Abstract
Rifampin resistance is a key prognostic marker for treatment success in tuberculosis patients. Recently, Wang et al. demonstrated that Rv2629 A191C mutations were present in 99.1% of rifampin-resistant and 0% of rifampin-susceptible clinical Mycobacterium tuberculosis isolates and that overexpression of the Rv2629 191C allele in Mycobacterium smegmatis produced an eightfold increase in rifampin resistance. These results suggested that Rv2629 could be a cause of rifampin resistance and a valuable target for rifampin resistance detection assays. We developed a molecular-beacon assay to study the association between Rv2629 191 alleles and rifampin resistance in 246 geographically and phylogenetically diverse clinical M. tuberculosis isolates. The 191C allele was present in 30/98 (30.6%) rifampin-resistant isolates and 25/148 (16.9%) rifampin-susceptible isolates and was more common in isolates from Asia. Phylogenetic analysis demonstrated complete overlap between the 191C allele and single nucleotide polymorphism cluster group 2 (SCG-2), a phylogenetic lineage that corresponds to the Beijing-W clade of M. tuberculosis. All 55 (100%) 191C isolates were SCG-2, while none of the 191 191A isolates were SCG-2 (P < 0.001). No association was found between the 191C allele and rifampin resistance in an analysis that included the SCG type (P = 1.0). Also, in contrast to the findings of Wang et al., we found that overexpression of either Rv2629 191 allele in M. smegmatis did not produce an increase in rifampin resistance. We conclude that the Rv2629 191C allele is not associated with rifampin resistance and that the allele cannot be used as a molecular target to detect rifampin resistance. The allele appears to be an excellent marker for the Beijing-W clade/SCG-2 phylogenetic group.
Resistance to the drug rifampin complicates treatment of Mycobacterium tuberculosis infections and is a key component of both multidrug-resistant (MDR) and extensively drug-resistant tuberculosis (5, 14). Rifampin resistance has been used as a surrogate marker for MDR tuberculosis, and several widely used molecular assays have been developed to detect this type of resistance (7, 17, 18). Approximately 95% of all clinical rifampin-resistant M. tuberculosis isolates contain mutations in a short region of the rpoB gene termed the rpoB “core region.” Rapid assays that detect mutations in this region have excellent sensitivity and specificity for identifying rifampin-resistant and MDR tuberculosis (7, 17, 19, 25). The causes of rifampin resistance in the approximately 5% of clinical M. tuberculosis isolates without rpoB mutations have remained a mystery for over a decade. Recently, Wang et al. (26) reported that 111 of 112 rifampin-resistant M. tuberculosis isolates contained an A-to-C mutation at position 191 of the Rv2629 gene, while none of 30 rifampin-susceptible isolates contained mutations at this site. Interestingly, Rv2629 A191C mutations were also detected in rifampin-resistant isolates that did not contain any rpoB mutations. This suggested that the Rv2629 191C allele might be a better indicator of rifampin resistance than the detection of rpoB core mutations. The same group also showed that the Rv2629 protein was induced by treating M. tuberculosis with rifampin and that overexpression of the 191C mutant, but not the wild-type M. tuberculosis Rv2629 gene, conferred an eightfold increase in the rifampin MIC of Mycobacterium smegmatis.
The results reported by Wang et al. (26) were highly significant in that they suggested that Rv2629 was a major cause of rifampin resistance and an outstanding diagnostic marker. However, the study had been performed on a relatively small number of isolates, which were all of Asian origin. It was important to validate these findings on a more geographically and biologically diverse set of M. tuberculosis isolates. We designed a molecular-beacon-based assay to rapidly differentiate between the Rv2629 191A and -C alleles. We then applied this assay to a global reference set of drug-resistant and drug-susceptible M. tuberculosis isolates assembled by the World Health Organization (WHO) and by our laboratory to further investigate the association between Rv2629 alleles and rifampin resistance. Here, we report that Rv2629 A191C mutations are not associated with rifampin resistance in this global strain set. We further report that overexpression of the Rv2629 gene containing the 191C allele does not increase the rifampin MIC of M. smegmatis. Instead, we present evidence that this mutation is deeply rooted in the M. tuberculosis Beijing-W clade-single nucleotide polymorphism (SNP) cluster group 2 (SCG-2) phylogenetic lineage and should serve as an excellent marker for this important group of strains. These results have important implications for the development of rifampin resistance detection assays and for our understanding of the causes of rifampin resistance.
MATERIALS AND METHODS
M. tuberculosis isolates.
The 245 strains analyzed in this study included a pedigreed collection of 211 highly characterized M. tuberculosis isolates established by the United Nations Children's Fund/UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases. The culture isolates, originating in 88 different countries and five continents, were regrown from single colonies to ensure clonality; characterized for isoniazid, rifampin, ethambutol, and streptomycin resistance phenotype by the proportions method to determine the MIC; and characterized for genotype by sequencing the katG, inhA, rpoB, embB, rpsL, and rrs genes, which have known resistance-associated mutations. An additional phylogenetically diverse set of 35 drug-susceptible M. tuberculosis isolates from a previous study (8) were also analyzed. Details of the antibiotic susceptibility profiles of the study isolates are shown in Table 1.
TABLE 1.
Resistance phenotypes of the tested clinical DNA samples
| Resistance phenotypea | Frequency of recovery |
|---|---|
| R | 9 |
| I | 8 |
| S | 12 |
| E | 10 |
| RI | 17 |
| RS | 5 |
| RE | 0 |
| IS | 16 |
| IE | 7 |
| SE | 2 |
| RIS | 15 |
| RIE | 16 |
| RSE | 1 |
| ISE | 10 |
| RISE | 35 |
| Pansusceptible | 83 |
| Total | 246 |
R, rifampin; I, isoniazid; S, streptomycin; E, ethambutol.
Molecular-beacon assay.
The real-time PCR assay to detect Rv2629 191A and -C alleles amplified a 113-bp region of the Rv2629 gene spanning nucleotides 136 to 248 by PCR using primers Rv2629MB-F (5′-GAAAGCCGCGACGCGAAGCAGGAG-3′) and Rv2629MB-R (5′-ACTTGCTCGCCGGTCGCGATCA-3′). Two molecular-beacon probes, MB-191A (6-carboxyfluorescein-5′-CGCGTCGTCGAGAATCCCGCACCGGACGCG-DABCYL-3′) [where the underlined sequences represent the stems of the probes and DABCYL represents (4-(4-dimethylamino)phenylazo)benzoic acid] and MB-191C (tetrachloro-6-carboxyfluorescein [TET]-5′-CGCGTCGTCGAGAAGCCCGCACCGGACGCG-DABCYL-3′) (Sigma-Aldrich), were used to detect the presence of the 191A and the 191C alleles, respectively, in a multiplex assay format. Each assay contained 1× PCR buffer, 300 μM deoxynucleoside triphosphates, 4 mM MgCl2, 1.0 μM of each primer, 0.06 U/μl of Jumpstart Taq polymerase (Sigma-Aldrich), 4 ng/μl of MB-191A, 3 ng/μl MB-191C, and 2 μl of M. tuberculosis DNA (approximately 2 to 4 ng per reaction) in a final reaction volume of 20 μl. PCRs were performed in 384-well microtiter plates in an ABI 7900 prism spectrofluorimetric thermal cycler (Applied Biosystems) according to the following parameters: initial denaturation at 95°C for 1 min, followed by 45 cycles of denaturation at 95°C for 15 seconds, annealing at 64°C for 30 seconds, and extension at 72°C for 15 seconds. The fluorescence was recorded during the annealing step of the assay.
SCG assignment.
Each M. tuberculosis isolate was assigned to one of 10 SCGs or SNP cluster subgroups as described previously (1) and then mapped onto a previously described phylogenetic tree (8) using nine SNP markers developed for the purpose (1). The allele of each SNP marker was identified using hairpin primer PCR assays as described previously (12).
Cloning of the Rv2629 191A and 191C alleles and overexpression in M. smegmatis.
The cloning and overexpression of the two Rv2629 alleles in M. smegmatis strain mC2155 was carried out using the same primer sequences and the same pMV261 vector described by Wang et al. (26). Briefly, oligonucleotide primers that incorporate the BamHI and HindIII digestion sites were used to PCR amplify both the 191A and 191C alleles of the Rv2629 gene from clinical M. tuberculosis DNA. The genes were cloned into the mycobacterial overexpression shuttle vector pMV261 to generate constructs pMV261::Rv2629A and pMV261::Rv2629C. These new plasmids were then electroporated into M. smegmatis mc2155 using Gene Pulsar X-Cell (Bio-Rad) to create the M. smegmatis mc2155(pMV261::Rv2629A) strain NJS30A and the M. smegmatis mc2155(pMV261::Rv2629C) strain NJS30C, respectively. The M. smegmatis transformants were selected on 7H10 medium (Becton Dickinson) containing 25 μg/ml of kanamycin sulfate (Sigma) and 10% albumin dextrose catalase. The sequences and orientations of the inserts, including the entire sequence of the cloned Rv2629 genes, were confirmed by PCR sequencing of the constructs isolated from the transformed M. smegmatis strains.
Drug susceptibility testing.
Drug susceptibility testing for rifampin, isoniazid, and ethambutol was performed for all the recombinant M. smegmatis clones and the parental mc2155 strain using the proportions method as described by the CLSI (16). MIC tests for rifampin were performed twice to confirm the results.
Reverse transcriptase and quantitative real-time PCR.
Total RNA was isolated from each log-phase M. smegmatis strain cultured to an optical density of 0.6, using the FastRNA Pro RNA isolation kit (Q-Biogene). The isolated RNA was subjected to DNase treatment and purified using the RNeasy mini kit (Qiagen). Reverse transcriptase (RT) PCR was carried out using the C. therm Polymerase 1-Step RT-PCR kit (Roche), followed by quantitative real-time PCR for the Rv2629 gene using the molecular-beacon assay as described above. The expression of the cloned Rv2629 genes was quantified by normalizing it against 16S rRNA gene expression as described previously (6). The primer and the molecular-beacon sequences targeting the 16S rRNA gene of M. smegmatis were as follows: Msm16SRT primer, 5′-ACCGCGGCTGCTGGCACGTAGTTGG-3′; cDNA amplification primers, Msm16SF (5′-CTACGGGAGGCAGCAGTGGGGAATA-3′) and Msm16SR (5′-CACGTAGTTGGCCGGTCCTTCTTCTGC-3′); and molecular-beacon probe targeting the 16S rRNA gene, Bcn.16S.smeg (5′-TET-ACGCGCCAAGCCTGATGCAGCGACGCGCGCGT-DABCYL-3′).
Statistical analysis.
Tests of association, both overall and for the various stratified analyses, were carried out using either chi-square tests or Fisher exact tests, depending on the relevant sample size. SAS (version 8.0) was used for all calculations.
RESULTS
Detection of Rv2629 alleles by molecular-beacon assay.
We developed a molecular-beacon assay to rapidly test large numbers of M. tuberculosis DNA samples for the presence of either the Rv2629 191A or -C allele. One molecular beacon (MB-191A) was designed to be specific for the Rv2629 191A allele and was labeled with the fluorophore 6-carboxyfluorescein, and a second molecular beacon (MB-191C) was designed to be specific for the Rv2629 191C allele and was labeled with the fluorophore TET. Both molecular beacons were then placed in the same reaction well, enabling each DNA sample to be tested simultaneously for both of the Rv2629 191 alleles. Reaction wells containing DNA with the 191A allele caused MB-191A to fluoresce brightly while MB-191C did not fluoresce, and PCR wells containing DNA with the 191C allele caused MB-191C to fluoresce brightly while MB-191A did not fluoresce. Reaction wells containing no DNA did not produce a significant signal for either molecular beacon. All 246 of the M. tuberculosis DNA samples tested with this assay produced an unambiguous allele call, identifying the presence of either the 191A or -C allele, when adequate amounts of DNA were placed in the PCR well. One hundred ninety-one (77.6%) of the samples were found to carry the 191A allele, and 55 (22.3%) of the samples were found to contain the 191C allele. All negative control wells remained negative throughout the course of the study. The entire Rv2629 gene was sequenced from six M. tuberculosis samples found to contain the 191A allele and six samples found to contain the 191C allele by the molecular-beacon assay. The DNA sequencing confirmed that all of the molecular-beacon allele calls were correct.
Rv2629 alleles, drug resistance, and continent of origin.
Wang et al. (26) described a very strong association between the Rv2629 191C allele and rifampin resistance. The molecular-beacon assay was used to test our study set for a similar association between either Rv2629 191 allele type and phenotypic drug resistance. Univariate analysis (Table 2) showed a statistically significant association between the 191C allele and both rifampin resistance (P = 0.011) and isoniazid resistance (P = 0.005). However, the 191C allele was present in both rifampin-resistant (30/98; 30.6%) and rifampin-susceptible (25/148; 16.9%) isolates, suggesting that this statistical association was due to confounding rather than causality. Analysis by continent of origin showed that the 191C allele was also much more common in isolates from Asia (40/86; 46.5%) than in those from other continents (15/160; 9.4%) (P < 0.001). Analysis of the association between the 191C allele, the drug resistance phenotype, and the country of origin showed that the significant association with rifampin and isoniazid resistance was present only in the Asian samples (P = 0.003 and 0.025, respectively) and not in non-Asian samples (P = 0.875 and 0.873, respectively) (Table 3).
TABLE 2.
Rv2629 191A/C allele distribution
| Characteristica | Rv2629 191C [no. (%)] | Rv2629 191A [no. (%)] | P |
|---|---|---|---|
| RIF resistant | 30 (30.6) | 68 (69.4) | 0.011 |
| RIF susceptible | 25 (16.9) | 123 (83.1) | |
| INH resistant | 37 (29.8) | 87 (70.2) | 0.005 |
| INH susceptible | 18 (14.8) | 104 (85.2) | |
| EMB resistant | 21 (25.9) | 60 (74.1) | 0.347 |
| EMB susceptible | 34 (20.6) | 131 (79.4) | |
| STR resistant | 26 (27.1) | 70 (72.9) | 0.155 |
| STR susceptible | 29 (19.3) | 121 (80.7) | |
| Pansusceptible | 11 (13.3) | 72 (86.7) | 0.001 |
| Continent of origin | <0.001 | ||
| Asia | 40 (46.5) | 46 (53.5) | |
| Africa | 1 (2.0) | 50 (98.0) | |
| Europe | 1 (5.6) | 17 (94.4) | |
| North America | 4 (16.7) | 20 (83.3) | |
| South America | 5 (12.2) | 36 (87.8) | |
| Unknown | 4 (15.4) | 22 (84.6) | |
| SCG | <0.001 | ||
| 1 | 0 (0.0) | 23 (100.0) | |
| 2 | 55 (100.0) | 0 (0.0) | |
| 3a | 0 (0.0) | 11 (100.0) | |
| 3b | 0 (0.0) | 35 (100.0) | |
| 3c | 0 (0.0) | 7 (100.0) | |
| 4 | 0 (0.0) | 7 (100.0) | |
| 5 | 0 (0.0) | 83 (100.0) | |
| 6a | 0 (0.0) | 18 (100.0) | |
| 6b | 0 (0.0) | 7 (0.0) | |
| 7 | 0 (0.0) | 0 (0.0) |
RIF, rifampin; INH, isoniazid; EMB, ethambutol; STR, streptomycin.
TABLE 3.
Rv2629 A191C allele and drug resistance in Asian and non-Asian isolates
| Susceptibility profilea | Rv2629 191 allele
|
P | |
|---|---|---|---|
| C [no. (%)] | A [no. (%)] | ||
| Asian | |||
| RIF resistant | 24 (64.9) | 13 (35.1) | 0.003 |
| RIF susceptible | 16 (32.7) | 33 (67.3) | |
| INH resistant | 31 (55.4) | 25 (44.6) | 0.025 |
| INH susceptible | 9 (30.0) | 21 (70.0) | |
| EMB resistant | 18 (51.4) | 17 (48.6) | 0.449 |
| EMB susceptible | 22 (43.1) | 29 (56.9) | |
| STR resistant | 19 (45.2) | 23 (54.7) | 0.817 |
| STR susceptible | 21 (47.7) | 23 (52.3) | |
| Non-Asian | |||
| RIF resistant | 6 (9.8) | 55 (90.2) | 0.875 |
| RIF susceptible | 9 (9.1) | 90 (90.9) | |
| INH resistant | 6 (8.8) | 62 (91.2) | 0.837 |
| INH susceptible | 9 (9.8) | 83 (90.2) | |
| EMB resistant | 3 (6.5) | 43 (93.5) | 0.432 |
| EMB susceptible | 12 (10.5) | 102 (89.5) | |
| STR resistant | 7 (13.0) | 47 (87.0) | 0.266 |
| STR susceptible | 8 (7.6) | 98 (92.5) | |
RIF, rifampin; INH, isoniazid; EMB, ethambutol; STR, streptomycin.
Rv2629 alleles, phylogenetic group, and drug resistance.
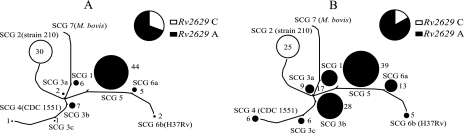
The association between the Rv2629 191C allele and M. tuberculosis isolates of Asian origin suggested that the allele might be associated with a particular phylogenetic linage rather than drug resistance. In fact, there is a known association between particular M. tuberculosis phylogenetic groups and the country of origin (13, 21, 23). Phylogenetic analysis using SNP markers has been established as a useful method to unambiguously establish the phylogenetic lineages of M. tuberculosis isolates in a number of regional and global studies (1, 8, 9, 11). An analysis of nine key SNP markers enabled us to assign all 246 of the DNA samples to one of the known SCGs and SNP cluster subgroups. Analysis of Rv2629 191 alleles by SCG revealed a very strong association between the 191C allele and SCG-2. The results showed that 55/55 (100%) of the isolates containing the 191C allele belonged to SCG-2 and that all SCG-2 isolates contained the 191C allele. The complete colocalization of the 191C allele and SCG-2 existed for both rifampin-resistant (Fig. 1A) and rifampin-susceptible (Fig. 1B) isolates. The remaining 191 isolates in our study carrying the 191A allele were distributed in five different SCGs, including both the rifampin-resistant (Fig. 1A) and rifampin-susceptible (Fig. 1B) isolates, indicating that our sample set was phylogenetically diverse.
FIG. 1.
Phylogenetic distribution of M. tuberculosis isolates by SCG. (A) Distribution of rifampin-resistant isolates. (B) Distribution of rifampin-susceptible isolates. The circle sizes reflect the numbers of M. tuberculosis isolates in each SCG. ○, isolates containing the Rv2629 191C allele; •, isolates containing the Rv2629 191A allele.
Our results suggested that the 191C allele might be independently associated with the SCG type only and not with the drug resistance phenotype or continent of origin. Stratifying our analysis by SCG-2 confirmed that the 191C allele was not associated with any type of drug resistance or continent of origin when the phylogenetic groups of the isolates were taken into account (P = 1.0 in all cases). We examined the known genome sequence and drug susceptibility of the M. tuberculosis 210 strain as a final test for the association between the Rv2629 191C allele and SCG-2 rather than rifampin resistance (Table 4). This strain has been the subject of whole-genome sequencing studies (3). It is known to be SCG-2 and is at least as rifampin susceptible (MIC 0.05 μg/ml) as the M. tuberculosis SCG-6A laboratory strain H37Rv (MIC = 0.08 μg/ml) (20). As predicted, the 210 strain also had an Rv2629 191C allele and was fully susceptible to isoniazid, rifampin, and ethambutol (Table 4).
TABLE 4.
MICs of M. tuberculosis and M. smegmatis strains
| Strain | Rv2629 191 allele | MIC (μg/ml)
|
||
|---|---|---|---|---|
| Rifampin | Isoniazid | Ethambutol | ||
| M. tuberculosis | ||||
| H37Rv | A | 0.08-2a | 0.03a | 2.5a |
| 210 | C | 0.05a | 0.05a | 2a |
| M. smegmatis | ||||
| mc2155 | 30b | 2.5c | 0.5b | |
| NJS31d | 40b | 2.5c | 0.5b | |
| NJS30Ae | A | 40b | 2.5c | 0.5b |
| NJS30Cf | C | 40b | 2.5c | 0.5b |
Performed by the Bactec method.
Performed by the proportions method using 7H10 medium.
Performed by the proportions method using 7H11 medium.
M. smegmatis NJS31 contains empty pMV261 vector control.
M. smegmatis NJS30A is an Rv2629 191A allele overexpression strain.
M. smegmatis NJS30C is an Rv2629 191C allele overexpression strain.
Effect of Rv2629 overexpression in M. smegmatis on rifampin susceptibility.
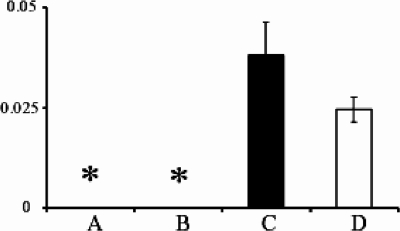
One of the key findings by Wang et al. (26) suggesting a role for the Rv2629 191C allele in rifampin resistance was their observation that overexpression of the Rv2629 191C allele, but not the 191A allele, in M. smegmatis strain mc2155 increased rifampin resistance eightfold. Our negative-association studies caused us to question this result. We overexpressed both Rv2629 191A and -C alleles in M. smegmatis mc2155, being careful to use a cloning strategy and an expression vector identical to those described by Wang et al. (26). These efforts created the M. smegmatis mc2155(pMV261::Rv2629A) strain NJS30A, containing the 191A allele, and the M. smegmatis mc2155(pMV261::Rv2629C) strain NJS30C, containing the 191C allele. DNA sequencing of plasmid DNAs extracted from both strains confirmed that each Rv2629 gene was properly inserted into the vector, that the expected 191A or -C allele was present, and that no additional mutations in the Rv2629 genes had been introduced during the cloning process. Real-time RT-PCR studies (Fig. 2) also confirmed that the Rv2629 genes were strongly overexpressed in NJS30A and NJS30C compared to the M. smegmatis mc2155 parental strain or to an M. smegmatis mc2155 control containing the empty pMV261 vector (NJS31).
FIG. 2.
Expression levels of Rv2629 mRNA in M. smegmatis strains. The mean ± 1 standard deviation of three real-time RT-PCR experiments is shown for each strain. The values were normalized to 16S mRNA levels. (A) Parental strain mc2155. (B) Vector-only control strain NJS31. (C) Rv2629 191A allele overexpression strain NJS30A. (D) Rv2629 191C allele overexpression strain NJS30C. *, fewer than 10 molecules of Rv2629 mRNA.
We tested the MIC of each M. smegmatis mc2155 strain to rifampin, isoniazid, and ethambutol using the proportions method to assess the effect of overexpressed Rv2629 alleles and drug resistance. Our results failed to show any effect of Rv2629 191A or -C allele overexpression on the MIC to any drug (Table 4). Notably, the rifampin MIC was 30 μg/ml for M. smegmatis mc2155 and only 40 μg/ml for all three recombinant strains, NJS30A, NJS30C, and the empty-vector control strain NJS31. These in vitro data further confirm our association studies, suggesting that there is no significant relationship between Rv2629 and rifampin resistance.
DISCUSSION
Rifampin resistance is a key prognostic marker for treatment success in patients with tuberculosis. Most patients diagnosed with rifampin-resistant tuberculosis have difficult-to-treat MDR or extensively drug-resistant tuberculosis (4, 22). Even rifampin-monoresistant tuberculosis responds poorly to standard therapy (15). Rapid assays for rifampin resistance may improve treatment outcomes in patients with rifampin-resistant tuberculosis by quickly identifying the need for alternate therapy. Currently available molecular markers for rifampin resistance fail to identify approximately 5% of rifampin-resistant clinical M. tuberculosis isolates. Furthermore, most of the mutations that are currently used to identify rifampin resistance are spread out over an 81-bp region of the rpoB gene, which complicates the design of a molecular diagnostic assay. The possibility, suggested by Wang et al. (26), that virtually all rifampin-resistant M. tuberculosis isolates might be identified by a single SNP in the Rv2629 gene was of great importance in this setting. The suggestion that Rv2629 191C allele overexpression could increase the rifampin MIC was also of great potential interest, as this observation might suggest ways to overcome or inhibit rifampin resistance that could be associated with Rv2629 induction or codon 191 mutation.
Our study failed to confirm the observations made by Wang et al. related to rifampin resistance (26). The Rv2629 191C allele appeared to be associated with rifampin and isoniazid resistance in Asian strains, but many Asian strains containing the 191C allele were rifampin and/or isoniazid susceptible. Phylogenetic analysis confirmed that the 191C allele was in fact associated with SCG-2 isolates rather than rifampin resistance or the continent of origin. Furthermore, we failed to detect a significant increase in the rifampin MIC when we overexpressed the Rv2629 191C gene in M. smegmatis. It is important to note that we did discover that the Rv2629 191C allele is likely to be an excellent marker for M. tuberculosis members of the SCG-2 lineage. SCG-2 status is strongly correlated with membership in the Beijing-W clade of M. tuberculosis as defined by spoligotyping (8). Beijing-W clade isolates have been implicated as potentially more pathogenic and more likely to become drug resistant (2, 10). Therefore, our molecular-beacon assay, which easily distinguishes between the Rv2629 191A and -C alleles, may prove to be a useful method to identify Beijing-W clade strains for epidemiological or clinical purposes.
What are the possible explanations for the discrepancies between our study and that of Wang et al. (26)? It is possible that there was a serious and consistent error in rifampin MIC determinations in our study sample. Molecular analysis of the rpoB core region of our study strains suggests that this possibility is highly unlikely. A total of 94/98 rifampin-resistant isolates in the WHO collection had a mutation in the rpoB core region that is currently considered diagnostic for rifampin resistance, while none of the 113 rifampin-susceptible isolates in the collection had any rpoB core region mutation. The nearly perfect correlation between an established genotypic marker and phenotypic rifampin resistance rules out a fundamental error in rifampin susceptibility testing. Furthermore, we found that the M. tuberculosis 210 strain, which is well established to be rifampin susceptible, also contained the Rv2629 191C allele. Repeat MIC testing of the 210 strains in our laboratory confirmed a low rifampin MIC consistent with rifampin susceptibility. Second, it is possible that our molecular-beacon assay provided inaccurate identification of the Rv2629 191 alleles. This also appears to be highly unlikely, as DNA sequencing of 12 isolates (6 with the 191A and 6 with the 191C allele) provided results identical to those of the molecular-beacon assay.
It is possible that the clinical drug resistance association study by Wang et al. suffered from a selection bias. We suggest that all of the 111 rifampin-resistant Rv2629 191C allele clinical M. tuberculosis isolates described by Wang et al. were members of the Beijing-W clade/SCG-2 phylogenetic lineage. Beijing-W clade/SCG-2 isolates are commonly identified in M. tuberculosis isolates of Asian origin; therefore, it is easy to see how this type of sampling bias might have occurred (8, 24). In contrast, we suggest that all of the control drug-susceptible isolates in the same study must have inadvertently been members of other non-SCG-2 phylogenetic lineages. Should this have been the case, it is possible that different selection criteria were applied to rifampin-resistant and rifampin-susceptible isolates, which resulted in an unintentional “phylogenetic bias.”
It is more difficult to understand the differences between our results and those of Wang et al. in regard to the Rv2629 overexpression studies and the rifampin MIC in M. smegmatis. We were careful to use the same cloning strategy, cloning vector, and M. smegmatis strain described in their investigation. We confirmed the correctness of our constructs by DNA sequencing, and we confirmed overexpression of the Rv2629 gene by RT-PCR assays. One possibility is that Wang et al. accidentally introduced rifampin resistance at another chromosomal site during the cloning and selection process. Additional experiments may need to be performed by others to confirm our findings. However, our overexpression results are entirely consistent with our clinical-association study showing no association between the Rv2629 191C allele and rifampin resistance.
In conclusion, we failed to confirm any association between the Rv2629 191C allele and rifampin resistance or any other type of drug resistance. This suggests that the mechanisms underlying rifampin resistance in the 5% of M. tuberculosis isolates that do not contain rpoB mutations remain to be identified. However, we did discover that the Rv2629 191C allele appears to be an excellent molecular marker for Beijing-W clade/SCG-2 M. tuberculosis isolates. Our results demonstrate the power of phylogenetic analysis in sorting out the biological relevance of specific mutations. Phylogenetic analysis can also be used as an important criterion to ensure full representation of the M. tuberculosis species in survey studies. The molecular-beacon assay presented here is likely to be a useful tool for rapidly identifying Beijing-W clade or SCG-2 M. tuberculosis isolates.
Acknowledgments
This work was supported by NIH grants AI52523 and AI065663 and a grant from the Foundation for Innovative Diagnostics.
We gratefully acknowledge the United Nations Children's Fund/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases for supplying the banked M. tuberculosis strains.
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 4539-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 1045-52. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9452-458. [DOI] [PubMed] [Google Scholar]
- 4.Caminero, J. A. 2006. Treatment of multidrug-resistant tuberculosis: evidence and controversies. Int. J. Tuberc. Lung Dis. 10829-837. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2007. Extensively drug-resistant tuberculosis—United States, 1993-2006. MMWR Morb. Mortal. Wkly. Rep. 56250-253. [PubMed] [Google Scholar]
- 6.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbon, R. Harbacheuski, N. J. Megjugorac, W. R. Jacobs, Jr., A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 551829-1840. [DOI] [PubMed] [Google Scholar]
- 7.El-Hajj, H. H., S. A. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 394131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7328-337. [DOI] [PubMed] [Google Scholar]
- 10.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193121-128. [DOI] [PubMed] [Google Scholar]
- 12.Hazbon, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 421236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 1014871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao, W. W., I. Mokrousov, G. Z. Sun, M. Li, J. W. Liu, O. Narvskaya, and A. D. Shen. 2007. Molecular characteristics of rifampin and isoniazid resistant Mycobacterium tuberculosis strains from Beijing, China. Chin. Med. J. 120814-819. [PubMed] [Google Scholar]
- 15.Mitchison, D. A., and A. J. Nunn. 1986. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am. Rev. Respir. Dis. 133423-430. [DOI] [PubMed] [Google Scholar]
- 16.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes. Approved standard M24-A. NCCLS, Wayne, PA. [PubMed]
- 17.Ozkutuk, N., H. Gazi, S. Surucuoglu, A. Gunduz, and B. Ozbakkaloglu. 2007. Characterization of rpoB mutations by line probe assay in rifampicin-resistant Mycobacterium tuberculosis clinical isolates from the Aegean region in Turkey. Jpn. J. Infect. Dis. 60211-213. [PubMed] [Google Scholar]
- 18.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigouts, L., O. Nolasco, P. de Rijk, E. Nduwamahoro, A. Van Deun, A. Ramsay, J. Arevalo, and F. Portaels. 2007. Newly developed primers for comprehensive amplification of the rpoB gene and detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45252-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safi, H., B. Sayers, M. H. Hazbon, and D. Alland. 2008. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob. Agents Chemother. 522027-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53680-689. [DOI] [PubMed] [Google Scholar]
- 22.Talay, F., S. Kumbetli, and S. Altin. 2008. Factors associated with treatment success for tuberculosis patients: a single center's experience in Turkey. Jpn. J. Infect. Dis. 6125-30. [PubMed] [Google Scholar]
- 23.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 1014865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 333234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varma-Basil, M., H. El-Hajj, R. Colangeli, M. H. Hazbon, S. Kumar, M. Bose, M. Bobadilla-del-Valle, L. G. Garcia, A. Hernandez, F. R. Kramer, J. S. Osornio, A. Ponce-de-Leon, and D. Alland. 2004. Rapid detection of rifampin resistance in Mycobacterium tuberculosis isolates from India and Mexico by a molecular beacon assay. J. Clin. Microbiol. 425512-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Q., J. Yue, L. Zhang, Y. Xu, J. Chen, M. Zhang, B. Zhu, and H. Wang. 2007. A newly identified 191A/C mutation in the Rv2629 gene that was significantly associated with rifampin resistance in Mycobacterium tuberculosis. J. Proteome Res. 64564-4571. [DOI] [PubMed] [Google Scholar]