipavirus | ||
| Hendra virus | Hendra | Paul Rota |
| Nipah virus | Nipah | Paul Rota |
| Morbillivirus | ||
| Measles virus | MV | Paul Rota |
| Canine distemper virus | CDV | Randy Renshaw |
| Phocine distemper virus | PDV | Bert Rima |
| Dolphin morbillivirus | DMV | Bert Rima |
| Porpoise morbillivirus | PMV | Bert Rima |
| Respirovirus | ||
| Bovine parainfluenza virus 1 | BPIV1 | Dean Erdman |
| Bovine parainfluenza virus 3 | BPIV3 | Randy Renshaw |
| Human parainfluenza virus 1 | HPIV1 | Dean Erdman |
| Human parainfluenza virus 3 | HPIV3 | Dean Erdman |
| Sendai virus | Sendai | Lela K. Riley |
| Unclassified Respirovirus | ||
| Pacific salmon paramyxovirus | PSPV | Gael Kurath |
| Guinea pig parainfluenza virus 3 | CaVPIV3 | Lela K. Riley |
| Rubulavirus | ||
| Mumps virus | Mumps | Paul Rota |
| Human parainfluenza virus 2 | HPIV2 | Dean Erdman |
| Canine parainfluenza virus 2 | CaPIV2 | Randy Renshaw |
| Simian virus 5 | SV5 | Richard W. Compans |
| Simian virus 41 | SV41 | Yasuhiko Ito |
| Unclassified Paramyxovorinae | ||
| Fer-de-Lance virus | FDLV | Gael Kurath |
| Menangle virus | Menangle | Paul Rota |
| Salem virus | SalV | Randy Renshaw |
| Pneumovirinae | ||
| Metapneumovirus | ||
| Avian metapneumovirus type C | AMPV-C | Jack King, Qingzhong Yu |
| Human metapneumovirus 75 | HMPV75 | Dean Erdman |
| Human metapneumovirus 83 | HMPV83 | Dean Erdman |
| Pneumovirus | ||
| Bovine respiratory syncytial virus | BRSV | Randy Renshaw |
| Human respiratory syncytial virus A | hRSVa A | Dean Erdman |
| Human respiratory syncytial virus B | hRSV B | Dean Erdman |
| Pneumonia virus of mice | PVM | Joseph Domachowske |
hRSV, human RSV.
Broadly reactive oligonucleotide primer selection.
Conserved amino acid sequences for the family, subfamily, and genera were selected from alignment of deduced L-protein-coding sequences recorded in GenBank (National Institutes of Health, Bethesda, MD). A total of 33 nonredundant paramyxovirus L-protein sequences were used and aligned using the Clustal W program. We selected highly conserved domains between 8 and 10 amino acids in length and back translated into degenerate nucleotide sequences to represent all possible codons for the corresponding amino acids. To minimize the number of primers, primers were designed with mixed degenerate bases restricted to between 9 and 12 nucleotides in the 3′ portion of the primers, and inosines (maximum of four) and consensus nucleotides were used for the remaining middle and 5′ portion of the primers. In addition, primers were designed to achieve similar reaction conditions and an amplicon size between 200 and 500 bp. To minimize the potential for nonspecific cross-reactivity, a Blastn search analysis of GenBank was performed for similarities with known sequences. The primers were synthesized at the Biotechnology Core Facility, Division of Scientific Resources of the Centers for Disease Control and Prevention (CDC).
RT-PCR and nested RT-PCR amplification.
To maximize sensitivity and specificity, we nested or seminested PCR assays and optimized reaction conditions, including primer concentration, magnesium (Mg2+) concentration, and thermal cycling temperatures and profiles. For the first PCR in the seminested assay, we used the SuperScript III One-Step reverse transcription-PCR (RT-PCR) kit (Invitrogen, Carlsbad, CA). The optimized PCR mixtures contained 50 pmol each of forward and reverse primers, 1× buffer with a final concentration of 2.0 mM MgSO4 and 200 μM (each) deoxynucleoside triphosphates, 20 U of RNase inhibitor, a 5-μl aliquot of RNA/DNA extracts, and 1 U of SuperScript III RT/Platinum Taq mix. Water was then added to achieve a final volume of 50 μl. The RT-PCR mixture was sequentially incubated at 60°C for 1 min for denaturing, 44 to 50°C for 30 min (for RT), 94°C for 2 min (for hot start), and then 40 cycles at 94°C for 15 s, 48 to 50°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 7 min. For the second amplification in the seminested PCR assay, we used 1× buffer (Platinum Taq kit; Invitrogen), 2 mM MgCl2, 200 μM (each) of deoxynucleoside triphosphates, 50 pmol (each) of forward and reverse primers, 1 U Platinum Taq, one 2-μl aliquot from the first reaction, and water to achieve a final volume of 50 μl. The mixture was first heated to 94°C for 2 min. The cycling conditions were 40 cycles with the same conditions as for the first amplification: 94°C for 15 s, primer annealing at 48 to 50°C for 30 s, and 72°C for 30 s. A final extension was carried out at 72°C for 7 min. The final nested or seminested PCR products were visualized by UV light after electrophoresis on a 2% agarose gel containing 0.5 μg/ml ethidium bromide in 0.5× Tris-borate buffer (pH 8.0). A DNA VIII marker (Roche, Indianapolis, IN) was run in the gels to estimate amplicon size.
Specificity and sensitivity.
Initial primer validation and selections were performed using one reference RNA template from the subfamily or genera for which the PCR assay was designed. Mumps virus RNA was used for the Rubulavirus-Avulavirus genus subgroup-specific primers and the Paramyxovirinae subfamily primers, Hendra RNA for the Morbillivirus-Respirovirus-Henipavirus genus subgroup-specific primers, and respiratory syncytial virus A RNA for the Pneumovirinae subfamily-specific primers. Following the initial screening, the selected primer pairs were then tested against the representative paramyxoviruses listed in Table 1. Finally, to test for unanticipated nonspecific reactivity, the PCR assays were tested against pooled nucleic acids of influenza A and B viruses, rhinoviruses, adenovirus, two distinct human coronaviruses, human coronavirus 229E, and human coronavirus OC-43 and bacteria (Chlamydia pneumoniae, Haemophilus influenzae, Streptococcus pneumoniae, and Mycoplasma pneumoniae).
The sensitivities of the PCR assays were determined using two sources of RNA: RNA that was extracted from each dilution of a 10-fold dilution series of virus-infected cell culture with known infectivity titers (PFU) and serial dilutions of synthetic RNA that was transcribed in vitro from cloned genome fragments as previously described (18).
Sequencing.
Amplicons from the final round of PCR were purified using the QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA). Both strands of the amplicons were sequenced with a BigDye Terminators v1.1 ready reaction cycle sequencing kit on an ABI Prism 3100 automated sequencer (Applied Biosystems, Foster City, CA) using the corresponding PCR primers. The remaining reaction conditions were according to the manufacturer's instructions.
RESULTS
Development of consensus degenerate primers.
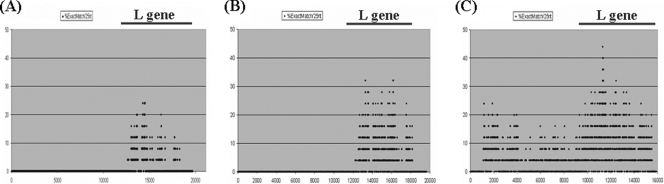
To design family- or subfamily-specific primers, all full-length genome sequences were obtained from the Paramyxoviridae family viral sequences archived in the GenBank viral database. Viral sequences with more than 95% similarity were treated as identical; the resulting 29 nonredundant full-length genome sequences were used initially to design primers for this study. First, a multiple full-length genome sequence alignment was performed by using Clustal W and was scanned according to a program written in house to identify genes that have the most-conserved multiple regions from the broadest possible grouping in the family Paramyxoviridae. The plot (Fig. 1) showed that the RNA polymerase (L) coding sequences were the most conserved, and thus, this gene was selected as the locus for the broadly reacting PCR assays. For the final primer design, we used an additional four L-gene sequences available in GenBank for a total of 33 L-gene-specific nonredundant sequences. Since the subfamilies, Paramyxovirinae and Pneumovirinae, are so genetically distinct, primers were designed for each separately. The highly conserved amino acid regions a with minimal length of six amino acids were checked for possible presence in other nontarget organisms by a BLAST search to avoid nonspecific amplification in PCR. Primers were designed from these highly conserved regions using all codon possibilities for 9 to 12 bases at the 3′ end of the primer and consensus sequences or inosines at positions of fourfold degeneracy for a “consensus clamp” at the 5′ end. After comparative analysis including sizes of amplicons, the similarity of reaction kinetics, nonspecific cross-reactivity, and experimental evaluation, three consensus degenerate primers (two for the first PCR and the third for the second, seminested PCR) corresponding to the most-conserved motifs were selected as the pan-Paramyxovirinae primers and three consensus degenerate primers corresponding to the most-conserved motifs were selected as the pan-Pneumovirinae primers, as shown in Table 2. Since the subfamily Paramyxovirinae has greater diversity among its members than the subfamily Pneumovirinae, we further divided the Paramyxovirinae into two subgroups of genera based on RNA polymerase gene relatedness, the Morbillivirus-Respirovirus-Henipavirus subgroup and the Rubulavirus-Avulavirus subgroup, and further developed primers to achieve less degeneracy and greater sensitivity, as noted in Table 2.
FIG. 1.
Similarity plots of the aligned paramyxoviruses' genomes. The plots were obtained using an in-house program based on multiple alignments of viral genomes from 29 different strains in the Paramyxoviridae (A), 22 different strains in the Paramyxovirinae (B), and 7 different strains in the Pneumovirinae (C). The identity percentage score given on the y axis was calculated based on the exact-match percentage with a window of 25 nucleotide positions, and the window was progressively moved across the alignment in 1-nucleotide-position steps. The x axis shows the first position of the window in the multiple alignments of the viral genomes.
TABLE 2.
Consensus degenerate primers used for detection of paramyxoviruses
| Primer name | Amino acid motif in RNA-dependent DNA polymerase | Targeted group |
|---|---|---|
| PAR-F1 | GAA GGI TAT TGT CAI AAR NTN TGG AC | Paramyxovirinae |
| PAR-F2 | GTT GCT TCA ATG GTT CAR GGN GAY AA | Paramyxovirinae |
| PAR-R | GCT GAA GTT ACI GGI TCI CCD ATR TTN C | Paramyxovirinae |
| RES-MOR-HEN-F1 | TCI TTC TTT AGA ACI TTY GGN CAY CC | Respirovirus, Morbillivirus, Henipavirus |
| RES-MOR-HEN-F2 | GCC ATA TTT TGT GGA ATA ATH ATH AAY GG | Respirovirus, Morbillivirus, Henipavirus |
| RES-MOR-HEN-R | CTC ATT TTG TAI GTC ATY TTN GCR AA | Respirovirus, Morbillivirus, Henipavirus |
| AVU-RUB-F1 | GGT TAT CCT CAT TTI TTY GAR TGG ATH CA | Avulavirus, Rubulavirus |
| AVU-RUB-F2 | ACA CTC TAT GTI GGI GAI CCN TTY AAY CC | Avulavirus, Rubulavirus |
| AVU-RUB-R | GCA ATT GCT TGA TTI TCI CCY TGN AC | Avulavirus, Rubulavirus |
| PNE-F1 | GTG TAG GTA GIA TGT TYG CNA TGC ARC C | Pneumovirinae |
| PNE-F2 | ACT GAT CTI AGY AAR TTY AAY CAR GC | Pneumovirinae |
| PNE-R | GTC CCA CAA ITT TTG RCA CCA NCC YTC | Pneumovirinae |
Standardization of optimized conditions.
Different combinations of RT and PCR steps including one tube/one step, one tube/two steps, and two tubes/two steps, have a significant effect on the outcome of the assays. By comparative testing, we observed that a one-tube/one-step RT-PCR protocol was not only more convenient and more sensitive (data not shown) but also minimized risk of handling cross-contamination. RT-PCRs were optimized for band intensity and low background by evaluating combinations of primer concentration, Mg2+ concentration, and annealing temperatures.
Broad reactivity of consensus degenerate primers.
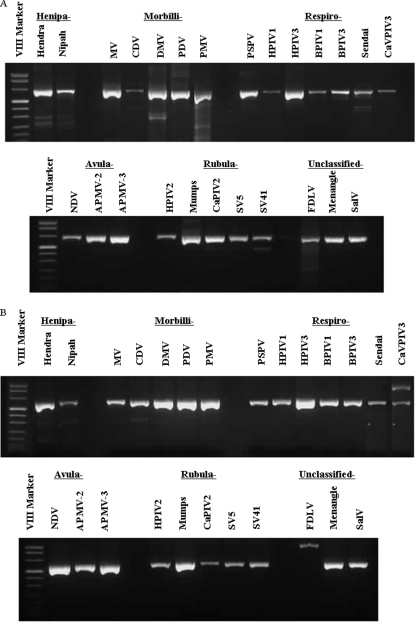
As shown in Fig. 2A and B, all of the 25 reference viruses representing five genera from the Paramyxovirinae subfamily were successfully detected by pan-Paramyxovirinae primers. As shown in Fig. 3 and 4, members of the two groups of genera within this subfamily were also detected by the appropriate subgroup-specific primers. The observed variation in amplicon intensity was probably due at least in part to differences in amount of template RNA for the respective viruses. No amplicons were detected for the PCR assays against the pooled “other respiratory pathogen genomes” or the negative controls (see Fig. 6). Two previously unclassified paramyxoviruses, FDLV and Salem virus (SalV), were amplified only by the Morbillivirus-Respirovirus-Henipavirus genus subgroup-specific primers and Menangle virus only by the Rubulavirus-Avulavirus genus subgroup-specific primers. These results suggest that FDLV and SalV strains are more closely related to the Morbillivirus-Respirovirus-Henipavirus subgroup and the Menanagle strain to the Rubulavirus-Avulavirus subgroup.
FIG. 2.
Amplification of RNAs from 25 different viral members in the subfamily Paramyxovirinae by one-step RT-PCR using the pan-PAR-F1/PAR-R primer pair (A) or the pan-PAR-F2/PAR-R primer pair (B). Viral names are abbreviated as shown in Table 1.
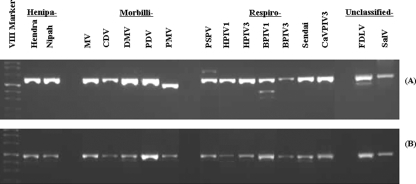
FIG. 3.
Amplification of RNAs from 14 different viral members in the genera of Henipavirus, Morbillivirus, and Respirovirus and two unclassified viral members in the subfamily Paramyxovirinae by one-step RT-PCR using the pan RES-MOR-HEN-F1/RES-MOR-HEN-R primer pair (A) or the pan RES-MOR-HEN-F2/RES-MOR-HEN-R primer pair (B). Each viral name is abbreviated as shown in Table 1.
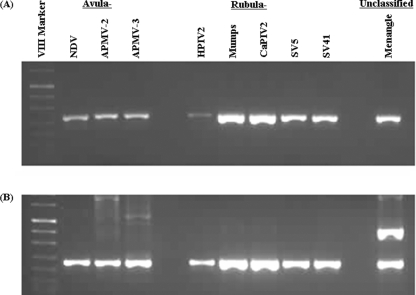
FIG. 4.
Gel electrophoresis of amplification products of a one-step RT-PCR assay against RNA from eight different members of Avulavirus and Rubulavirus genera and one previously unclassified member of the subfamily Paramyxovirinae. The pan AVU-RUB-F1/AVU-RUB-R primer pair (A) or the pan AVU-RUB-F2/AVU-RUB-R primer pair (B) was used. Each virus gives an appropriately sized band and is identified by its abbreviation as shown in Table 1.
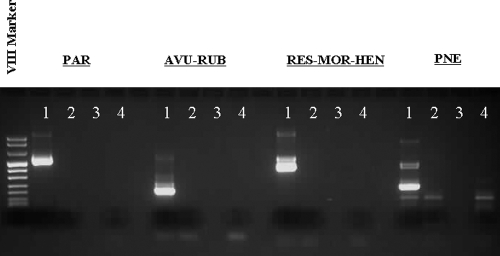
FIG. 6.
Gel electrophoresis of amplification products with subfamily and genus group seminested RT-PCR assays showing the appropriately sized positive band for the positive control material for the respective RT-PCR assay and no appropriately sized band for negative-control material or a pool of RNA from other common respiratory pathogens, as described in Materials and Methods. The seminested RT-PCR assays are pan-PAR (Paramyxovirinae subfamily), pan RES-MOR-HEN (group of Respirovirus, Morbillivirus, and Henipavirus genera), pan-AVU-RUB (group of Avulavirus and Rubulavirus genera), and pan-PNE (Pneumovirinae subfamily). Lanes: 1, positive control; 2, negative control (water); 3, blank; 4, pool of RNA from other common respiratory pathogens.
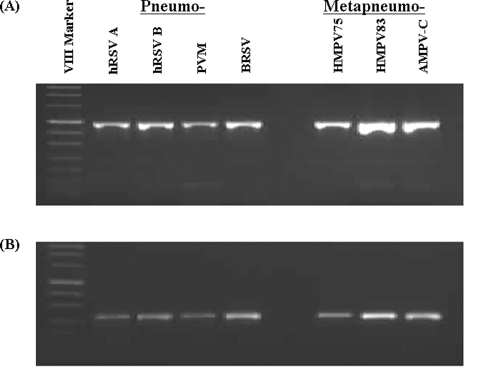
As shown in Fig. 5, the Pneumovirinae seminested PCR assay detected all of the seven tested reference viruses, and the specificity of the amplification was confirmed by sequence studies. These primers did not amplify RNA from other common respiratory viruses or the negative controls (Fig. 6).
FIG. 5.
Gel electrophoresis of amplification products of one-step RT-PCR assays against RNA from seven different members of the subfamily Pneumovirinae. (A) Pan PNE-F1/PNE-R primer pair. (B) Pan PNE-F2/PNE-R primer pair. Each virus gives an appropriately sized band and is identified by its abbreviation as shown in Table 1.
Sensitivity of consensus degenerate PCR assay.
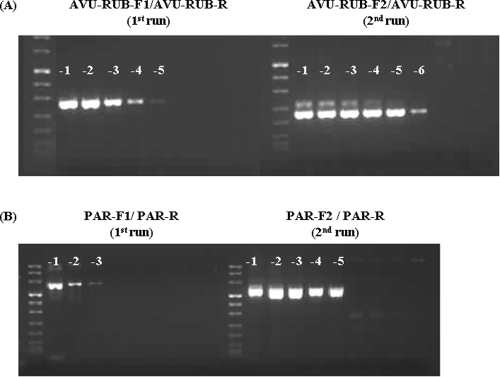
To test the sensitivity of the PCR assays, we used mumps virus RNA for both the Paramyxovirinae subfamily-specific primers and the Rubulavirus-Avulavirus genus subgroup-specific primers. Hendra viral RNA was used to evaluate the Morbillivirus-Respirovirus-Henipavirus genus subgroup-specific primers and respiratory syncytial virus RNA for the Pneumovirinae subfamily-specific primers. Generally, sensitivity was improved at least 10-fold with the addition of the nested or seminested step (Fig. 7). The same 10-fold serial dilutions of mumps virus RNA were used to compare the sensitivity between the Paramyxovirinae subfamily-specific RT-PCR and the Rubulavirus-Avulavirus genus subgroup-specific RT-PCR. As expected, the Rubulavirus-Avulavirus genus subgroup-specific primers with less degeneracy resulted in a 10-fold- to 100-fold-higher sensitivity than the Paramyxovirinae subfamily-specific primers. By using serial dilutions of RNA transcripts, the sensitivity limit was calculated as between 10 and 100 RNA copies for the Rubulavirus-Avulavirus subgroup-specific PCR, the Morbillivirus-Respirovirus-Henipavirus subgroup-specific PCR, and the Pneumovirinae subfamily-specific PCR and between 500 and 1,000 copies for the Paramyxovirinae subfamily-specific PCR(data not shown).
FIG. 7.
Improved detection sensitivity by seminested RT-PCR and by genus subgroup primers with less degeneracy. (A) Seminested RT-PCR amplification of RNA extracted from 10-fold serial dilution of mumps virus stock using the pan-AVU-RUB-F1/AVU-RUB-R primer pair (1st run) and the pan-AVU-RUB-F2/AVU-RUB-R primer pair (2nd run). (B) Seminested RT-PCR amplification of RNA extracted from 10-fold serial dilution of mumps virus stock using the pan-PAR-F1/PAR-R primer pair (1st run) and the pan-PAR-F2/PAR-R primer pair (2nd run).
Validation with clinical specimens.
To validate further the specificity and sensitivity of each of these group-specific PCRs with clinical samples, we tested a panel of 14 blinded samples provided by Dean Erdman (CDC), consisting of paramyxovirus-containing clinical samples, non-paramyxovirus-containing clinical samples, and cell culture isolates in different dilutions, which had been tested by agent-specific assays. As noted in Table 3, the appropriate subfamily-specific or genus subgroup-specific PCR assays correctly identified each of the specimens with the correct specificity.
TABLE 3.
Validation with clinical specimensa
| Specimen ID | PCR result
|
Sequence confirmation result | Blinded result | |||
|---|---|---|---|---|---|---|
| Paramyxovirinae | Rubulavirus- Avulavirus subgroup | Respirovirus- Morbillivirus- Henipavirus subgroup | Pneumovirinae | |||
| 2005495091 | Pos | Neg | Pos | Neg | HPIV3 | HPIV3 |
| 2005495105 | Pos | Neg | Pos | Neg | HPIV3 | HPIV3 |
| 2005495106 | Pos | Neg | Pos | Neg | HPIV3 | HPIV3 |
| 2002023415 | Neg | Neg | Neg | Pos | hRSV-B1 | hRSV-B1c |
| 2002023422 | Neg | Neg | Neg | Pos | hRSV-B1 | hRSV-B1 |
| 2002023428 | Neg | Neg | Neg | Neg | Neg | FLU A |
| 2002023434 | Neg | Neg | Neg | Neg | Neg | FLU A |
| 2002023436 | Pos | Neg | Pos | Neg | HPIV3 | HPIV3 |
| 2002023437 | Pos | Neg | Pos | Neg | HPIV3 | HPIV3 |
| 2002023644b | Neg | Neg | Neg | Neg | Neg | HMPV |
| 2002029001 | Neg | Neg | Neg | Pos | HMPV | HMPV |
| 2002029002 | Neg | Neg | Neg | Pos | HMPV | HMPV |
Pos, positive; Neg, negative. For other abbreviations, see Table 1.
The specimen 2002023644 had a low viral load (<10 copies/5 μl template) which is below the detection limit of the pan-Pneumovirinae RT-PCR.
hRSV-B1, human RSV-B1.
DISCUSSION
In this report, we describe successful development of a set of broadly reactive PCR assays for Paramyxoviridae. These assays were developed by identifying conserved sequences among members of various groups in the family Paramyxoviridae, using degenerate and inosine-containing primers to account for mismatches, a second, nested, PCR to improve sensitivity, and the consensus-degenerate hybrid oligonucleotide primer strategy (13) to improve sensitivity and specificity. In designing the broad-range primers, we first looked for conserved amino acid sequences in the RNA-dependent RNA polymerase protein coded by the L gene, the most conserved viral gene in the family Paramyxoviridae. The analysis of the RNA-dependent RNA polymerase proteins of paramyxoviruses indicated that they encompass three conserved domains (I, II, and III) separated by two nonconserved hinge regions, and strong selective constraints act against amino acid sequence changes in these three conserved domains (6, 11). They are predicted to be essential for the key functions of RNA binding, RNA replication, and protein kinase activity and may have retained the same structure, that of their putative common ancestor, as they have diverged in sequence. The invariance of these conserved sequences suggests that they may be ideal targets for the exploration of unidentified members in the Paramyxoviridae. The broad-range primers in this report were designed from the highly conserved domain I and II among paramyxovirus members.
The extensive variability within this family and the drop in sensitivity with increased primer degeneracy prevented us from developing a single assay for the family but instead led us to develop subfamily-specific and two genus subgroup PCR assays to achieve the desired level of sensitivity, <100 copies of RNA in the reaction mixture. The two genus subgroup assays took advantage of more closely related genera, i.e., a group of the Rubulavirus and Avulavirus genera and a group of the Morbillivirus, Respirovirus, and Henipavirus genera. The two genus subgroup assays reached the desired level of sensitivity, 10 and 100 copies, while the corresponding Paramyxovirinae subfamily assay achieved a sensitivity of 500 to 1,000 copies.
In summary, we have developed a set of seminested RT-PCR assays for detection of paramyxoviruses. The broad reactivity of these RT-PCR assays should allow us to detect known and novel members of the family Paramyxoviridae within genera described to date. The utility of these assays in discovery of novel members is supported by our ability to detect and classify eight recently isolated paramyxovirus species, Porpoise morbillivirus, Pacific salmon paramyxovirus, Bovine parainfluenza virus 1, Guinea pig parainfluenza virus 3, Canine parainfluenza virus 2, Canine parainfluenza virus 3, Menangle virus, and Salem virus, whose sequences were not available when the primers were designed. These pan-paramyxovirus PCR assays and similar assays for other viral families should enhance our ability to quickly identify, by virus family, subfamily, or genus, a wide range of novel viral pathogens and should enhance our ability to respond to and characterize outbreaks and diseases of unknown etiology.
Acknowledgments
We acknowledge many people who provided reference viruses and/or clinical specimens for this study. They are Jack King and Qingzhong Yu (U.S. Department of Agriculture); Paul Rota and Dean Erdman (CDC); Bert Rima (Queen's University of Belfast, Belfast, United Kingdom); Randy Renshaw (Cornell University); Lela K. Riley (University of Missouri); Gael Kurath (Western Fisheries Research Center, Seattle, WA); Yasuhiko Ito (Mie University School of Medicine, Japan); and Joseph Domachowske (State University of New York Upstate Medical University). A special acknowledgment is made for current and former lab personnel and their contributions, including Susan Ruone, Charryse Birdge, Wendy Tan, Bo Shu, Byron Cook, and Shannon Reed.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Anonymous. 1999. Outbreak of Hendra-like virus—Malaysia and Singapore, 1998-1999. MMWR Morb. Mortal. Wkly. Rep. 48265-269. [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Garcia-Sastre, and P. Palese. 2005. A novel paramyxovirus? Emerg. Infect. Dis. 11108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths, D. J., P. J. Venables, R. A. Weiss, and M. T. Boyd. 1997. A novel exogenous retrovirus sequence identified in humans. J. Virol. 712866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn, J. S. 2003. Human metapneumovirus: a newly emerging respiratory pathogen. Curr. Opin. Infect. Dis. 16255-258. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 6.McIlhatton, M. A., M. D. Curran, and B. K. Rima. 1997. Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected sequence). J. Gen. Virol. 78571-576. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery, S. M., D. L. Morris, R. E. Pounder, and A. J. Wakefield. 1999. Paramyxovirus infections in childhood and subsequent inflammatory bowel disease. Gastroenterology 116796-803. [DOI] [PubMed] [Google Scholar]
- 8.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262914-917. [DOI] [PubMed] [Google Scholar]
- 9.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3282-286. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier, G., P. Dery, Y. Abed, and G. Boivin. 2002. Respiratory tract reinfections by the new human Metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8976-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 711153-1162. [DOI] [PubMed] [Google Scholar]
- 12.Rima, B. K., and W. P. Duprex. 2006. Morbilliviruses and human disease. J. Pathol. 208199-214. [DOI] [PubMed] [Google Scholar]
- 13.Rose, T. M. 2005. CODEHOP-mediated PCR—a powerful technique for the identification and characterization of viral genomes. Virol. J. 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose, T. M., J. G. Henikoff, and S. Henikoff. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 313763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C. C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 714138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell, W. C. 1983. Paramyxovirus and morbillivirus infections and their relationship to neurological disease. Prog. Brain Res. 59113-132. [DOI] [PubMed] [Google Scholar]
- 17.Simons, J. N., T. P. Leary, G. J. Dawson, T. J. Pilot-Matias, A. S. Muerhoff, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1564-569. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki, T., M. Akimoto, M. Mandai, M. Takahashi, and N. Yoshimura. 2005. A new PCR-based approach for the preparation of RNA probe. J. Biochem. Biophys. Methods 62251-258. [DOI] [PubMed] [Google Scholar]