Abstract
The chaplins are a family of eight secreted proteins that are critical for raising aerial hyphae in Streptomyces coelicolor. These eight chaplins can be separated into two main groups: the long chaplins (ChpA to -C) and the short chaplins (ChpD to -H). The short chaplins can be further subdivided on the basis of their abilities to form intramolecular disulfide bonds: ChpD, -F, -G, and -H contain two Cys residues, while ChpE has none. A “minimal chaplin strain” containing only chpC, chpE, and chpH was constructed and was found to raise a substantial aerial mycelium. This strain was used to examine the roles of specific chaplins. Within this strain, the Cys-containing ChpH was identified as the major polymerization unit contributing to aerial hypha formation and assembly of an intricate rodlet ultrastructure on the aerial surfaces, and the two Cys residues were determined to be critical for its function. ChpC augmented aerial hypha formation and rodlet assembly, likely by anchoring the short chaplins to the cell surface, while ChpE was essential for the viability of wild-type S. coelicolor. Interestingly, the lethal effects of a chpE null mutation could be suppressed by the loss of the other chaplins, the inactivation of the twin arginine translocation (Tat) secretion pathway, or the loss of the rodlins.
The gram-positive soil-dwelling streptomycetes have a mycelial growth habit that culminates in the formation of dormant exospores that permit survival under adverse environmental conditions (13). Germinating spores produce one or more germ tubes that grow by tip extension to form a network of branching vegetative hyphae known as the vegetative mycelium. Antibiotics (and other secondary metabolites) are produced within the vegetative hyphae, and from this vegetative mycelial network emerge specialized reproductive structures known as aerial hyphae. These aerial hyphae undergo a number of maturation steps, including a synchronous round of cell division, to differentiate into chains of unigenomic spores.
The transition from vegetative growth in an aqueous environment to the emergence of aerial hyphae into the air requires significant adaptation of the cell surface: the surfaces of vegetative hyphae are hydrophilic, while those of aerial hyphae and spores are extremely hydrophobic. Three groups of proteins are known to be involved in the modulation of cell surfaces during aerial hypha formation in Streptomyces coelicolor: the chaplins, the rodlins, and SapB (reviewed in references 8, 16, and 36). These proteins are thought to collectively function like the fungal hydrophobins, which are important for surface modulation and aerial growth in the filamentous fungi (reviewed in reference 37). Hydrophobins are small secreted proteins that assemble into a distinctive “rodlet” layer on the fungal cell surface; this rodlet ultrastructure is a feature shared with a number of Streptomyces species, including S. coelicolor. As their name implies, hydrophobins impart hydrophobic characteristics to the fungal cell surface that help in attachment to other surfaces (host plants, nutrient sources, etc.). They are defined by a characteristic pattern of eight cysteine residues that interact to form four intramolecular disulfide bridges but otherwise do not share significant sequence similarity with each other or with other proteins. These four disulfide bridges serve to stabilize a compact, globular protein structure with amphiphilic properties (19, 26). Hydrophobins, like the chaplins and SapB, are highly surface active and are capable of dramatically reducing surface tension at the colony air-water interface (37).
SapB was the first morphogenetic protein discovered in S. coelicolor, and its structure has been elucidated: it is an amphiphilic, lantibiotic-like peptide that is the product of the ram gene cluster (25). SapB is produced during the vegetative phase of development, and available evidence suggests that it acts as a surfactant, coating both the nascent aerial hyphae and air-water interfaces to facilitate the emergence of aerial filaments into the atmosphere (35, 36).
The chaplins are secreted proteins that, like SapB, have strong surfactant properties and have similarly been shown to coat the surfaces of aerial hyphae and spores (9, 14). The eight chaplin proteins have Sec-dependent secretion signal sequences and share a region of extensive similarity termed the chaplin domain. Five of the chaplins (the short chaplins, ChpD to -H) have a single chaplin domain, while the remaining three long chaplins (ChpA to -C) have two chaplin domains and an extended C terminus that contains a predicted sorting signal for covalent attachment to the cell wall by a sortase enzyme. The chaplin domain itself is hydrophobic and includes two conserved Cys residues that are found in all chaplins, apart from ChpE. We have previously proposed a model for chaplin assembly in which the long chaplins act as cell wall anchors for the binding and polymerization of the short chaplins, and this polymerization ultimately results in the formation of a hydrophobic sheath encasing the aerial filaments, presumably in conjunction with SapB (5, 14). Deletion of either the SapB biosynthetic genes (the ram cluster) or the chaplin genes causes conditional defects in aerial hypha formation (5, 10, 28, 29), while deletion of both sets of genes results in a strain that is severely impaired in aerial development under all growth conditions (5). In addition to their role in aerial mycelium formation, the chaplins have also been implicated in the formation of the rodlet ultrastructure that decorates the surfaces of aerial hyphae and spores, together with the rodlin proteins (8, 10). Unlike the chaplins, however, the rodlins are dispensable for both aerial development and surface hydrophobicity; deletion of the rodlins simply results in a loss of the rodlet decoration on the surfaces of the aerial structures (11).
While the collective importance of the chaplins in aerial development has been firmly established, the roles played by individual chaplins are not well understood. In this work, we investigate the contributions made by the long and short chaplins to aerial hypha formation and the development of the rodlet ultrastructure, explore the importance of the conserved Cys residues, and demonstrate a unique role for ChpE in the viability of S. coelicolor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The Streptomyces strains used in this study are summarized in Table 1 and were cultured at 30°C on R5, MS, DNA, and minimal media plus glucose agar medium or in tryptone soya broth and yeast extract-malt extract liquid medium (24). Escherichia coli strains were typically grown at 37°C and included DH5α (for plasmid construction and routine subcloning), XL-1 Blue (for site-directed mutagenesis), ET12567/pUZ8002 (for generation of methylation-free DNA and conjugation into Streptomyces) (27, 31), and BW21153/pIJ790 (for PCR-targeted gene disruptions; grown at 30°C) (18). Descriptions of the plasmids used and constructed in this work are included in Table 1.
TABLE 1.
S. coelicolor strains and plasmids used in this study
| Strain/plasmid | Genotype, description, or function | Reference |
|---|---|---|
| S. coelicolor | ||
| M145 | SCP1− SCP2− | 24 |
| M600 | SCP1− SCP2− | 6 |
| J3149A | M600 ΔchpAD ΔchpCH::aadA ΔchpB ΔchpF ΔchpG (7× chp) | 5 |
| TP3 | M145 ΔtatB::apr | 34 |
| TP4 | M145 ΔtatC | 34 |
| E101 | M600 ΔchpE::apr tatB::IS1649 | This work |
| E102 | TP4 ΔchpE | This work |
| E103 | M600 ΔrdlAB::vph | This work |
| E104 | M600 ΔrdlAB::vph ΔchpE::apr | This work |
| J3287 | M600 ΔramR::vph | 5 |
| J3288 | M600 ΔramCSAB::vph | 5 |
| S. lividans | ||
| 1326 | SLP2+ SLP3+ | 24 |
| 10-164 | 1326 msiK | 21 |
| Plasmids | ||
| BT340 | Temperature-sensitive FLP recombination plasmid | 12 |
| pUC19 | E. coli cloning vector | |
| pBluescript KS | E. coli cloning vector | Stratagene |
| pGEM-T | E. coli cloning vector | Promega |
| pIJ2925 | E. coli cloning vector | 24 |
| pSET152 | Integrative cloning vector; ori pUC18 apr oriT RK2 int φC31 attP φC31 | 4 |
| pIJ82 | pSET152 derivative (hygromycin resistant) | |
| pMS82 | Integrative cloning vector; hyg oriT int φBT1 attP φBT1 | 17 |
| pIJ6916 | pIJ2925+pvanJ+chpE | This work |
| pIJ6917 | pIJ82+pvanJ+chpE | This work |
| pIJ6937 | pSET152+chpH | This work |
| pIJ6936 | pSET152+chpH* | This work |
| pIJ6935 | pSET152+chpCH | 14 |
| pIJ6934 | pSET152+chpCH* | This work |
| pIJ6933 | pMS82+chpDA | This work |
| pMC101 | pMS82+chpABC | This work |
| pTDW47 | pSET152+dagA | 34 |
| pTDW46 | pSET152+dagA (signal peptide sequence replaced with aadA) | 34 |
Protoplast transformation and conjugation from E. coli into Streptomyces.
Introduction of DNA from E. coli into S. coelicolor requires the DNA to be passaged through the E. coli strain ET12567 (dam dcm hsd) in order to circumvent the methyl-specific restriction system of S. coelicolor. Protoplast generation and transformation, and plasmid conjugation, were carried out as described by Kieser et al. (24).
Construction of chaplin and rodlin mutants.
chpE mutants and an rdlAB mutant were constructed as described by Elliot et al. (14, 15) according to the methods of Gust et al. (18).
Construction of “minimal chaplin” strains.
The integrating plasmid vector pSET152 containing chpC and chpH was constructed as described previously (14) and was introduced into the 7× chp mutant (J3149A) (Table 1) by conjugation. chpH was obtained by digesting the plasmid pSET152+chpCH with EcoRI, followed by gel purification of the resulting chpH fragment. This DNA fragment was then ligated with pSET152, which had also been digested with EcoRI, and dephosphorylated, creating pIJ6937. This construct was also introduced into the 7× chp mutant (J3149A) by conjugation. chpDA were PCR amplified using Pfu DNA polymerase (Stratagene) with M600 wild-type chromosomal DNA as a template. The DNA fragment was cloned into the SmaI site of pIJ2925 before being excised with HindIII and KpnI and cloned into pMS82 digested with the same enzymes, creating pIJ6933. pIJ6933 was introduced into the 7× chp mutant strain alone and carrying pSET152+chpCH by conjugation.
Site-directed mutagenesis of chpH.
chpH was excised as an EcoRI fragment from pBluescript containing chpCH and was cloned into pUC19 digested with EcoRI. The resulting plasmid (pUC+chpH) was used as a template for the PCR-based mutagenesis of the two Cys residues in chpH (C56 and C74). The complementary primers Cys56Val 1 and Cys56Val 2 (Table 2) were used to amplify the entire plasmid using Pfu enzyme (Stratagene), changing the Cys (TGC) at amino acid position 56 to a valine (GTC). The PCR program was 95°C for 5 min and then 5 cycles of 95°C for 1 min, 65°C for 1 min, and 72°C for 10 min; 19 cycles of 95°C for 1 min, 62°C for 1 min and 72°C for 10 min; followed by a final elongation at 72°C for 15 min. Next, the template DNA was selectively cleaved by digestion with DpnI (which recognizes and cleaves methylated DNA) for 2 h at 37°C. Three microliters of the reaction mixture was then used for electroporation into E. coli XL-1 Blue, and positive transformants were selected by plating them on LB containing 100 μg/ml ampicillin. Four colonies were selected for further examination through plasmid isolation and sequencing. One of the four (pUC+H C56V) had the correct mutation introduced, with no additional sequence changes, so it was subjected to a second round of mutagenesis. The complementary primers Cys74Gly 1 and Cys74Gly 2 were used to amplify the entire pUC+H C56V plasmid, as outlined above, to change the Cys (TGC) at amino acid position 74 to a glycine (GGC) residue. Again, sequencing was conducted to ensure that only the desired mutation was introduced. The mutagenized chpH gene (chpH*) was then excised as an EcoRI fragment and was introduced into either pSET152 (creating pIJ6936) or pSET152+chpC cut with EcoRI (creating pIJ6934) before being conjugated into S. coelicolor J3149A.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence (5′ to 3′)a | Function |
|---|---|---|
| Cys56Val 1 | GGTGAACGTCGTCGGCAACACG | Mutagenesis of chpH |
| Cys56Val 2 | CGTGTTGCCGACGACGTTCACC | Mutagenesis of chpH |
| Cys74Gly 1 | CCTTCGGCAACGTCGGCATCAACAAGTGACG | Mutagenesis of chpH |
| Cys74Gly 2 | CGTCACTTGTTGATGCCGACGTTGCCGAAGG | Mutagenesis of chpH |
| chpC end | CGAGTACGGACACTGGGAG | Cloning of chpCH |
| chpH up | CGGAGTGGACGAGCGGGTGC | Cloning of chpCH |
| chpD up | GCTGTCGGCGAACGGCGAGG | Cloning of chpAD |
| chpA end | CGCCTCTAGACCCTGCACCTGGACCTGACC | Cloning of chpAD |
| tatB1 | CCCTCGACATGAACTACACGG | PCR amplification of tatB |
| tatB2 | GCGTCACGTCCTGGATGACC | PCR amplification of tatB |
| tatB3 | GCCTCCTCGCTCTCGTCGG | PCR amplification of tatB |
| chpE up | GGGCAGATCTGCGACTGCCGCGGCGATCG | Cloning of chpE |
| chpE end | GGAGCGGGGGCGGTGACCG | Cloning of chpE |
| chpE Fwd | GGCGACTGCCGCGGCGATCGCAAGGAGGGGTTGTAAGTG | Knockout of chpE |
| ATTCCGGGGATCCGTCGACC | ||
| chpE Rev | GCCCGCTGCTGAGGCGGCATTGGGGGGCGGCCTGCGTCA | Knockout of chpE |
| TGTAGGCTGGAGCTGCTTC | ||
| rdlAB Fwd | AAGTCAGCGGGCCGCCCGTACCGGGCTGGGCTGGGCTCA | Knockout of rdlAB |
| ATTCCGGGGATCCGTCGACC | ||
| rdlAB Rev | CCGCCGAAGTGCTCGGCGGCCCGCCCCGGGGCGATGTCA | Knockout of rdlAB |
| TGTAGGCTGGAGCTGCTTC | ||
| rdlA NdeI | GGCGCGCATATGCTCAAGAAGGCAATGGTC | Cloning of RdlA signal peptide |
| rdlA BamHI | GGCGCGGGATCCCACGGCCGGCCCGTTGTC | Cloning of RdlA signal peptide |
| rdlB NdeI | GGCGCGCATATGAGCTCCGGTGGACGGGGTTTC | Cloning of RdlB signal peptide |
| rdlB BamHI | GGCGCGGGATCCGCTGTCGTCGCCGATCGC | Cloning of RdlB signal peptide |
Engineered restriction endonuclease recognition sequences are underlined.
Construction of chpE complementation constructs.
The vancomycin-inducible promoter of vanJ (vanJp) was excised from pIJ6882 (20) with HindIII and SalI and was cloned into pIJ2925 digested with the same enzymes. chpE was PCR amplified using primers chpE up and chpE end (Table 2) and was cloned into the vector pGEM-T (Promega). chpE was removed using BglII and SacI and was cloned into pIJ2925+pvanJ digested with BamHI and SacI, creating pIJ6916. The vanJp-chpE fragment was excised from pIJ2925 using BglII and was introduced into pIJ82 (a kind gift from H. Kieser), which is a derivative of pSET152 in which the apramycin resistance gene has been replaced by a hygromycin resistance gene. This plasmid (pIJ6917) was then introduced into S. coelicolor M600. The chpE knockout cosmid was then introduced into the plasmid-containing strain, and the resulting colonies were screened for the creation of a chpE null mutant in the presence (10 μg/ml) or absence of the inducer, vancomycin.
Agarase assay.
The agarase assay for detection of Tat-dependent signal peptides was conducted as described by Widdick et al. (34). Sequences corresponding to the RdlA and RdlB signal peptides were introduced upstream of the leaderless dagA gene in pTDW46. These constructs were introduced into Streptomyces lividans 10-164 by conjugation. pTDW47, containing an intact dagA gene (with its associated leader peptide), was also introduced into S. lividans 10-164 as a positive control for agarase activity. Agarase production rates in S. lividans 10-164 alone and containing pTDW46-RdlA, pTDW46-RdlB, and pTDW47 were compared. Ten thousand spores of each strain were spotted on minimal medium plus glucose agar medium plates and were allowed to grow for 72 h before the plates were stained with Lugol solution (VWR) for 45 min. Zones of clearing indicated agarase activity.
Scanning electron microscopy.
For scanning electron microscopy, colonies were mounted on the surface of an aluminum stub with optimal cutting temperature compound (Miles Scientific), plunged into liquid nitrogen slush at approximately −210°C to cryopreserve the material, and transferred to the cryostage of an Alto 2500 cryotransfer system (Gatan, Oxford, England) attached to a Zeiss Supra 55 VP field emission gun scanning electron microscope (Zeiss SMT, Germany). The surface frost was sublimated at −95°C for 3 min before the sample was sputter coated with platinum for 2 min at 10 mA at below −110°C. Finally, the sample was moved onto the cryostage in the main chamber of the microscope, held at approximately −130°C, and viewed at 1.2 to 5.0 kV. Scanning electron microscope images were saved as TIF graphic files and manipulated in Adobe Photoshop 7.0.
RESULTS
Creation of a “minimal chaplin strain.”
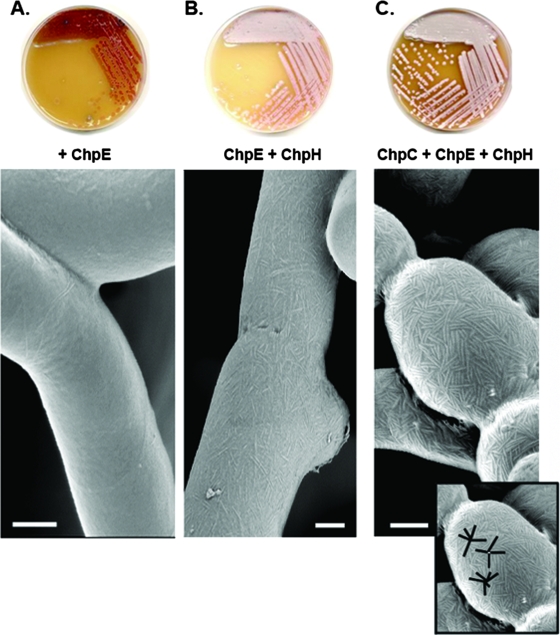
Deletion of individual chaplin genes (chpH or chpB) or pairs of genes (chpCH or chpAD) had no obvious phenotypic effect on colony growth or development (14), suggesting significant redundancy in chaplin function. To determine the extent of chaplin redundancy, we attempted to develop a “minimal chaplin strain” that would raise an abundant aerial mycelium by reintroducing individual chaplin genes into a chp mutant strain containing only chpE (the 7× chp mutant J3149A). chpC, chpE, and chpH are the only chaplin genes conserved in all Streptomyces species whose genome sequences are available (2, 22, 29a) (www.sanger.ac.uk/Projects/S_scabies/). In addition, previous transcript analyses showed that chpC, chpE, and chpH were the only chaplins expressed prior to the initiation of aerial hypha formation, with chpE and chpH expressed at continuously high levels throughout the developmental cycle (9, 14). As such, it seemed likely that these three chaplins would make a major contribution to aerial hypha development. A strain containing only chpE (Fig. 1A) had a bald colony phenotype identical to that of the complete chaplin (8× chp) mutant strain (5), as did a strain containing only the genes for the long chaplins (chpA, chpB, and chpC) (data not shown). Introduction of chpH into the chpE-containing mutant background, however, resulted in a strain that could raise a sparse aerial mycelium on MS medium after extended incubation (Fig. 1B). In contrast, when we introduced chpC and chpH together into the chpE-containing 7× mutant, the resulting strain developed a robust, sporulating aerial mycelium (Fig. 1C), albeit with growth kinetics similar to those of the chpE-and chpH-containing strain (i.e., at a lower rate than the wild-type strain). Thus, expression of the long chaplin ChpC, in combination with the short chaplins ChpE and ChpH, was sufficient to promote the growth of an abundant aerial mycelium, demonstrating redundancy among the chaplins but suggesting that both long and short chaplins play important roles in aerial development. To determine whether the presence of any long chaplin and short Cys-containing chaplin was sufficient to promote aerial hypha formation, we introduced chpA (encoding a long chaplin) and chpD (encoding a Cys-containing short chaplin) into the strain containing only chpE. We observed significant aerial hypha formation, although not as robust as with chpC and chpH (data not shown), likely due to the fact that chpA and chpD are not as highly expressed at all times as chpC and chpH (14).
FIG. 1.
The introduction of additional chaplin genes into the 7× chp mutant strain (J3149A), which contains only chpE, results in increased aerial hypha formation (plate images) and increased surface ultrastructure (scanning electron micrographs). (A) J3149A. (B) J3149A with chpH introduced. (C) J3149A with chpC and chpH introduced. (Inset) Starburst rodlet pattern on the spore surface. Images were taken after 7 days of incubation. Bars = 250 nm.
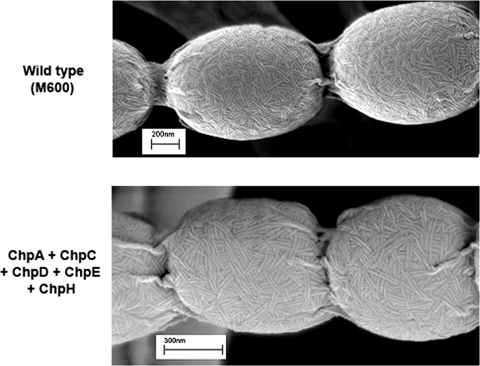
As the loss of all eight chaplin genes eliminated rodlet formation (5, 10), we examined the surfaces of the aerial hyphae and spores of strains containing chpEH and chpCEH to see if they had regained the characteristic rodlet ultrastructure. We found that chpE and chpH together restored the formation of sparse but well-defined individual surface fibers (Fig. 1B), although we did not observe any of the paired rodlet filaments typical of the wild type. The addition of chpC significantly enhanced the formation of both individual and paired surface fibers, and these were frequently arranged in a “starburst-like” pattern, with multiple fibers appearing to emanate from a central point (Fig. 1C). This provides direct experimental evidence that the chaplins are major contributors to the rodlet ultrastructure on aerial surfaces. Furthermore, we found that the introduction of either two copies of chpCH (one copy on each of two independently integrating plasmid vectors) (data not shown) or chpCH together with chpAD (Fig. 2) into the chpE-containing J3149A strain resulted in near-wild-type abundance of paired rodlets, in contrast to the minimal chaplin strain; however, these rodlets did not show the same degree of organization as those of the wild type.
FIG. 2.
Scanning electron micrographs comparing the surface ultrastructure of wild-type S. coelicolor M600 (top) with that of a 7× chp mutant strain (containing only chpE) to which chpAD and chpCH were introduced on integrating plasmid vectors (bottom).
The formation of aerial hyphae and surface fibers depends upon conserved Cys residues in ChpH.
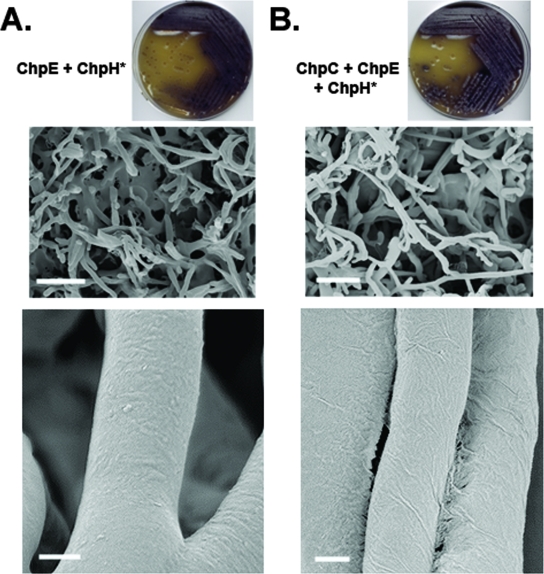
Given that the chpCEH-containing strain could raise an abundant aerial mycelium and assemble a rodlet ultrastructure on aerial surfaces, we decided to use the strain to probe the role of the short chaplins and, in particular, the Cys-containing short chaplins. The chaplin domains of all short chaplins, apart from ChpE, contain two highly conserved Cys residues (Fig. 3). Previous work suggested that these Cys residues formed intramolecular disulfide bonds in the mature extracellular chaplin proteins (14), and it is well established that analogous disulfide bonds in the fungal hydrophobins stabilize their amphiphilic structure and are critical for function (19, 26). To investigate the importance of these residues in chaplin function, we replaced the two Cys residues in ChpH with a glycine and a valine (these residues are found in place of the Cys residues in ChpE in S. coelicolor and thus were deemed unlikely to disrupt chaplin stability) (Fig. 3). The Cys-free ChpH-encoding gene (chpH*) was then introduced into the chpE-containing mutant strain either alone or in conjunction with chpC. The two resulting strains formed a very sparse aerial mycelium (Fig. 4) compared with the otherwise identical strains carrying the wild-type chpH gene (Fig. 1) (see above). Examination of the aerial surfaces of these two mutant strains also revealed a complete absence of rodlet ultrastructure (Fig. 4). This suggested that the Cys residues in ChpH were necessary both for aerial hypha formation and for assembly of the rodlet ultrastructure. To exclude the possibility that ChpH* had a dominant-negative effect on aerial hypha formation, we compared the effects of introducing chpH* versus chpH into a wild-type strain. No phenotypic differences were observed (data not shown), suggesting that ChpH* does not inhibit aerial morphogenesis.
FIG. 3.
Alignment of the short-chaplin domains from S. coelicolor (sc), S. avermitilis (av), S. griseus (sg), and S. scabies (ss). The sequences shown represent the processed (signal peptide removed), mature chaplin form. Identical amino acid residues are highlighted in black, and similar amino acid residues are shown in gray. The arrows indicate the sites of conserved Cys residues.
FIG. 4.
The phenotypic effect of removing the two Cys residues in ChpH. (A) A strain producing only ChpE and ChpH* is largely incapable of raising aerial hyphae (top and middle) and exhibits no rodlet ultrastructure (bottom). (B) A strain producing ChpC, ChpE, and ChpH* is capable of raising slightly more hyphae than the strain shown in panel A but is still largely defective in aerial hypha formation (top and middle) and is also devoid of rodlet fibers on the aerial surfaces (bottom). Bars: middle, 5 μm; bottom, 200 nm.
chpE is essential.
ChpE differs from the other short chaplins in that it lacks the two highly conserved Cys residues important for ChpH function (Fig. 3). In addition, ChpE has a different hydrophobicity profile, with alanine replacing valine or leucine at multiple positions (e.g., at residues 18, 25, and 33 in the mature S. coelicolor ChpE). These characteristics are conserved in the ChpE orthologues from Streptomyces avermitilis (SAV6478), Streptomyces griseus (SGR5696) and Streptomyces scabies (Fig. 3), suggesting that ChpE has a unique function among the chaplins. Our creation of the minimal chaplin strain did not reveal a function for ChpE, as a strain containing only ChpE looked identical to one lacking all the chaplins. Furthermore, ChpE could not substitute for ChpH* in stimulating aerial hypha formation. Therefore, we decided to investigate ChpE function in a wild-type background. We tried to create a chpE mutant in the wild-type M600 strain by replacing the chromosomal chpE gene with an apramycin resistance cassette, using the robust Redirect PCR-targeting method of Gust et al. (18), but were repeatedly unable to do so. We and others had successfully deleted chpE in strains already lacking other chp genes (5, 9), which suggested either a technical problem or that chpE was essential in a wild-type genetic background. To distinguish between these two possibilities, we introduced a second copy of chpE, under the control of a vancomycin-inducible promoter, into the wild-type S. coelicolor strain M600. In the presence of vancomycin, we were readily able to disrupt the native chromosomal copy of chpE (108/736 colonies screened), whereas in the absence of vancomycin, no double-crossover gene replacements were obtained (0/522 colonies screened), providing strong evidence that chpE is essential in a wild-type genetic background. Attempts to grow the knockout strains on solid medium in the absence of vancomycin induction resulted in the frequent appearance of colonies that had acquired compensatory suppressor mutations. These colonies were heterogeneous in their appearance: many had a wild-type appearance, while others had an unusual small-colony phenotype (see below).
Suppressors of chpE mutation.
The above observations suggested that chpE was essential in a wild-type background; however, chpE was successfully deleted from a strain that was missing all of the other chaplin genes (5, 10) and also from a strain that was missing all of the chaplin genes apart from chpF and chpG (9). This suggested that the essential nature of chpE might be conditionally dependent upon the presence of particular chp genes. One model that could account for these observations is that ChpE is involved in coordinating the assembly or polymerization of the other chaplins. To test this hypothesis, we focused on chpC and chpH, two of the most highly expressed and most conserved (apart from chpE itself) of the chaplin genes, and tested whether it was possible to introduce chpC and chpH together into an 8× chp mutant strain lacking chpE. We found that we were able to readily construct this strain, implying that the essential nature of chpE is not tied to the expression and function of chpC and chpH. We also tested the relative ease with which we could create a chpE mutant in a variety of other developmental mutant strains, including ΔramR and ΔramCSAB strains, which are both unable to produce SapB, and a ΔrdlAB strain, which is unable to produce the two rodlin proteins, RdlA and RdlB. We were unable to create a chpE mutant in either of the ram mutant strains, but we could easily knock out chpE in the rodlin mutant background (8/25 colonies screened).
Second-site suppressors of chpE are found in the Tat secretion system.
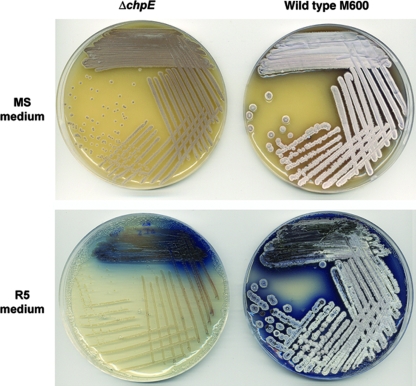
During the course of our attempts to create a chpE knockout in the wild-type genetic background, we identified several colonies that had the correct antibiotic resistance profile for a chpE knockout and confirmed that these colonies represented chpE null mutations using PCR and Southern blot analyses (data not shown). Interestingly, however, these mutants had a phenotype distinct from those of wild-type strains (Fig. 5) and characterized rdl and chp mutant strains (5, 9, 10, 11). On MS medium, the mutants had a small-colony phenotype and did not produce the secreted hydrolytic enzyme agarase (which, in the wild-type strain, gives rise to craters in the agar surrounding the colonies); on rich (R5) medium, the colonies were “bald” (devoid of aerial hyphae) and produced reduced levels of the blue-pigmented antibiotic actinorhodin; in high-sucrose liquid culture (yeast extract-malt extract) the mutants failed to grow; and in rich liquid medium (tryptone soya broth), the mutants grew in a very dispersed manner, more reminiscent of E. coli-type growth than the pellet-like mycelial growth typical of Streptomyces. None of these mutant phenotypic characteristics could be complemented by the introduction of a wild-type copy of chpE, suggesting that the unusual phenotypes were due to a second-site suppressor mutation rather than the chpE null mutation itself.
FIG. 5.
Phenotypic comparison of wild-type S. coelicolor M600 with the constructed chpE null mutant (carrying an insertion element in tatB). The top panel shows the two strains grown on MS medium, while the bottom panel shows the two strains grown on rich R5 medium.
Intriguingly, the mutant phenotype was very similar to that of strains carrying mutations in the twin arginine (Tat) secretion pathway (32, 34), with agarase being a known Tat substrate (34). Unlike the Sec secretion pathway, which translocates proteins in an unfolded conformation, the Tat pathway is dedicated to the translocation of folded substrates, many of which contain cofactors. Secretion through the Tat pathway requires three membrane-localized proteins: TatA, TatB, and TatC. The mutation of genes encoding any of these three proteins confers a phenotype seemingly identical to that of the chpE suppressor mutants.
To determine whether the chpE mutant had defects in any of the Tat translocation components, we PCR amplified tatA, tatB, and tatC from the chpE suppressor mutants and sequenced each of the resulting products. For several independent mutant isolates, there was an insertion element (IS1649) disrupting the coding sequence of tatB. This suggested that inactivation of the Tat pathway suppressed the lethality of the chpE null mutation and, additionally, that there may be an insertion “hot spot” within the 5′ end of tatB, given the isolation of several independent suppressor strains carrying IS1649 inserted at an identical location, 26 nucleotides downstream from the start of the coding sequence.
Loss of a functional Tat secretion system permits deletion of chpE.
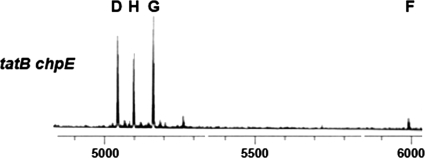
To further examine the connection between the Tat secretion system and chpE, we attempted to create a chpE gene knockout in a Tat mutant background, using a tatC null mutant (34). We found that chpE null mutants could be readily constructed in this mutant background (6/32 colonies screened). This confirmed that inactivation of the Tat pathway suppressed the lethality of the chpE null mutation and, furthermore, showed that the connection to chpE was not specific to tatB. There is no obvious direct link between the chaplins and Tat secretion, given that all the chaplins have typical Sec-dependent signal sequences. However, it was formally possible that a tat mutation might indirectly prevent secretion of the other chaplin proteins, thereby suppressing the lethality of chpE disruption. To address this possibility, we isolated cell wall fractions of sporulating cultures of a tatB chpE double mutant and followed the chaplin purification procedure used previously for the isolation and identification of the short chaplins (14). Using matrix-assisted laser desorption ionization-time of flight mass spectrometry, we successfully identified all of the four remaining short chaplins in these cell surface fractions, suggesting that chaplin secretion is not impaired in a tatB chpE double mutant (Fig. 6).
FIG. 6.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry of cell wall extracts isolated from the tatB chpE double mutant grown on MS medium. The peaks corresponding to ChpD, ChpH, ChpG, and ChpF are labeled. The x axis represents the mass (m)/charge (z) ratio, where z = 1.
RdlA and RdlB are not Tat substrates.
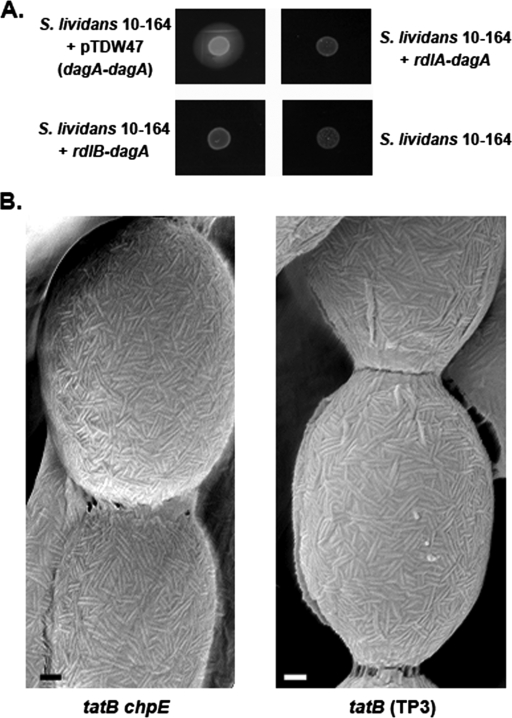
As it was straightforward to create a chpE knockout in a rdlAB mutant background, we examined the sequences of the signal peptides of RdlA and RdlB to determine whether either was a potential Tat substrate, thus providing a potential link between the Tat system and chpE. The RdlB leader sequence revealed a potential Tat-dependent signal peptide, as predicted by the TatP 1.0 server (1), although it had a degenerate “Tat motif” at its extreme N terminus (G-R-X-F-L as opposed to the prototypical R-R-X-[FGAVML]-[LITMVF]). The same program did not predict a Tat-dependent signal peptide for RdlA. To test whether RdlB or RdlA was a Tat substrate, we used an agarase assay that had been developed to differentiate proteins secreted via the Sec secretion system from those secreted via the Tat secretion system (34). Agarase, encoded by the dagA gene, is functional only when secreted by the Tat system in S. coelicolor. The agarase assay involves replacing the native DagA signal peptide with a signal peptide of interest (in this case, the RdlA and RdlB signal peptides) and monitoring the secretion of functional agarase upon introduction into a non-agarase-producing strain (S. lividans 10-164). The production and secretion of agarase are monitored using Lugol solution, an iodine-based dye that stains agar a dark-brown color, apart from areas of agarase activity, which are detected as zones of clearing around producing colonies. We introduced the RdlA-DagA and RdlB-DagA fusion constructs, along with DagA carrying its native signal peptide as a positive control, into S. lividans 10-164. We found that there was no agarase activity detected when it was fused to the RdlA or RdlB signal peptides (Fig. 7A). This suggested that neither rodlin signal peptide was capable of driving agarase secretion through the Tat system and that RdlA and RdlB were instead likely substrates for the Sec secretion system.
FIG. 7.
(A) Agarase assay in S. lividans 10-164 comparing secretion of agarase with its native signal peptide (top left), RdlA signal peptide (top right), and RdlB signal peptide (bottom left). S. lividans 10-164 alone served as a negative control (bottom right). (B) Scanning electron micrographs showing the surfaces of the tatB chpE double mutant and the tatB (TP3) mutant. Abundant rodlet ultrastructure is evident for both strains. Bars = 100 nm.
To further investigate whether RdlA or RdlB was a Tat substrate, we examined the surface of a tatB mutant using high-resolution scanning electron microscopy. RdlA and RdlB are not functionally redundant, as both proteins are required for the assembly of a rodlet ultrastructure on aerial surfaces (10). We found that both a tatB mutant strain and a tatB chpE double-mutant strain had significant rodlet ultrastructure on their surfaces, albeit appearing slightly less organized than that of a wild-type strain (Fig. 7B). This implied that the Tat secretion system plays a novel, as yet uncharacterized role in chaplin fiber assembly, independent of chaplin export to the surface and interaction with the rodlins.
DISCUSSION
Model for the activity of long and short chaplins in aerial hypha and rodlet formation.
In our original model of chaplin activity (14), we proposed that the long chaplins, believed to be covalently attached to the cell wall by a sortase enzyme(s), serve as primary (but not exclusive) anchors for the short chaplins on the cell surface through the heteropolymerization of chaplin domains. This was predicted to result in the formation of a hydrophobic surface layer that would help to break surface tension during the erection of aerial hyphae and would also help to prevent desiccation of the aerial filaments. In addition to modulating surface hydrophobicity, work by Claessen et al. (10) has suggested that the chaplins also interact with the rodlin proteins to generate the characteristic rodlet pattern found on the surfaces of aerial structures. We used our minimal chaplin strain to examine the roles played by the long and the short chaplins and to test our model of chaplin activity.
In the minimal chaplin strain, the short chaplin ChpH appears to be the main polymerization unit driving aerial hypha and rodlet formation. Neither ChpE nor ChpC contributes significantly to either of these processes in the absence of a functional ChpH; however, ChpC greatly enhances both the formation of aerial hyphae and the assembly of a rodlet ultrastructure when introduced in conjunction with ChpH and ChpE. This would be consistent with a role for ChpC as an anchor, rather than a significant polymerization unit—a role that is further supported by our observation that a strain containing only the long chaplins has a phenotype very similar to that of an 8× chp mutant. Examination of the aerial structures of strains containing ChpC, ChpE, and ChpH revealed increased rodlet fibrils and a greater organization of these fibrils than was found in the absence of ChpC. A striking feature of these ChpC-containing strains is the appearance of foci from which many fibers can be seen to emanate. This, too, is consistent with an anchoring role for ChpC in the polymerization of the short chaplins. We suggest that these functions of ChpC as a cell wall anchor and ChpH as a polymerization unit are likely to be representative of the functions of the other long chaplins and the other Cys-containing short chaplins, respectively. In support of this, a strain expressing ChpADE (where ChpA is a long chaplin and ChpD is a Cys-containing short chaplin) could also raise aerial hyphae and assemble a rodlet ultrastructure (data not shown).
Cysteines and disulfide bond formation.
An important difference between ChpE and the remaining short chaplins is the absence of two Cys residues in ChpE that are conserved in the other four. These Cys residues are integral to the function of ChpH, as their removal resulted in greatly reduced aerial hypha formation and a complete abrogation of rodlet assembly in the minimal chaplin strain. Our previous work suggested that these Cys residues form intramolecular disulfide bonds in the mature extracellular ChpH and in the other Cys-containing short chaplins (14). We propose that these disulfide bonds play a critical role in the stabilization of chaplin structure, perhaps by locking the proteins in an amphipathic conformation; the amino acids between the two Cys residues are predominantly hydrophobic and could form a hydrophobic patch having the capacity to confer amphipathic properties on the chaplin domain.
Hydrophobins, which are involved in rodlet assembly and aerial hypha formation in the filamentous fungi, have eight highly conserved Cys residues that form four intramolecular disulfide bonds. Loss of the eight Cys residues from the MPG1 hydrophobin of Magnaporthe grisea results in defects in aerial hypha formation (23) that are similar to those we observed after loss of the Cys residues from ChpH. In contrast to the ChpH*-containing strain, however, loss of the Cys residues in the fungal hydrophobins did not disrupt rodlet assembly, suggesting that, unlike for the chaplins, disulfide bond formation is not a prerequisite for fungal hydrophobin self-assembly. The disulfide bonds formed in the fungal hydrophobins are known to be important structural determinants: they reduce conformational flexibility and stabilize the globular, amphiphilic structure of the proteins (19, 26). A similar compact, amphiphilic structure is adopted by the Streptomyces morphogenetic surfactant peptide SapB. Instead of disulfide bonds, however, SapB forms lanthionine bridges, which impose flexibility constraints and result in the exposure of surface-localized hydrophobic side chains, thus forming an amphipathic molecule (25).
ChpE serves a unique function among the chaplins.
The inability of ChpE to form intramolecular disulfide bonds, due to the absence of the conserved Cys residues, suggested that it would have a unique structure relative to the other chaplins. The fact that chpE is essential in a wild-type genetic background implies that ChpE also has a unique function. As chpE is expressed early in development, it is possible that ChpE has a vital role, independent of the other chaplins, in early colony development. However, ChpE is dispensable in tat mutants, an rdlAB mutant, and strains lacking the other chaplin genes, suggesting that vegetative growth is not dependent upon ChpE function. Instead, we propose that ChpE coordinates the assembly and/or polymerization of the other chaplins, possibly by mediating their interaction with the rodlin proteins, and that loss of this coordination is lethal to the developing Streptomyces colony. The abundance of other chaplin proteins appears to be key, as chpE disruption can be accomplished in a variety of chaplin mutant backgrounds, as shown here and in previous work (5, 9). It is interesting that, although the number of chaplin genes present in Streptomyces species is variable, ChpE is conserved in all sequenced Streptomyces genomes available thus far.
The apparent functional difference, but extensive sequence similarity, between ChpE and the other short chaplins is reminiscent of the curli system in E. coli. Curli fibers, like chaplin fibers, are amyloid-like structures (7, 33) that are formed through the polymerization of two homologous proteins: CsgA and CsgB. These two proteins share significant sequence similarity; however, CsgA has been identified as the main curli polymerization unit (30), while CsgB functions primarily as the nucleator for the polymerization of CsgA (3). Whether such functional differentiation exists between ChpE and ChpH remains to be seen.
The connection between ChpE and the Tat secretion system is intriguing but obscure. Neither the chaplins nor the rodlins are Tat substrates, and their secretion is not affected in a tatB mutant, yet mutations in the tat genes suppress the lethality of the chpE null mutation in an otherwise wild-type genetic background. The most likely explanation for these observations is that a Tat-dependent substrate(s) contributes to ChpE-dependent colony viability; however, the nature of this connection awaits further investigation.
Acknowledgments
We are extremely grateful to T. Palmer and D. Widdick for providing us with tat mutant strains and for many helpful discussions.
This work was supported by the Canada Research Chairs program (to M.A.E.), the Canadian Institutes of Health Research (to M.A.E.; grant no. MOP-77553), and the Biotechnology and Biological Sciences Research Council of the United Kingdom (to M.J.B.; grant no. 208/EGH16080).
Footnotes
Published ahead of print on 17 June 2008.
REFERENCES
- 1.Bendtsen, J., L. Kiemer, A. Fausboll, and S. Brunak. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bian, Z., and S. Normark. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 165827-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 5.Capstick, D. S., J. M. Willey, M. J. Buttner, and M. A. Elliot. 2007. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 64602-613. [DOI] [PubMed] [Google Scholar]
- 6.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1795854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claessen, D., W. de Jong, L. Dijkhuizen, and H. A. B. Wösten. 2006. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 14313-319. [DOI] [PubMed] [Google Scholar]
- 9.Claessen, D., R. Rink, W. de Jong, J. Siebring, P. de Vreugd, F. G. H. Boersma, L. Dijkhuizen, and H. A. B. Wösten. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 171714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claessen, D., I. Stokroos, H. J. Deelstra, N. A. Penninga, C. Bormann, J. A. Salas, L. Dijkhuizen, and H. A. B. Wösten. 2004. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53433-443. [DOI] [PubMed] [Google Scholar]
- 11.Claessen, D., H. A. B. Wösten, G. von Keulen, O. G. Faber, A. M. C. R. Alves, W. G. Meijer, and L. Dijkhuizen. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 441483-1492. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliot, M. A., M. J. Buttner, and J. R. Nodwell. 2007. Multicellular development in Streptomyces, p. 419-438. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 14.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 171727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliot, M. A., T. R. Locke, C. M. Galibois, and B. K. Leskiw. 2003. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol. Lett. 22535-40. [DOI] [PubMed] [Google Scholar]
- 16.Elliot, M. A., and N. J. Talbot. 2004. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 7594-601. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, M. A., R. Till, and M. C. M. Smith. 2003. Integration site for Streptomyces phage φBT1 and development of site-specific integrating vectors. J. Bacteriol. 1855320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. 1001541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakanpää, J., A. Paananen, S. Askolin, T. Nakari-Setälä, T. Parkkinen, M. Penttilä, M. B. Linder, and J. Rouvinen. 2004. Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile. J. Biol. Chem. 279534-539. [DOI] [PubMed] [Google Scholar]
- 20.Hong, H. J., M. I. Hutchings, J. M. Neu, G. D. Wright, M. S. B. Paget, and M. J. Buttner. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 521107-1121. [DOI] [PubMed] [Google Scholar]
- 21.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17367-377. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21526-531. [DOI] [PubMed] [Google Scholar]
- 23.Kershaw, M. J., C. R. Thornton, G. E. Wakley, and N. J. Talbot. 2005. Four conserved intramolecular disulphide linkages are required for secretion and cell wall localization of a hydrophobin during fungal morphogenesis. Mol. Microbiol. 56117-125. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 25.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. 10111448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan, A. H. Y., R. D. Winefield, M. Sunde, J. M. Matthews, R. G. Haverkamp, M. D. Templeton, and J. P. Mackay. 2006. Structural basis for rodlet assembly in fungal hydrophobins. Proc. Natl. Acad. Sci. 1033621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 11161-68. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, K. T., J. M. Willey, L. D. Nguyen, L. T. Nguyen, P. H. Viollier, and C. J. Thompson. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 461223-1238. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 4545-57. [DOI] [PubMed] [Google Scholar]
- 29a.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. The genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO13350. J. Bacteriol. 1904050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsén, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7523-536. [DOI] [PubMed] [Google Scholar]
- 31.Paget, M. S. B., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaerlaekens, K., L. Van Mellaert, E. Lammertyn, N. Geukens, and J. Anné. 2004. The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology 15021-31. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., D. R. Smith, J. W. Jones, and M. R. Chapman. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2823713-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widdick, D. A., K. Dilks, G. Chandra, A. Bottrill, M. Naldrett, M. Pohlschröder, and T. Palmer. 2006. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. 10317927-17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willey, J., R. Santamaria, J. Guijarro, M. Geistlich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65641-650. [DOI] [PubMed] [Google Scholar]
- 36.Willey, J. M., A. Willems, S. Kodani, and J. R. Nodwell. 2006. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 59731-742. [DOI] [PubMed] [Google Scholar]
- 37.Wösten, H. A. B. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55625-646. [DOI] [PubMed] [Google Scholar]