Abstract
Pili have been observed on the surface of several gram-positive bacteria, including Streptococcus pneumoniae. The S. pneumoniae strain TIGR4 pilus is composed of three structural subunit proteins encoded in the rlrA pathogenicity islet, RrgA, RrgB, and RrgC. RrgB comprises the pilus backbone, RrgA is observed at intervals along surface pili, while RrgC is found in a loosely defined relationship with RrgA. We investigated the incorporation of each subunit into pili and the reliance of such placement on each of the other subunits. Both accessory subunits RrgA and RrgC are present in similar quantities in pili of all sizes. However, neither protein is required for the polymerization of RrgB, suggesting a nonessential role for RrgA and RrgC in the initiation of pilus assembly. Additionally, the rlrA islet encodes three sortases, SrtC-1, SrtC-2, and SrtC-3 (formerly SrtB, SrtC, and SrtD), which are divergent in sequence from the housekeeping sortase, SrtA. We determined the contributions of these four sortases to pilus assembly and found that SrtA is dispensable for pilus assembly and localization to the cell wall. Instead, SrtC-1, SrtC-2, and SrtC-3 are responsible for pilus assembly and exhibit functional redundancy with respect to backbone assembly and cell wall localization. A level of specificity and coordination among the class C sortases was revealed by the finding that SrtC-1 and SrtC-3 are required for the incorporation of the accessory subunits and by showing a deleterious effect on pilus assembly upon alteration of the cell wall sorting signals of the accessory subunit proteins.
Streptococcus pneumoniae is a human commensal and pathogen that can infect various areas of the body, resulting in otitis media, meningitis, pneumonia, and septicemia. The bacterium's diversity of infectious routes results in much interest in the surface proteins of S. pneumoniae, as they are often involved in the switch from a harmless commensal to a virulent pathogen and also play a role in determining the type of disease that is caused.
Recently, a group of surface proteins was found to form a multisubunit pilus on the surface of S. pneumoniae (3, 18). With this finding, S. pneumoniae joins a group of eight gram-positive bacteria that have been reported to encode pili. Although a comprehensive model of pilus assembly is lacking, the requirements for this process are beginning to be elucidated. Work done with Corynebacterium diphtheriae has determined that conserved genetic requirements necessary for pilus formation include the pilin motif (WxxxVxVYPK), E box domain (YxLxETxAPxGY), and cell wall sorting signal (CWSS) (LPxTG, followed by a stretch of hydrophobic and then charged amino acids), each present within the major pilin subunit (26, 28, 29). These findings, in addition to the recently solved crystal structure of the Streptococcus pyogenes pilus (15), which elucidated the peptidyl linkage of one major pilin subunit to the next, lend support to the “sortase-pilus” hypothesis proposed by Ton-That and Schneewind as a model of pilus assembly as a sortase-mediated event (17). According to this hypothesis, the major subunit is assembled into a pilus by a cis-encoded sortase that catalyzes the covalent attachment between the conserved pilin motif lysine residue of one subunit with the conserved threonine residue of the CWSS of another subunit. In addition, one or more accessory subunits are incorporated into the pilus by a mechanism that is unknown but requires one or more cis-encoded sortases, as well as the conserved E box domain within the major pilin protein. This sortase-mediated assembly pathway results in a pilus that consists of a major subunit and is decorated with one or more accessory subunits. We sought to establish this model further in S. pneumoniae, as the mechanism of assembly of the pilus in this organism has not been extensively addressed.
The S. pneumoniae pilus is encoded by the rlrA pathogenicity islet, a 12-kb locus flanked by IS elements, which contains seven genes (11). The first gene in the islet is rlrA, which positively regulates itself and other genes in the islet (12). The other six genes code for three surface proteins, RrgA, RrgB, and RrgC, and three sortases, SrtC-1, SrtC-2, and SrtC-3. A housekeeping sortase, SrtA, is encoded elsewhere in the genome. In the last several years, it has been shown that the three surface proteins are polymerized into a surface-localized pilus (3, 18). RrgB contains a pilin motif, E box, and CWSS and is the major pilin subunit forming the shaft or backbone of the pilus. In an rrgB-null strain, no pilus is formed. Additionally, RrgB is not dependent on either of the two accessory Rrg proteins for polymerization; however, RrgA and RrgC are both found in the pilus. Both RrgA and RrgC contain a CWSS and E box-like motifs where the conserved threonine in the latter is instead an asparagine and leucine, respectively. By immuno-gold transmission electron microscopy (TEM), RrgA is detected at regular intervals along surface pili (18), and although RrgC's location is unclear, it was recently reported to be located within purified pili both in single units and clustered with RrgA (13).
Three other genes within the rlrA islet encode class C sortases, which have been shown to be involved in pilus formation (9). Because they are classified as such, we propose here that the previous nomenclature for these genes (srtB, srtC, and srtD) be changed to the more appropriate nomenclature of srtC-1, srtC-2, and srtC-3 (9). Sortases are a family of enzymes found in gram-positive bacteria that act as both proteases and transpeptidases (19, 25, 27). They have a conserved catalytic site sequence, TLxTC, which is found in all four of the sortases found in the S. pneumoniae genome. There is one sortase found in the genome of almost every gram-positive bacterium looked at to date, with the exception of Mycobacterium and Microplasma (22), referred to as the housekeeping sortase, which covalently links proteins to the cell wall. Sortase-mediated cell wall localization requires that the protein that is linked to the cell wall have two domains. The first is a signal sequence that targets the protein for export out of the cell. The second is a sortase-recognized CWSS typically found in the carboxy-terminal region of the protein. It is this LPxTG motif that interacts with the sortase and is the location of the covalent attachment of the protein to the cell wall.
The S. pneumoniae pilus cluster is similar to the widely studied C. diphtheriae pilus in that there are three subunit proteins that make up the pilus. However, there is a distinction between the two bacteria as the rlrA islet encodes three sortases and C. diphtheriae has two sortases per pilus cluster (with three clusters in the genome) (29). Additionally, the CWSSs of the pilin subunits in C. diphtheriae have the canonical SrtA-recognized LPxTG motif; however, the three S. pneumoniae Rrg proteins that assemble into the pilus each have a motif that is divergent from this canonical CWSS motif. The CWSS LPxTG motifs of RrgA, RrgB, and RrgC are YPRTG, IPQTG, and VPDTG, respectively, each with a variant first amino acid. Intrigued by the variant CWSSs, we used mutants that have either an altered first amino acid of the CWSS or a replacement of the entire CWSS in order to investigate whether the CWSS confers specificity on the sortases that assemble each Rrg protein into the pilus or whether the first amino acid of the LPxTG motif is the specificity determinant.
In addition to the pilus-associated sortases, a role has been described for the housekeeping sortase in pilus assembly in Bacillus cereus, C. diphtheriae, and Streptococcus agalactiae (6, 21, 24). Therefore, we were interested in determining if the housekeeping sortase of S. pneumoniae, SrtA, is involved in the assembly of the pilus and localization of the pilus to the cell wall. We hypothesized, due to their variant CWSSs, that the Rrg proteins would not be substrates for SrtA for either assembly into pili or localization to the cell wall. Indeed, we show here that an srtA mutant has a wild-type amount of pili in the cell wall, suggesting that the housekeeping sortase is not necessary for pilus polymerization or localization. By addressing details of the assembly of the RlrA pilus, we have started to uncover a complex mechanism by which the three proteins are constructed into a pilus and to determine the individual roles of the sortases in this process.
MATERIALS AND METHODS
Strains and bacterial growth.
The S. pneumoniae strains used in this study are listed in Table 1. Cultures were grown in Todd-Hewitt broth plus 0.5% yeast extract supplemented with 5 μl ml−1 Oxyrase (Oxyrase, Inc., Mansfield, OH). The antibiotic concentrations used were as follows: streptomycin, 100 μg ml−1; chloramphenicol, 4 μg ml−1; spectinomycin, 200 μg ml−1. DNA was introduced into S. pneumoniae strain TIGR4 by natural transformation as previously described (5), and all mutations were confirmed by PCR and DNA sequencing. Except for the RrgC V307Y mutant strain, the triple sortase deletion (srtC1-C3) strain, and the rrgA rrgC double mutant, all strains were as previously reported (11, 18). Briefly, the srtC-1, srtC-2, and srtC-3 mutants contain a mariner transposon derivative (magellan2 or magellan5 [11]) inserted to create a disruption of the gene or its expression; srtC-1 (magellan5 insertion at nucleotide 336 of the coding region), srtC-2 (magellan5 insertion 27 nucleotides upstream of the start codon), and srtC-3 (magellan2 insertion at nucleotide 137 of the coding region). These transposon insertions were shown not to exert a polar effect on downstream gene expression by RNase protection assays (D. Hava and A. Camilli, unpublished data). The srtA mutant was generated by insertion-duplication with a derivative of the suicide vector pAC1000 (14) containing bases 20 to 402 of srtA, which correspond to TIGR4 genome coordinates 13791 to 14173 (http://cmr.jcvi.org). The 5′ and 3′ ends of this sequence are flanked by the extra sequences 5′-GATT-3′ and 5′-AATC-3′, respectively, introduced by subcloning, and the entire sequence was inserted into the SacII and SpeI polylinker sites in pAC1000.
TABLE 1.
S. pneumoniae strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| TIGR4 | Wild-type serotype 4 encapsulated | 11 |
| AC353 | Smr derivate of TIGR4 | 5 |
| AC1215 | AC353 rrgA::magellan5 Smr Spcr | 11 |
| AC1216 | AC353 rrgB::magellan5 Smr Spcr | 11 |
| AC1217 | AC353 rrgC::magellan5 Smr Spcr | 11 |
| AC1218 | AC353 srtC1::magellan5 Smr Spcr | 11 |
| AC1219 | AC353 srtC2::magellan5 Smr Spcr | 11 |
| AC1214 | AC353 srtC3::magellan2 Smr Cmr | 11 |
| AC1335 | AC353 ΔsrtA Smr | 18 |
| AC1441 | rrgA::rrgCCWSS Smr | 18 |
| AC1443 | rrgA::LPXTGCWSS Smr | 18 |
| AC1446 | AC1442 ΔsrtA Smr Cmr | 18 |
| JL50 | rrgA-HA ΔrrgB ΔrrgC Smr | 18 |
| JL51 | ΔrrgC rrgA::magellan5 Smr Spcr | This study |
| JL52 | AC353 Δislet Smr | 18 |
| JL53 | AC353 RrgC (V307Y) Smr | This study |
| JL54 | AC353 ΔsrtC1-srtC3 Smr Cmr | This study |
Construction of strains.
The RrgC V307Y point mutant was constructed by splicing by overlap extension PCR (23) with primers that alter the amino acid valine at position 307 (GTG) to a tyrosine (TAT). The primers that were used for this are (5′ to 3′) as follows: V307YF2, ACGGATTGATTATCCAGATACAGGGGAAGAAAC; V307YR1, CTGTATCTGGATAATCAATCCGTGGTCGCTTGT; V307YF1, GAGAATCAGATTGAAGTATCTC; V307YR2, TCAGCAGTACCAGCATAAAC.
The rrgA rrgC double mutant was constructed by transforming the magellan5 transposon insertion from strain AC1215 into a previously constructed strain containing a clean deletion of rrgC (18). The deletion of rrgC was confirmed by PCR after transposition.
The triple sortase deletion was constructed in the manner described previously (14). The entire coding region of the three sortases was replaced with a chloramphenicol resistance cassette, cat, which has its own promoter. The cat cassette was PCR amplified from the suicide vector pAC1000 with the already described primers catF1 and NcatR1. Fragments were PCR amplified from the S. pneumoniae TIGR4 chromosome that were 1 kb upstream of the start of srtC-1 with primers (5′ to 3′) srtC1F (AACAGATACAATGACAACAAAG) and srtC1R (GAAGAAGGTTTTTATATTACAGCTCCACCCTTTATCTTCAAACT CAT) and 1 kb downstream of the stop of srtC-3 with primers srtC3F (CAAGCTTATCGATACCGTCACCTTTTCTTTATCTTTGAG) and srtC3R (AACCAATTGAAGAAATATGAC). The primers srtC1R and srtC3R each contain a region of homology with the cat cassette, allowing the fusion of each of these 1-kb fragments of DNA to the cat cassette by splicing by overlap extension. This final 3-kb PCR product of the cat cassette flanked by 1 kb of DNA upstream and downstream of the three sortases was transformed into S. pneumoniae TIGR4 as previously described (5), and the double recombination event was selected for with medium containing 4 μg/ml chloramphenicol.
Production of anti-RrgA and anti-RrgC antibodies.
The portion of rrgC encoding amino acids 28 to 361 was cloned into the pTYB12 vector, and the IMPACT-CN protein purification strategy of New England BioLabs was used in order to express and purify the polypeptide. An amino-terminally hexahistidine-tagged, truncated form of RrgA (amino acids 499 to 866) was cloned into the expression vector pQE30. The cloned polypeptides were purified, dialyzed into phosphate-buffered saline, and injected into 8-week-old BALB/c female mice in combination with an adjuvant. Two weeks later, the mice were boosted and antisera were obtained after another 2 weeks. The antisera were used directly for detection of the proteins via Western blotting. Antiserum against RrgB was produced as previously described (18).
Fractionation of S. pneumoniae.
The fractionation of S. pneumoniae cultures into cell wall, protoplast, and culture supernatant was done as previously described by Bender et al. (4). Cultures were grown to an optical density of 0.5 to 0.6, and the cells were centrifuged at 4,000 × g for 15 min at 4°C. Ten milliliters of supernatant was removed and concentrated to a volume of 1 ml by centrifugation with an Amicon Ultra centrifugal filter device (3,000-molecular-weight cutoff). The pellet was washed once in protoplast buffer (20% sucrose, 50 mM MgSO4, 50 mM Tris [pH 7.4]) and spun at 16,100 × g for 15 min at 4°C. The pellet was resuspended in 1 ml protoplast buffer containing 40,000 U of mutanolysin (Sigma). The cell wall was digested for 2 h at 37°C. The lysates were centrifuged at 16,100 × g at 4°C for 10 min. The supernatant containing the cell wall fraction was removed, and the protoplast pellet was resuspended in 1 ml protoplast buffer.
Measuring pilus assembly by immunoblot analysis.
Fractionation samples were boiled in sodium dodecyl sulfate (SDS)-gel loading buffer (50 mM Tris-Cl [pH 6.8], 2% SDS, 0.5% bromophenol blue, 10% glycerol, 100 mM β-mercaptoethanol) for 10 min before gel electrophoresis. After cooling to room temperature (RT), the samples were loaded onto a NuPAGE 3 to 8% Tris-acetate gel (Invitrogen) and run at 150 V for 80 min and the gel was transferred to a nitrocellulose membrane (Invitrogen). The membrane was blocked with 5% nonfat milk in phosphate-buffered saline with 0.1% Tween (PBS-T) for 1 h at RT. All membranes were left in primary antisera overnight at 4°C. After four successive washes in PBS-T, the membranes were left in secondary antibody (sheep anti-mouse immunoglobulin G-horseradish peroxidase [Amersham]) for 1 h at RT. Protein was detected with the ECL-Plus horseradish peroxidase Western blotting detection kit (Amersham).
Quantification of pili with a 125I-labeled secondary antibody.
In order to quantify the amount of RrgB protein in pili, the same protocol was followed as described above for measuring pilus assembly by immunoblot analysis. However, for incubation of the secondary antibody, 10 μCi of radioactive secondary antibody (125I-labeled goat anti-mouse immunoglobulin G [Perkin-Elmer]) was added to 10 ml PBS-T and the membrane was incubated for 2 h at RT. The membranes were washed three times, and RrgB was visualized by phosphorimaging. Quantitation was analyzed with ImageQuant TL software (Amersham).
RESULTS
The requirements of the accessory pilin subunits for incorporation and the covalent association of RrgA and RrgC.
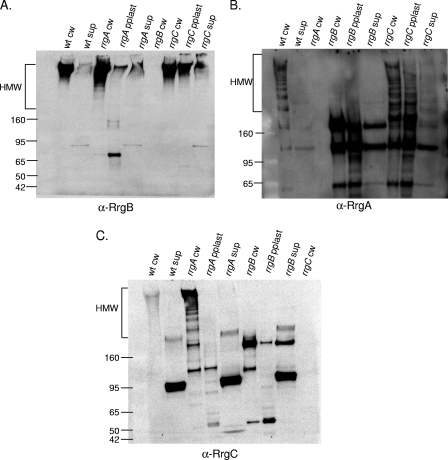
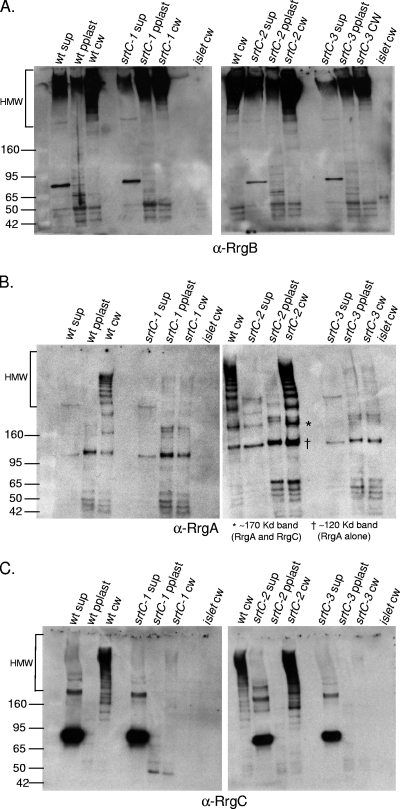
The incorporation of accessory subunits into the pilus backbone proceeds by an unknown mechanism. We wanted to determine the requirement for each of the three pilus subunits for this incorporation. In order to investigate this mechanism, the TIGR4 wild-type strain and isogenic rrg mutants were grown to the same optical density and the cultures were separated into three fractions, i.e., the culture supernatant, protoplast, and cell wall. Each of the fractions was electrophoretically separated under reducing conditions and probed separately by Western blotting with antisera against the individual pilin subunits. In the cell wall fraction of the wild-type strain, all three subunits are detected within high-molecular-weight (HMW) bands, where the bands represent pili of different lengths. The predicted molecular masses of the mature Rrg subunit proteins are as follows: RrgA, 92.7 kDa; RrgB, 64.9 kDa; RrgC, 38.9 kDa. Interestingly, the intensity of the labeling varied among the subunit-specific antisera. Specifically, the intensity of RrgB staining increased dramatically as the molecular weight of the pilus bands increased (Fig. 1A, lane 1). This is consistent with the presence of increasing amounts of this pilus backbone subunit as pilus length increases. In contrast, the intensity of RrgA labeling is constant, even for lower-molecular-weight pilus bands predicted to contain only one or a few subunits (Fig. 1B, lane 1). This suggests that the same amount of RrgA is present within each band, which leads us to hypothesize that each pilus species contains one or, at most, two RrgA subunits.
FIG. 1.
Incorporation of pilin subunits into pili in the absence of individual subunits. Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of the wild type (wt) and an rrgA, rrgB, or rrgC mutant strain were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antisera against RrgB (A), RrgA (B), and RrgC (C). As a negative control, the cell wall fraction of a mutant strain corresponding to the antiserum that was used in each panel is included. The HMW bands represent pili of differing lengths. The values on the left of each panel are molecular weight marker sizes in kilodaltons.
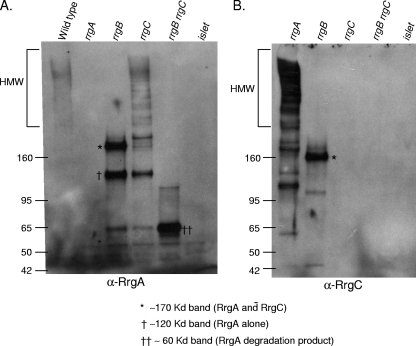
In an rrgC mutant strain, RrgA is still incorporated into pili, suggesting that incorporation of RrgA is not dependent on RrgC (Fig. 1B, lane 7). The reverse is also true; i.e., RrgC is incorporated into the pilus in an rrgA mutant strain (Fig. 1C, lane 3). However, in an rrgB mutant strain, in which no pilus backbone is formed, both RrgA and RrgC remain in the cell wall fraction, where they comigrate in a nonladder form at a molecular mass of ∼170 kDa (asterisks in Fig. 2A, lane 3, and B, lane 2). A second RrgA-positive band of ∼120 kDa is observed in the rrgB mutant strain cell wall (cross in Fig. 2A, lane 3); however, this band does not contain RrgC as it is not recognized by the anti-RrgC antiserum. This was determined by stripping and reprobing the blot (data not shown). It is possible that this RrgA-positive band is a monomer of RrgA that is running at a higher molecular mass (∼120 kDa) than its sequence would predict (92.7 kDa). The B. cereus pilin subunit BcpA has also been noted to appear larger via gel electrophoresis than would be expected (6). However, it cannot be ruled out that this band, which is larger than the predicted monomeric size of RrgA, is a multimer of RrgA proteins. The formation of RrgA multimers would be consistent with prior studies that detected RrgA clusters by immuno-gold TEM (13, 18).
FIG. 2.
RrgA and RrgC form a complex in the cell wall compartment. The bacterial cell wall (cw) fractions of the wild type; rrgA, rrgB, and rrgC mutants; and an rrgB rrgC double mutant were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-RrgA (A) or anti-RrgC (B) antiserum. The band representing an RrgA/RrgC multimer is indicated with an asterisk, and the large and small RrgA bands are represented by one or two crosses, respectively, with a key below the panel. The values on the left of each panel are molecular weight marker sizes in kilodaltons.
In an rrgB rrgC double mutant, an RrgA-positive species is observed at ∼60 kDa (Fig. 2A, lane 5), which is substantially lower than the predicted molecular masses of both mature RrgA and the RrgA-positive species that is observed in either the rrgB or the rrgC single mutant (Fig. 2A, lanes 3 and 4). This shift to a lower molecular mass of ∼60 kDa suggests that this form of RrgA is monomeric with additional processing events taking place or is a degradation product. In a prior report examining the localization of RrgA in the cell wall of an rrgB rrgC double mutant, a faint ∼60-kDa putative degradation product is observed in addition to a major band of ∼100 kDa that is presumably monomeric RrgA (20). Because a major band of this size is not visualized in our data, we hypothesize that there are strain differences or variations in the growth conditions used in the two studies that led to these contrasting results.
The observation that the ∼170-kDa RrgA- and RrgC-positive bands comigrate in the rrgB strain cell wall fraction, even after boiling in SDS and denaturing electrophoresis, suggests that RrgA is covalently attached to RrgC and thus that they may be found together in the pilus. To examine this further, the membrane that was probed with anti-RrgC was stripped and reprobed with anti-RrgA antiserum. The ∼170-kDa band was detected with the anti-RrgA antiserum (data not shown), indicating that it contains both RrgC and RrgA. The presence of RrgA together with RrgC within native pili is supported by immuno-gold TEM data showing that RrgA is typically found in clusters in surface pili, with RrgC found either colocalized with these RrgA clusters or in what appear to be single copies in pili (13). Interestingly, this same 170-kDa RrgA-RrgC species can be seen in the culture medium (supernatant) of an rrgB strain when it is probed with both anti-RrgA and anti-RrgC antisera (Fig. 1B, lane 6, and C, lane 8). This indicates that the RrgC and RrgA multimer is released into the culture medium when RrgB is absent.
RrgB polymerization occurs in the absence of both RrgA and RrgC.
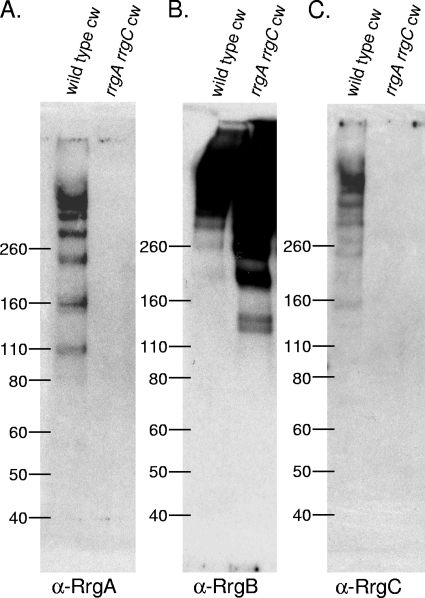
To further investigate the requirements for the backbone protein, RrgB, to polymerize, we tested whether RrgA or RrgC is necessary for RrgB polymerization to occur. Although we knew from previous results that one of the two could be absent and still result in RrgB polymerization (Fig. 1), we wanted to test the hypothesis that the polymerization of RrgB would not occur in the absence of both accessory proteins. Therefore, we constructed an rrgA rrgC double mutant and used Western blotting to determine if RrgB could still polymerize. This mutant strain retained the ability to polymerize RrgB into HMW pili localized in the cell wall fraction (Fig. 3B). Therefore, we conclude that neither accessory subunit is required for the processes of RrgB polymerization and localization to the cell wall.
FIG. 3.
RrgB polymerization occurs in the absence of both RrgA and RrgC. The bacterial cell wall (cw) fraction of the wild type and an rrgA rrgC strain was separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antisera against RrgA (A), RrgB (B), and RrgC (C). The values on the left of each panel are molecular weight marker sizes in kilodaltons.
The housekeeping sortase, SrtA, is dispensable for pilus assembly and cell wall localization.
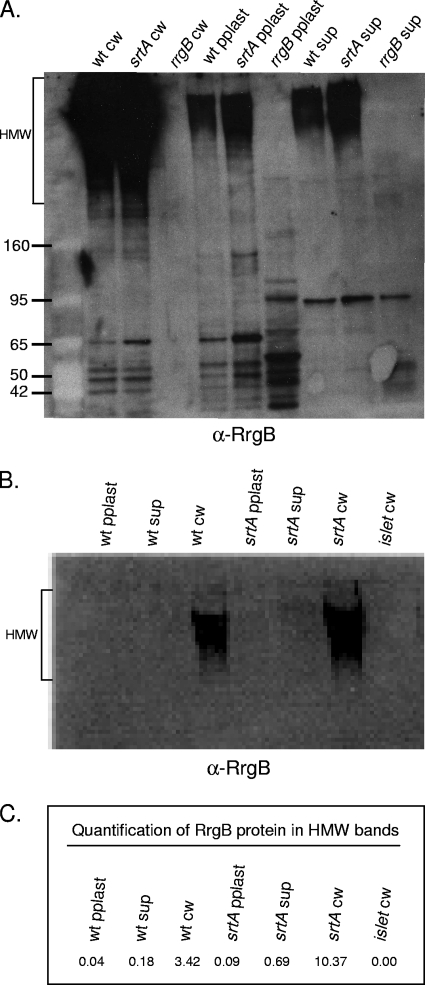
In order to investigate the sortase requirements for RrgB polymerization and localization to the cell wall, we constructed strains lacking each of the four sortase genes present in TIGR4. We first investigated a possible role for SrtA in pilus biogenesis and cell wall localization since, in three distinct pilus systems, it has been shown that the housekeeping sortase facilitates the anchoring of pili to the cell surface. A strain lacking srtA was able to assemble RrgB into HMW pili and, surprisingly, was unaffected in its ability to localize pili to the cell wall (Fig. 4A, lanes 1 and 2). If anything, the srtA mutant appeared to produce more cell wall-localized pili than the wild type. In order to investigate this possible difference in expression level further, the fractions were analyzed by quantitative Western blotting with a radioactive secondary antibody (Fig. 4B and C). The values are the averages of two separate experiments that gave similar results. Although there is threefold more RrgB detected in the srtA cell wall fraction, there is a similar increase in RrgB detected in the other two fractions of this mutant. Thus, the amount of RrgB released into the culture supernatants was similar for the wild-type and srtA mutant strains, 5 and 6%, respectively. The small fraction of RrgB that is found in the culture supernatant may be due to peptidoglycan turnover. Based on these results, we hypothesize that SrtA is dispensable in S. pneumoniae for the processes of pilus polymerization and cell wall localization.
FIG. 4.
SrtA is not required for pilus polymerization or localization to the cell wall. (A) Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of the wild type (wt) and an srtA strain were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antiserum against RrgB. As a negative control, an rrgB strain was included. The values on the left are molecular weight marker sizes in kilodaltons. (B) Quantification of RrgB in HMW pili with a 125I-labeled secondary antibody. (C) The amount of HMW RrgB protein in each sample is shown. These values are the averages of two separate experiments, one of which is the blot shown in panel B.
Pilus assembly in class C sortase mutants.
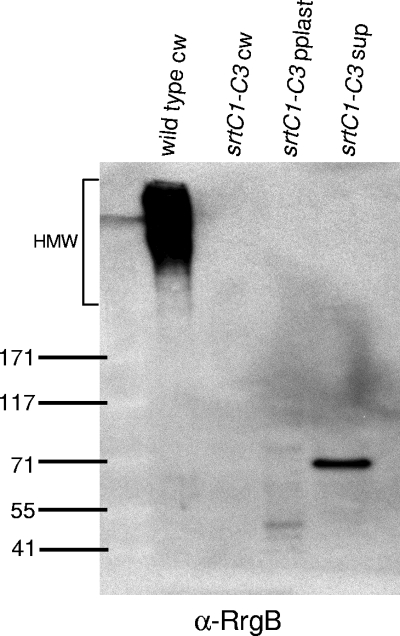
Having found no detectable role for SrtA in pilus biogenesis, we turned our attention to the three class C sortases encoded in the rlrA pilus locus. We first asked whether RrgB polymerization would occur in the absence of all three class C sortases by constructing a triple sortase deletion strain (srtC1-C3). RrgB polymerization was undetectable in this strain (Fig. 5), indicating a requirement for one or more of these sortases in pilus assembly.
FIG. 5.
The class C sortases encoded in the islet are required for pilus polymerization. The bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of an srtC1-C3 triple sortase deletion mutant strain were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antiserum against RrgB. The bacterial cell wall fraction of the wild type was run as a positive control. The values on the left are molecular weight marker sizes in kilodaltons.
We next investigated the requirement of the three individual class C sortases for pilus assembly by probing individual sortase mutants for the ability to polymerize RrgB into pili (Fig. 6A). The Western blot results show that RrgB polymerization does not rely on any one class C sortase. That is to say that RrgB is polymerized into cell wall-localized HMW pili in an srtC-1, srtC-2, or srtC-3 mutant strain (Fig. 6A, lanes 6, 11, and 14). To investigate the incorporation of the accessory pilin subunits in these sortase mutants, the same cell wall fractions were probed with antisera to RrgA and RrgC. In an srtC-2 mutant strain, both RrgA and RrgC are present in HMW pili, showing that srtC-2 is not required for either of these subunits to be incorporated into pili (Fig. 6B and C, lane 11 in each panel). In contrast, the srtC-1 and srtC-3 mutants show greatly reduced RrgC and RrgA incorporation into pili (Fig. 6B and 6C, lanes 6 and 14 in each panel). These data indicate that there is redundancy in the sortases for incorporation of RrgB into pili but specificity in the recognition and/or processing of the accessory subunits RrgA and RrgC.
FIG. 6.
Redundancy of the sortases for pilus assembly. Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of the wild-type (wt) and srtC-1, srtC-2, and srtC-3 mutant strains were separated by gel electrophoresis, transferred to nitrocellulose, and probed with antiserum against RrgB (A), RrgA (B), or RrgC (C). As a negative control, a cell wall fraction of a strain in which the entire islet has been deleted was run on each gel. In panel B, the band corresponding to an RrgA/RrgC multimer is indicated with an asterisk, the RrgA band is represented by one cross, and a key is provided below the panel. The values on the left of each panel are molecular weight marker sizes in kilodaltons.
There is an abundant band (∼75 kDa) in the culture supernatant recognized by the RrgC antisera, as shown in Fig. 1C, 6C, 7C, 8, and 9C. However, this band is a cross-reactive species as it is detected at the same intensity in the supernatant fraction of an rrgC mutant strain (Fig. 8, lane 6). It is also seen in the supernatant fractions, although to a lesser extent, when they are probed with antisera to RrgB and RrgA. It is unclear if this cross-reactive band represents a component of the culture medium or is a secreted protein of S. pneumoniae.
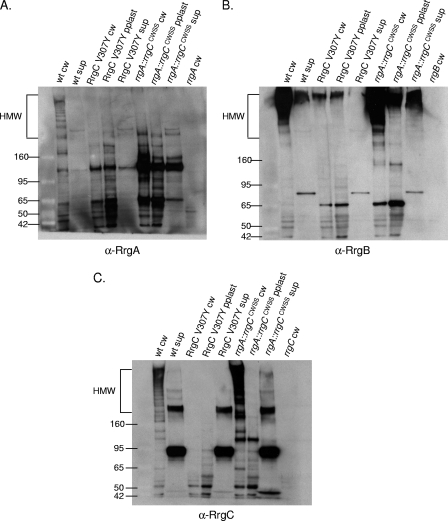
FIG. 7.
The variant CWSSs of the Rrg proteins generate specificity in the pilus assembly process. Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of wild-type (wt) and RrgC V307Y and rrgA::rrgCCWSS mutant strains were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-RrgA (A), anti-RrgB (B), and anti-RrgC (C) antisera. The values on the left of each panel are molecular weight marker sizes in kilodaltons.
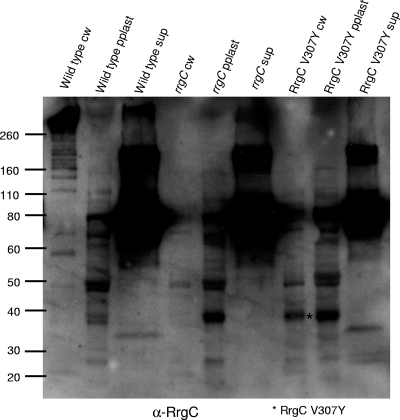
FIG. 8.
The RrgC V307Y mutant protein is stably expressed. Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of the wild-type and rrgC and RrgC V307Y mutant strains were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-RrgC antiserum. The RrgC V307Y mutant protein is indicated by an asterisk and the key at the bottom. The values on the left are molecular weight marker sizes in kilodaltons.
FIG. 9.
RrgA containing a canonical LPxTG CWSS cannot enter the pilus assembly pathway. Bacterial and culture fractions (cell wall [cw], protoplast [pplast], and culture supernatant [sup]) of the wild-type (wt) and rrgA::SP0082CWSS mutant strains and the cell wall fraction of the rrgA mutant strain were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-RrgA (A), anti-RrgB (B), and anti-RrgC (C) antisera. The values on the left of each panel are molecular weight marker sizes in kilodaltons.
The variant CWSSs of the Rrg proteins determine sortase specificity.
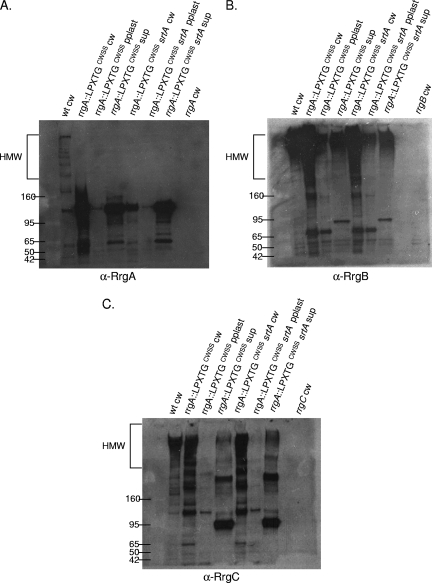
In most of the gram-positive pilus clusters examined to date, the pilin subunits have a CWSS that begins with the canonical leucine of the LPxTG motif. S. pneumoniae TIGR4, in contrast, has three pilin subunits, RrgA, RrgB, and RrgC, with variant first amino acids of Y, I, and V, respectively. In order to test if these amino acids or, alternatively, the entire CWSS determines the specificity of assembly by the sortases, three mutant strains with altered CWSSs were examined for subunit incorporation into the pilus.
The first strain, RrgC V307Y, is isogenic with the wild type except for a change of the first amino acid of the CWSS of RrgC to that of RrgA. When this mutant strain was fractionated and probed with RrgB, RrgA, and RrgC antisera, it was apparent that it had a profound defect in pilus assembly (Fig. 7, lane 3 in each panel). There is a small amount of HMW pili found in the cell wall and protoplast fractions (Fig. 7B, lanes 3 and 4), suggesting that pilus assembly is not completely blocked. When incorporation of the accessory subunits was investigated in this mutant, it followed that they were no longer seen in an HMW ladder form due to a defect in RrgB polymerization (Fig. 7A and C, lane 3 in both panels).
Although it was not incorporated into the pilus, the analysis of RrgA in the RrgC V307Y strain yielded two interesting results. Probing with RrgA antiserum revealed that RrgA is retained in the cell wall in the previously mentioned ∼120-kDa species (Fig. 7A, lane 3). This indicates that the localization of RrgA to the cell wall and, if the ∼120-kDa species is a RrgA dimer, linkage to itself are not dependent on polymerization of RrgB. In contrast, the ∼170-kDa RrgA/RrgC multimer was no longer detected in the RrgC V307Y mutant cell wall (Fig. 7C, lane 3). Due to the lack of the RrgC V307Y protein in the cell wall fraction, either in the monomeric form or in the ∼170-kDa RrgA/RrgC multimer, we hypothesize that a specific interaction between RrgC and one or more sortases has been disrupted or altered in the RrgC V307Y mutant, resulting not only in inhibition of pilus assembly but also inhibition of the linkage of RrgC to RrgA. We hypothesize that this blockage of the sortase complex, and potential jamming of the Sec translocon, results in the degradation and/or a block in translation of the Rrg proteins. An alternative possibility is that the point mutant is unstable in the cell and is therefore not detected. To test this second possibility, we examined the expression of the RrgC V307Y mutant protein by Western blotting with a 4 to 12% gel in order to visualize lower-molecular-weight proteins. The RrgC V307Y mutant protein was expressed and stable, as it is seen in the cell wall fraction in an ∼38-kDa form (Fig. 8, lane 7), which is in agreement with the predicted molecular mass of the mature form of RrgC (38.9 kDa). In the cell wall fraction of a wild-type strain (Fig. 8, lane 1), RrgC is not found in this presumably monomeric form but is instead present in HMW pili. Based on this result, we favor the first hypothesis, i.e., that lack of RrgB polymerization in this strain is a result of the RrgC V307Y point mutant “poisoning” the pilus assembly sortase complex.
Additionally, we wanted to investigate whether or not pili would be formed in a strain where the entire CWSS of an accessory subunit protein was replaced. A previously constructed strain was used in which amino acids including and downstream of the YPxTG sequence from RrgA were precisely replaced with the CWSS of RrgC. In this chimera, referred to as rrgA::rrgCCWSS, RrgB was polymerized into a pilus (Fig. 7B, lane 6). Additionally, RrgC is incorporated into the pilus (Fig. 7C, lane 6). In contrast, there is no RrgA chimera found in the pilus, although the protein was produced since it can be identified at a low molecular mass (∼120 kDa) in large amounts in all three fractions (Fig. 7A, lanes 6 to 8). Notably, the ∼170-kDa RrgA/RrgC multimer is not detected in this strain, indicating that the chimera inhibits the linkage of RrgA to RrgC.
Interestingly, there is a large amount of low-molecular-weight RrgA chimera found in the supernatant fraction, where RrgA does not normally accumulate in the wild type (Fig. 7A, lane 2 compared with lane 8). This suggests that a specific interaction between RrgA and one or more sortases has been altered in the rrgA::rrgCCWSS mutant, resulting in missorting of a significant fraction of the chimera which is released into the culture supernatant in monomeric and/or dimeric form. However, this missorting does not inhibit RrgB and RrgC pilus assembly.
The canonical LPxTG CWSS cannot substitute for a pilus-specific CWSS.
We wanted to further investigate the role of the CWSS and, more specifically, if altering the RrgA CWSS to a canonical SrtA-mediated CWSS would affect its ability to be incorporated into pili and/or its localization to the cell wall in both the wild type and an srtA mutant. Although we demonstrated above that SrtA is not required for pilus assembly or localization to the cell wall, we were interested in the possibility that adding a canonical CWSS would create a situation in which SrtA gained the function of incorporating RrgA into the pilus.
In order to address this, we fractionated a previously reported chimeric strain that has the RrgA CWSS precisely replaced by the CWSS of the LPxTG motif protein SP0082. The chimeric RrgA protein, referred to as rrgA::LPXTGCWSS, was not incorporated into pili but was found in the cell wall and supernatant fractions in an ∼120-kDa species (Fig. 9A, lanes 2 and 4). Additionally, the srtA mutation resulted in reduced levels of this band in the cell wall fraction (Fig. 9A, lane 5). These data are consistent with the model in which specificity in the sorting process is determined by the CWSS.
Similar to the result obtained with the rrgA::rrgCCWSS chimera (Fig. 7), the rrgA::LPXTGCWSS chimera does not inhibit pilus polymerization as RrgB is still polymerized into an HMW ladder when visualized via Western blotting (Fig. 9B, lane 2). Additionally, the loss of SrtA in this strain has no effect on RrgB polymerization (Fig. 9B, lane 5). Similarly, RrgC incorporation into the pilus was not interrupted by the presence of the chimeric protein or the deletion of srtA in the chimeric strain (Fig. 9C, lanes 2 and 5). These data suggest that missorting of RrgA, by either SrtA or the pilus-associated sortases, does not interfere with polymerization of RrgB or with the incorporation of the accessory subunit RrgC into pili.
DISCUSSION
In gram-positive bacteria, the processes of multisubunit pilus assembly and localization to the cell wall is incompletely understood. Namely, the mechanism of incorporating the accessory subunits into pili and the sortase functions that distinguish between backbone assembly and accessory subunit incorporation remain black boxes. To address this complex process, and building upon a previous study (18) that showed that incorporation of RrgA into pili required SrtC-3, we sought to determine the requirements of each of the pilus subunits and of each of the sortases for pilus biosynthesis.
It was shown that RrgB requires neither RrgA nor RrgC for polymerization, but both of these accessory subunits require the backbone pilin, RrgB, for incorporation into the pilus. It was also seen, through analysis of a mutant lacking RrgB, that RrgA and RrgC are covalently linked and are localized to the cell wall. Although we have no data to suggest a mechanism for RrgA linkage to RrgC, it is possible that the E box-like motifs found in both of these proteins play a role. We do not have an understanding of how the E box motif functions in incorporating accessory subunits into pili. However, it has been shown that the glutamic acid within the E box is required for this incorporation to occur. Because RrgA and RrgC have the conserved glutamic acid in their E box-like motifs, it is plausible that these two subunits are linked in a manner similar to how accessory subunits are incorporated into pili.
There are many roles that can be proposed for the RrgA and RrgC accessory proteins, both separately and together. Although RrgA has been shown to mediate adherence to host cells in vitro, it is likely that it serves multiple roles, some of which may rely on its association with RrgC. As RrgB is known to function as the pilus backbone subunit, we postulate roles for RrgA and RrgC in the assembly or regulation of the assembly of the pilus. For example, in the wild-type strain, these accessory proteins could initiate the pilus polymerization process. Additional or alternative possible roles include the accessory subunits acting as stabilizers or, in contrast, adding flexibility to surface pili or pilus bundles. All of these possibilities require further experimentation for verification and clarification.
One model that we favor, and which our data support, is that the accessory subunits, either singly or in an RrgA/RrgC multimer form, are found at the tip of each pilus. This idea is supported by Western blot analysis showing that these two accessory proteins are found in similar quantities within each “rung” of the ladder of bands that we believe represent pili of different lengths. These data indicate that there is at least one (and possibly multiple) RrgA or RrgC subunit in each pilus, even low-molecular-weight pili that are near the bottom of the ladder and which likely consist of only two or three subunit proteins. This could only be possible if these proteins were involved in the initiation of pilus assembly. This model is also parsimonious with the sortase-pilus hypothesis proposed by Ton-That and Schneewind, which hypothesizes that the lysine found in the pilin motif of the major pilin is the nucleophile that mediates subunit incorporation at the base of the growing pilus. Specifically, new subunit incorporation is thought to occur through the pilin motif lysine amino group performing a nucleophilic attack of a sortase/pilin subunit thioester bond—a bond that exists between the conserved sortase active-site cysteine and the pilin subunit CWSS threonine. Notably, RrgA and RrgC lack a consensus pilin motif and thus should only be present at the tips of pili. In this model, RrgB would perform the initial nucleophilic attack on an RrgA- or RrgC-sortase thioester bond and then additional RrgB monomers would polymerize at the base of the pilus to extend its length. We note that the presence of RrgA and RrgC only at the tips of pili conflicts with the observed clustering of RrgA at many points along surface pili as visualized by immuno-gold TEM. However, based on the propensity of S. pneumoniae pili to bundle together (13), we speculate that the observed clustering of RrgA at many points along surface pili is due to clustering of RrgA subunits that are at the tips of different-length pili which are bundled together.
Another piece of the pilus assembly process in S. pneumoniae that our data shed light on is the role of the sortases. In teasing out which sortases are necessary to polymerize and localize pili to the cell wall, we considered the housekeeping sortase, SrtA, which is responsible for covalently attaching many surface proteins to the cell wall. There are three recent reports showing that housekeeping sortases are required for anchoring of pili to the cell wall. These reports were on different bacteria, B. cereus, C. diphtheriae, and S. agalactiae, all of which have one or more pilin subunits that have the canonical LPxTG motif. The model suggests that pili are polymerized by cis-encoded sortases and then the fully assembled pilus is attached to the cell wall in the final step by the trans-encoded housekeeping sortase. We suggest that this model does not apply to the S. pneumoniae RlrA pilus because our data show that in an srtA strain, the pilus was found polymerized and localized to the cell wall. However, we cannot rule out the possibility that SrtA does, in fact, catalyze the localization of RlrA pili to the cell wall but that, for unknown reasons, the pili are nevertheless copurifying with the cell wall fraction in our mutanolysin digestion and centrifugation protocol.
We observed an overall increase in pilus expression in the srtA mutant strain. A similar result has been reported for Enterococcus faecalis pilin subunits EbpB and EbpC (16). In a result contrasting to that obtained with S. pneumoniae and E. faecalis, SrtA has been shown to be involved in the positive transcriptional regulation of a pilus cluster in S. agalactiae (8, 21). Although we have not measured the transcript level in our system, the consistently higher level of RrgB protein in the srtA mutant lends itself to the hypothesis that, in S. pneumoniae, SrtA may be repressing islet expression. It would be informative to measure the transcript level of the islet in an srtA mutant during different points in the growth phase, as the islet has been shown to be differentially regulated in early log phase compared to late log phase.
Kharat and Tomasz previously reported a role for SrtA in anchoring LPxTG motif proteins to the S. pneumoniae cell wall and also reported the lack of a role for SrtC-1 and SrtC-3 in this process (17). Although SrtC-2 was not included in their analysis, we predict that the result would be the same. The lack of a role for SrtA in pilus assembly and cell wall localization in S. pneumoniae is consistent with this previous report, since the Rrg pilin proteins lack a canonical LPxTG motif. Instead, we show that the class C sortases encoded in cis are necessary and sufficient for pilus assembly and localization to the cell wall. For reasons that are not understood, these three sortases act in a redundant manner in the processes of RrgB polymerization and localization to the cell wall since RrgB pili were observed in each single-mutant strain.
In contrast, there is a drastic reduction in the incorporation of accessory subunits RrgA and RrgC into pili in an srtC-1 or srtC-3 mutant strain, which indicates that there is specificity in the sortases for the recognition and processing of these accessory subunits. This result is analogous to what has been reported for the SpaDEF cluster of C. diphtheriae, in which two sortases, SrtB and SrtC, are present. In this system, SrtB or SrtC can polymerize the major pilin subunit, SpaD, but SrtB alone is needed for the accessory subunit SpaE to be incorporated into the pilus (10). Additionally, these authors show that less SpaF is found in the HMW bands in an srtC mutant strain, revealing another specific interaction between an accessory subunit and a sortase. These parallel findings suggest that, at least for these two pilus systems, the roles of the sortases in pilus biogenesis differ with respect to backbone assembly versus accessory subunit incorporation. That is to say that there appears to be redundancy among the sortases for the recognition, cleavage, and assembly of the major pilin subunit into the pilus backbone but specificity among the sortases for the recognition and incorporation of the individual accessory subunits.
Although the results are similar between the two different bacterial pilus clusters, there is a distinct difference between them. All of the pilin subunit proteins of C. diphtheriae encode a CWSS with the canonical leucine as the first amino acid of the LPxTG motif, whereas the pilus proteins of S. pneumoniae have a variant amino acid at that position.
The importance of the leucine residue of the LPxTG motif has been investigated in some depth in S. pyogenes (1, 2). In this organism, a mutation was made in the housekeeping sortase, SrtA, which altered the localization of four out of the five surface proteins tested but did not have an effect on T6, which has been shown subsequently to be the major subunit of the S. pyogenes pilus. Interestingly, all five of the proteins, M6 (LPSTG), protein F (LPATG), ScpA (LPTTN), GRAB (LPTTG), and T6 (LPSTG), encode canonical LPxTG CWSS motifs. In addition, they showed that loss of another sortase, SrtB, had an effect only on T6 localization. This sortase is now known to be a member of the class C sortase family, the members of which are known to polymerize pili. These data indicate that a strict one-to-one correspondence between class A housekeeping sortases and CWSSs with an LPxTG motif is not universal but that other sequences outside this motif are also important in sortase recognition.
The experimental approach of altering the CWSS has been used to determine if the housekeeping sortase of S. pyogenes can properly process a protein with a variant CWSS (1). The surface protein T3 (formerly referred to as orf100 or Fct3), which has been shown to be a major pilin backbone subunit (30), has a CWSS of QVPTG, which was shown to be recognized by a class C sortase, SrtC2. However, a mutant protein in which the QVPTG CWSS was altered to LPSTG was no longer properly localized to the cell wall by SrtC2, indicating that SrtC2 is specifically recognizing the QVPTG CWSS. Interestingly, it also was not properly localized by the housekeeping sortase, SrtA, although it now had the canonical SrtA CWSS motif. This suggested that other parts of the T3 CWSS are involved in recognition and processing by SrtC2.
In the present study, we used two different approaches to test the importance of the CWSS in sortase substrate recognition. The first was to replace the first amino acid of the CWSS of the accessory subunit RrgC with that of accessory subunit RrgA. Making this change results in a surprising reduction of pilus formation, suggesting that the mutant protein is poisoning the pilus assembly complex. This finding suggests that the first amino acid of the CWSS contributes to, but is not the sole determinant of, sortase specificity. Together with the data on T3 in S. pyogenes, this finding leads us to hypothesize that specificity for the recognition and processing of pilin subunits by sortases requires multiple sequences within the CWSS. In light of this hypothesis, we speculate that the RrgC point mutant is recognized by a different sortase(s) than that which recognizes wild-type RrgC, which results in the mutant protein getting stuck in the sortase complex, thus blocking assembly. We further speculate that such a block results in a potential jamming of the Sec translocon and a subsequent degradation and/or block in the translation of the Rrg proteins, which is consistent with the decreased amount of pilin proteins detected in all fractions.
In the second approach, we analyzed chimeras in which the entire CWSS of RrgA was replaced with that of RrgC or a canonical LPxTG CWSS. Both mutants, in contrast to the point mutant above, still polymerized RrgB and incorporated RrgC as well. However, although both RrgA chimeric proteins were found in the cell wall fractions, in neither strain was the altered protein incorporated into pili. This result indicates that these chimeras were recognized and processed by sortases but were unable to be incorporated into pili. The question of why neither chimera is incorporated into pili remains unanswered. One possibility is that RrgA and RrgC are normally found together in the pilus and that the association between these two subunits requires RrgA sequences both inside and outside of its CWSS. In both RrgA chimeras, the native RrgA CWSS sequence would be missing, resulting in missorting. In support of this possibility, in the rrgB mutant, in which the pilus backbone is not assembled, we detect a cell wall-localized species that contains RrgA and RrgC, which may be a precursor that would normally be incorporated at the tips of pili. Although in this hypothesis we propose that these two accessory subunits are attached to one another, our data clearly show that this association is not a prerequisite, since either subunit can be incorporated into pili in the absence of the other. The biological role of this apparent plasticity remains to be determined.
Precisely how the specificity of SrtC-1 and SrtC-3 for the incorporation of the accessory subunits RrgA and RrgC into pili is determined remains unknown. We propose that differences in structure, conformation, and/or active-site sequences between the class C sortases mediate this specificity. All three sortases are classified as class C (or class 3) by the analyses of Dramsi et al. (9) and Comfort and Clubb (7). However, although SrtC-1 and SrtC-2 are 68% similar and 20% identical, SrtC-3 has a much lower degree of overall similarity to either SrtC-1 (37% similarity and 15% identity) or SrtC-2 (37% similarity and 14% identity). In addition, their hydrophobic characteristics suggest that SrtC-1 and SrtC-2 are anchored in the membrane via a mechanism separate from that of SrtC-3 (SrtC-1 and SrtC-2 have putative transmembrane anchors, whereas SrtC-3 is a putative lipoprotein), which might allow for a structural distinction that would lead to functional differences.
Despite the investigations presented here, we are left with many questions regarding pilus assembly that will undoubtedly be uncovered as future research continues to clarify this complicated and intricate biological process. Our analysis of the S. pneumoniae pilus cluster has revealed several interesting features, such as redundancy in the processes of assembly and localization of pili to the cell wall yet specificity in the processes of recognition and incorporation of the accessory subunits. Nevertheless, it is clear that much work remains to be done in order to fully understand the coordination and the level of complexity with which the three sortases work together.
Acknowledgments
This work was funded by the Howard Hughes Medical Institute and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
We thank Ernesto Munoz for critical reading of the manuscript.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Barnett, T. C., A. R. Patel, and J. R. Scott. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 1865865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 1842181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, M. H., R. T. Cartee, and J. Yother. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 1856057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172131-135. [DOI] [PubMed] [Google Scholar]
- 6.Budzik, J. M., L. A. Marraffini, and O. Schneewind. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66495-510. [DOI] [PubMed] [Google Scholar]
- 7.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 601401-1413. [DOI] [PubMed] [Google Scholar]
- 9.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 10.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 1881526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 451389-1405. [PMC free article] [PubMed] [Google Scholar]
- 12.Hava, D. L., C. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilleringmann, M., F. Giusti, B. C. Baudner, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and I. Ferlenghi. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of RrgA. PLoS Pathog. 4e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 1878340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang, H. J., F. Coulibaly, F. Clow, T. Proft, and E. N. Baker. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 3181625-1628. [DOI] [PubMed] [Google Scholar]
- 16.Kemp, K. D., K. V. Singh, S. R. Nallapareddy, and B. E. Murray. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 755399-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 712758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 742453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazmanian, S., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285760-763. [DOI] [PubMed] [Google Scholar]
- 20.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobbs, A. H., R. Rosini, C. D. Rinaudo, D. Maione, G. Grandi, and J. L. Telford. 9 June 2008. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect. Immun. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed]
- 22.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 997-102. [DOI] [PubMed] [Google Scholar]
- 23.Senanayake, S. D., and D. A. Brian. 1995. Precise large deletions by the PCR-based overlap extension method. Mol. Biotechnol. 413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ton-That, H., G. Liu, S. Mazmanian, K. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 9612424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ton-That, H., l. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53251-261. [DOI] [PubMed] [Google Scholar]
- 27.Ton-That, H., S. Mazmanian, K. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J. Biol. Chem. 2759876-9881. [DOI] [PubMed] [Google Scholar]
- 28.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in gram-positive bacteria. Trends Microbiol. 12228-234. [DOI] [PubMed] [Google Scholar]
- 29.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]
- 30.Zähner, D., and J. R. Scott. 2008. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J. Bacteriol. 190527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]