Abstract
ATP/ADP translocases are a hallmark of obligate intracellular pathogens related to chlamydiae and rickettsiae. These proteins catalyze the highly specific exchange of bacterial ADP against host ATP and thus allow bacteria to exploit their hosts' energy pool, a process also referred to as energy parasitism. The genome sequence of the obligate intracellular pathogen Lawsonia intracellularis (Deltaproteobacteria), responsible for one of the most economically important diseases in the swine industry worldwide, revealed the presence of a putative ATP/ADP translocase most similar to known ATP/ADP translocases of chlamydiae and rickettsiae (around 47% amino acid sequence identity). The gene coding for the putative ATP/ADP translocase of L. intracellularis (L. intracellularis nucleotide transporter 1 [NTT1Li]) was cloned and expressed in the heterologous host Escherichia coli. The transport properties of NTT1Li were determined by measuring the uptake of radioactively labeled substrates by E. coli. NTT1Li transported ATP in a counterexchange mode with ADP in a highly specific manner; the substrate affinities determined were 236.3 (± 36.5) μM for ATP and 275.2 (± 28.1) μM for ADP, identifying this protein as a functional ATP/ADP translocase. NTT1Li is the first ATP/ADP translocase from a bacterium not related to Chlamydiae or Rickettsiales, showing that energy parasitism by ATP/ADP translocases is more widespread than previously recognized. The occurrence of an ATP/ADP translocase in L. intracellularis is explained by a relatively recent horizontal gene transfer event with rickettsiae as donors.
Some of the most important bacterial pathogens of humans can only replicate within eukaryotic cells. These obligate intracellular bacteria have developed sophisticated mechanisms to interact with and exploit their hosts. Prime examples of obligate intracellular bacterial pathogens are members of the orders Chlamydiales and Rickettsiales (hereafter referred to as chlamydiae and rickettsiae, respectively). These phylogenetically largely unrelated groups of microorganisms employ nucleotide transport (NTT) proteins, which import nucleotides or allow parasitization of their hosts' energy pool by exchanging bacterial ADP for host ATP (6, 10, 16, 17, 26, 30, 45, 51). Among bacteria, NTT proteins are unique to chlamydiae and rickettsiae and were, in addition, only found in plastids of plants and algae (30, 39, 50, 57).
NTT proteins have been classified into the ATP/ADP antiporter family AAA by Saier and coworkers (TC number 2.A.12 in the Transport Classification Database [44]). Yet, recent studies showed that NTT proteins comprise transporters with highly dissimilar transport modes and substrate affinities. An alternative classification of NTT proteins according to transport mode was therefore proposed, subdividing the NTT protein family into three classes; class I contains nucleotide antiporters, class II contains proton-driven nucleotide symporters, and class III contains NAD+/ADP antiporters (17). Bacterial and plastidic NTT proteins are fundamentally different from the analogous ADP/ATP carriers of the mitochondrial carrier family with respect to structure and transport characteristics (25, 41-43, 57). In contrast to ATP/ADP translocases of NTT protein family class I, which enable bacterial energy parasitism, mitochondrial ADP/ATP carriers function in the reverse direction, exporting newly synthesized ATP from the mitochondrial matrix to the host cytosol in exchange for ADP.
Using BlastP (2) against the nonredundant protein sequences at GenBank/EMBL/DDBJ in order to find as-yet-unrecognized NTT proteins, we recently identified a gene coding for an NTT protein most similar to known chlamydial and rickettsial ATP/ADP translocases in the genome sequence of Lawsonia intracellularis PHE/MN1-00. L. intracellularis is a gram-negative, microaerophilic, obligate intracellular bacterium belonging to the Deltaproteobacteria. L. intracellularis enters the host cell via induced phagocytosis; the phagosome is quickly degraded, and Lawsonia resides directly in the cytoplasm (27). L. intracellularis is an important veterinary pathogen causing proliferative enteropathy (ileitis) in many mammals but mostly in pigs (27, 37, 46). Proliferative enteropathy is characterized by a progressive proliferation of immature intestinal epithelial cells (enterocytes) following infection with L. intracellularis. The disease, which can persist for several weeks, leads to anorexia, diarrhea, reduced growth of infected animals, and decreased reproductive performance (27, 34, 46). Although proliferative enteropathy is considered one of the most economically important diseases in the swine industry worldwide, causing hundreds of millions of U.S. dollars in extra costs annually (27, 29, 36), data on the molecular mechanisms important for the pathogenicity and interaction of L. intracellularis with its host cells are surprisingly scarce.
In this study, we characterized the predicted ATP/ADP translocase from L. intracellularis by using heterologous expression in Escherichia coli. We identified its biochemical transport properties and showed that it functions as an ATP/ADP antiporter, importing host ATP in exchange for bacterial ADP. Pathway reconstruction based on the L. intracellularis genome sequence, however, suggested that L. intracellularis is still able to regenerate ATP on its own. Phylogenetic analysis suggested that the ATP/ADP translocase from L. intracellularis was acquired from a rickettsial or a chlamydial donor by lateral gene transfer, possibly during infection of the same eukaryotic host cell.
MATERIALS AND METHODS
PCR and cloning.
L. intracellularis NCTC 12656 cells were used for DNA isolation with the DNeasy tissue kit (Qiagen, Vienna, Austria) according to the recommendations of the manufacturer. The gene Li0007, coding for the putative ATP/ADP transport protein, was amplified by using the High Fidelity PCR enzyme mix (MBI-Fermentas, St. Leon-Rot, Germany) according to the instructions of the manufacturer. A forward primer (5′-GAG AAC CTC GAG ATG AGT GAT AAA GGC AAG-3′) introducing an XhoI restriction site before the start codon and a reverse primer (5′-GAG AAC CTC GAG TTA GTT TGT GCA GAG CTC-3′) containing an XhoI restriction site after the stop codon were used. PCR conditions were as follows: denaturation at 94°C for 3 min, followed by 35 cycles of (i) denaturation at 94°C for 30 s, (ii) annealing at 56°C for 40 s, and (iii) elongation at 68°C for 90 s and a final elongation step at 68°C for 10 min. The resulting amplification products were gel purified and cloned into the cloning vector pCR-XL-TOPO by using the TOPO XL cloning kit (Invitrogen Life Technologies, Lofer, Austria). The resulting plasmid was digested with restriction endonuclease XhoI, gel purified, and inserted in frame into isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pET16b containing a promoter site for the T7 RNA polymerase (Novagen, Heidelberg, Germany). The newly constructed plasmid (pLiNTT1) was transformed into and maintained in E. coli XL1-Blue cells (Stratagene, Heidelberg, Germany). The integrity of the cloned gene was confirmed by sequencing on an ABI 3130 XL Genetic Analyzer by using the BigDye terminator kit v3.1 (ABI, Vienna, Austria).
Heterologous expression in E. coli.
For heterologous expression of pLiNTT1, E. coli strain BLR(DE3) was used. Synthesis of recombinant nucleotide transporters was conducted as previously described (16, 17). Briefly, E. coli cells harboring pLiNTT1 were induced with 1 mM IPTG at an optical density at 600 nm of 0.5. After 1 h, cells were pelleted (3,000 × g, 5 min, 8°C) and resuspended in 50 mM potassium phosphate buffer medium (pH 7.0) to an optical density at 600 nm of 5.0, kept at room temperature, and subsequently used for uptake experiments.
Analysis of substrate specificity and uptake kinetics.
To analyze the transport properties of NTT1Li, 100 μl of either induced E. coli cells harboring pLiNTT1 or noninduced cells (control) was added to 100 μl of phosphate buffer containing the indicated concentrations of α-32P-labeled substrates. Cells without pLiNTT1 but also induced by the addition of IPTG exhibited the same uptake as the control cells (data not shown). Uptake was allowed at 30°C for the indicated time spans and terminated by removal of the external substrate. For the latter purpose, cells were applied to nitrocellulose filters (0.45-μm pore size), prewetted with phosphate buffer medium, and set under a vacuum. The cells were subsequently washed three times with 4 ml phosphate buffer, and filters were transferred into 10-ml scintillation vessels containing 4 ml of water. Radioactivity in the samples was quantified in a scintillation counter (Tricarb 2500; Canberra-Packard, Heidelberg, Germany).
Back-exchange analysis and thin-layer chromatography.
To characterize the transport mode of recombinant NTT1Li, back-exchange studies were conducted. For preloading, pellets of 2 ml of IPTG-induced E. coli cells synthesizing NTT1Li were incubated in phosphate buffer containing 50 μM radioactively labeled [α-32P]ATP for 5 min. Subsequently, cells were harvested by centrifugation, washed two times, and resuspended in phosphate buffer medium containing 500 μM (10-fold excess) unlabeled nucleotides. Back exchange was carried out at 30°C for 2.5 min and terminated by rapid centrifugation. The remaining internal radioactivity in the pellet and the exported label in the supernatant were quantified in a scintillation counter. Additionally, radioactively labeled compounds exported by E. coli cells expressing NTT1Li were identified by thin-layer chromatography (32). For this, a 10-μl aliquot of the supernatant was loaded onto a 0.5-mm poly(ethylene amine) cellulose thin-layer chromatography plate and dried with a fan. Retardation factor values of radioactively labeled nucleotides and phosphate were determined after radioautography and corresponded to values of unlabeled nucleotides visualized under UV light and to radioactively labeled standards. Corresponding radioactively labeled positions were marked on the thin-layer plate, cut out, and quantified in a scintillation counter.
Phylogenetic analysis.
A database containing all of the nucleotide transport protein sequences available in the EMBL/GenBank/DDBJ public databases was established by using the ARB software package (31). For this, amino acid sequences were aligned automatically with MAFFT (24) and the alignment was imported into ARB. Phylogenetic trees were constructed with the PHYLIP distance matrix (Fitch) and maximum-parsimony methods (12), TREE-PUZZLE (using the VT model of amino acid substitution), and PROTml 2.3 (using the JTT amino acid replacement model) implemented in ARB. Bootstrap analysis was performed with 1,000 resamplings. A filter considering only those alignment positions that were conserved in at least 30% of all sequences (resulting in a total of 441 alignment columns) was used for all treeing calculations.
Comparative genome analysis.
Comparative sequence analysis of the publicly available L. intracellularis PHE-MN1-00 genome sequence was performed by using Entrez Genome at the National Center for Biotechnology Information website (56) and Integrated Microbial Genomes at the Joint Genome Institute website (33). Analysis of metabolic pathways was performed by using the Kyoto Encyclopedia of Genes and Genomes website (23). GC skew analysis was done with the GenSkew tool available at the Munich Information Center of Protein Sequences website (38).
Nucleotide sequence accession number.
The gene sequence of the ATP/ADP translocase from L. intracellularis NCTC 12656 was submitted to the EMBL/GenBank/DDBJ databases under accession number AM941722.
RESULTS
Identification and comparative sequence analysis of NTT1Li.
The biochemically well-characterized ATP/ADP translocase NTT1Pam (pc0250) of the amoeba symbiont Protochlamydia amoebophila UWE25 (9, 45, 53) was used as a query for BlastP searches (2) against the nonredundant protein sequence data set (nr) at the National Center for Biotechnology Information website (56). Using an E value cutoff of 1e−15, a putative nucleotide transport protein was identified in the genome sequence of L. intracellularis PHE/MN1-00. The identified protein (Li0007, GenBank accession number YP_594385) comprises 532 amino acids and shows a predicted molecular mass of 59 kDa and a predicted pI of 9.14, which corresponds well to known nucleotide transporters. Li0007 showed a match with PFAM family PF03219 (“TLC ATP/ADP transporter”) and was noted as a “nucleotide transport protein” in the automatically derived annotation.
The cloned gene encoding the putative ATP/ADP translocase from L. intracellularis NCTC 12656 was identical to Li0007 from L. intracellularis PHE/MN1-00. According to the nucleotide transporter nomenclature used by Haferkamp and coworkers (16, 17), we refer to this protein as NTT1Li. The amino acid sequence of NTT1Li was added to a data set containing all publicly available NTT proteins. Sequences were aligned and subjected to a detailed comparative analysis. NTT1Li showed the highest amino acid sequence identity to a nearly full-length sequence (423 amino acids) of a nucleotide transport protein from the rickettsial amoeba symbiont “Candidatus Paracaedibacter symbiosus” (NTT1Ps, 53.1%) and 47.6 and 46.5% sequence identity to NTT1Pam, the ATP/ADP transporter of Protochlamydia amoebophila UWE25, and NTT1Rp, the ATP/ADP translocase from Rickettsia prowazekii, respectively. Amino acid sequence identity to plastidic ATP/ADP transporters from plants and algae was 39 to 44%; sequence identity to functionally characterized NTT class II proteins was below 39%.
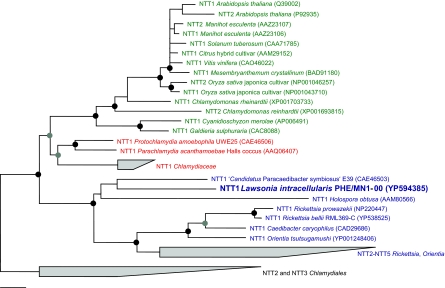
A number of highly conserved (charged) amino acid residues that are essential for the function of the ATP/ADP translocase of Arabidopsis thaliana (NTT1At) (52), i.e., K155, E245, E385, and K527 (referring to the NTT1At numbering), are present in NTT1Li. Using ConPred II (5), NTT1Li is predicted to contain 12 transmembrane alpha helices. This is consistent with the secondary-structure analysis of known ATP/ADP translocases and experimental evidence suggesting the presence of 12 transmembrane helices in NTT1Rp (1). Phylogenetic analysis using distance matrix, maximum-parsimony, and maximum-likelihood treeing methods demonstrated that NTT1Li consistently clustered together with rickettsial ATP/ADP translocases and their paralogues with high confidence (Fig. 1). The overall topology of the trees obtained was supported by all of the treeing methods applied and was similar to previously published analyses (3, 15, 45, 54).
FIG. 1.
Phylogenetic relationships of bacterial and plastidic ATP/ADP translocases and other nucleotide transport proteins. The amino acid-based tree was calculated by the TREE-PUZZLE method. Black circles indicate well-supported nodes showing greater than 90% TREE-PUZZLE support and parsimony bootstrap values (1,000 resamplings). Nodes that are supported by TREE-PUZZLE support values above 75% but which show only low-parsimony bootstrap values (less than 90%) are indicated by gray circles. TREE-PUZZLE support values below 75% and parsimony bootstrap values below 90% are not shown. Plastidic ATP/ADP translocases from plants and algae are in green, ATP/ADP translocases from chlamydiae are in red, and rickettsial ATP/ADP translocases and nucleotide transport proteins are in blue. GenBank accession numbers are in parentheses. The bar indicates a 10% estimated evolutionary distance.
Biochemical characterization of NTT1Li in E. coli.
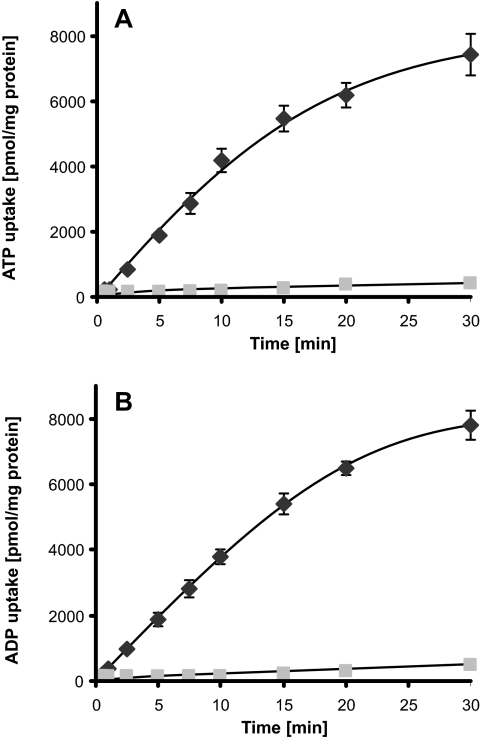
Based on the results of our comparative sequence analysis, we hypothesized that NTT1Li functions as an ATP/ADP translocase. We thus decided to investigate whether ATP and ADP are indeed substrates of NTT1Li by determining the time dependency of radioactively labeled ATP and ADP uptake by NTT1Li expressed in E. coli. These experiments also served to identify the linear phase of import, which is a prerequisite for the calculation of affinities and reaction velocities. In all cases, noninduced (control) cells showed no significant uptake of radioactivity (Fig. 2). ATP import by NTT1Li was linear for about the first 10 min (Fig. 2A); ADP import was linear for the first 10 to 15 min (Fig. 2B). Transport measurements with rising concentrations of labeled substrates (5 to 1,000 μM) allowed the calculation of Vmax values for ATP and ADP import by NTT1Li. ATP was imported at a rate of 139.2 (± 8.3) nmol mg protein−1 h−1, and ADP was imported at a rate of 149.2 (± 13.7) nmol mg protein−1 h−1. In addition, substrate affinities of NTT1Li for ATP and ADP were calculated. NTT1Li had an apparent substrate affinity of 236.3 (± 36.5) μM for ATP and 275.2 (± 28.1) μM for ADP.
FIG. 2.
Time dependency of ATP and ADP uptake into E. coli cells expressing the ATP/ADP translocase NTT1Li from L. intracellularis. Shown is the time-dependent uptake of [α-32P]ATP (A) and [α-32P]ADP (B) mediated by E. coli expressing NTT1Li (black diamonds) or into noninduced cells harboring pLiNTT1 (control, gray squares). Cells were incubated in phosphate buffer medium containing 50 μM labeled nucleotide for the indicated time periods. Data are the mean of at least three independent experiments. Standard errors are given.
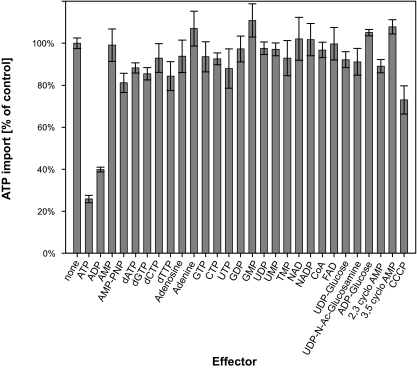
In order to verify that ATP and ADP are the preferred substrates of NTT1Li, effector studies measuring the uptake of [α-32P]ATP in the presence of unlabeled putative effectors (in 10-fold excess) were performed. Of the various putative substrates tested, only ATP (25.8% ± 1.7% residual activity) and ADP (39.9% ± 1.1% residual activity) significantly inhibited the uptake of [α-32P]ATP when compared to the transport without effector, indicating that ATP and ADP are indeed the only transported substrates for NTT1Li (Fig. 3). To analyze whether NTT1Li acts in a counterexchange mode of transport or as a proton-driven net import protein, we determined nucleotide uptake in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone. Addition of 100 μM carbonyl cyanide m-chlorophenylhydrazone reduced ATP import only to 73% (± 6.8%) residual activity (Fig. 3), indicating that NTT1Li-mediated ATP uptake is not driven by the proton gradient, a feature which is characteristic of known ATP/ADP translocases (53).
FIG. 3.
Effects of various metabolites on [α-32P]ATP uptake into E. coli cells expressing the ATP/ADP translocase NTT1Li from L. intracellularis. Uptake of [α-32P]ATP mediated by recombinant NTT1Li was measured at a substrate concentration of 50 μM and stopped after 3 min. Unlabeled effectors were present in 10-fold excess. Rates of nucleotide uptake are given as percentages of control rates (nonaffected transport = 100%). Data are the mean of three independent experiments. Error bars indicate the standard error. CoA, coenzyme A; FAD, flavin adenine dinucleotide; Ac, acetyl; CCCP, carbonyl cyanide m-chlorophenylhydrazone.
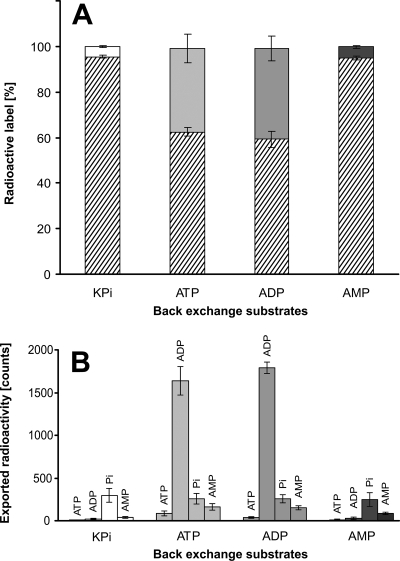
In order to further characterize NTT1Li as an ATP/ADP antiporter, back-exchange studies were performed. E. coli cells expressing NTT1Li were preloaded with labeled ATP, external radioactivity was removed, and the cells were resuspended in phosphate buffer (control) or transport buffer (containing putative counterexchange substrates). Back exchange was carried out at 30°C for 2.5 min. After centrifugation, the radioactivity in the cells and in the supernatant was quantified. Quantification of exported radioactivity allows differentiation between counterexchange (antiport) and unidirectional transport (symport) (17). E. coli cells expressing NTT1Li and preloaded with [α-32P]ATP released significant amounts of internal label (36 to 40% of the initial amount) only after resuspension in buffer medium supplemented with unlabeled ADP or ATP compared with the control (Fig. 4A). We verified by thin-layer chromatography that radioactive nucleotides exported in the presence of unlabeled substrates are mainly ADP, but also minor amounts of AMP and ATP were exported, whereas inorganic phosphate export was independent of the presence of counterexchange nucleotides (Fig. 4B).
FIG. 4.
Quantification and characterization of exported radioactively labeled nucleotides by E. coli cells expressing the ATP/ADP translocase NTT1Li from L. intracellularis. Preloading with radioactivity was achieved by incubation of induced E. coli cells in phosphate buffer containing 50 μM [α-32P]ATP for 5 min at 30°C. Washed cells were incubated in 500 μM nonlabeled adenine nucleotides or phosphate buffer (control) for 2.5 min at 30°C. Back-exchange substrates are indicated on the x axis, i.e., potassium phosphate buffer (KPi; white bars), ATP (light gray bars), ADP (dark gray bars), and AMP (black bars). (A) Fraction of exported label (white, gray, and black bars) and remaining radioactivity in the cells (hatched bars). (B) Nature of the exported label (indicated above the bars) as determined by thin-layer chromatography and quantification of the radioactive spots by scintillation counting. Data are the mean of three independent replicates.
DISCUSSION
Physiological significance of NTT1Li.
Biochemical characterization of the ATP/ADP translocase NTT1Li from L. intracellularis suggested that this pathogen, similar to chlamydiae and rickettsiae, is able to exploit its host cell by importing ATP in exchange for bacterial ADP. The determined transport properties of NTT1Li are in the range of reported Km and Vmax values of other functionally characterized ATP/ADP translocases from chlamydiae and rickettsiae (for an overview, see references 17 and 45). The affinity of NTT1Li should be sufficient to transport ATP at in vivo concentrations (see reference 17 for details).
Both rickettsiae and chlamydiae are capable of generating their own energy (4, 14, 18, 19, 21, 35). In order to survive as long as possible within a host cell, a general strategy of intracellular bacteria seems to be to limit the import of nutrients essential for the host in a way that the host is not impaired too severely because death of the host would lead to loss of their protective niche (22). By supplementing—but not replacing—bacterial energy production, ATP/ADP translocases might thus represent a fine-tuned and essential adaptation facilitating long-term survival within eukaryotic host cells. However, rickettsiae and chlamydiae are unable to synthesize nucleotides de novo (14, 19, 55), which is compensated for by two to five paralogous NTT proteins that are used for the net uptake of nucleotides, respectively (6, 16, 17, 51).
The genome sequence of L. intracellularis revealed that this pathogen, similar to chlamydiae and rickettsiae, should be able to generate its own energy by using a basic, probably microaerophilic, respiratory chain. The genome also encodes a complete glycolytic pathway; the ATP-dependent phosphofructokinase is notably absent but is probably complemented by the pyrophosphate-dependent phosphofructokinase (Li0052), allowing the glycolytic pathway to be functional. However, unlike chlamydiae and rickettsiae, L. intracellularis should be able to synthesize both purine and pyrimidine nucleotides de novo, which is consistent with the absence of additional nucleotide transporters for the net uptake of nucleotides in L. intracellularis. Like other obligate intracellular bacteria, L. intracellularis possesses a small, streamlined genome (1.76 Mb, including three plasmids). It is thus tempting to speculate that the genes that are retained are essential and fulfill a similar function in L. intracellularis as in rickettsiae and chlamydiae.
Phylogeny of nucleotide transport proteins and evolutionary implications.
The identification of a functional ATP/ADP translocase in L. intracellularis reveals a surprising conservation of energy parasitism among largely unrelated groups of bacterial pathogens belonging to the chlamydiae, rickettsiae, and Deltaproteobacteria. Protein phylogeny of nucleotide transport proteins revealed that NTT1Li consistently formed a deep branch with ATP/ADP translocases from “Ca. Paracaedibacter symbiosus” (NTT1Ps) and Holospora obtusa (NTT1Ho) and clustered together with other rickettsial nucleotide transport proteins (Fig. 1).
The evolution of nucleotide transport proteins, particularly of ATP/ADP translocases, has received considerable attention as phylogenetic analysis allowed insights into early events during the origin of the plant cell (3, 15, 20, 30, 45, 54, 58). In general, two evolutionary scenarios were postulated: First, ATP/ADP translocases may have been invented in an ancestor of mitochondria and Rickettsia. ATP/ADP translocase-encoding genes were subsequently transferred to chlamydiae and the nuclear genome of an early mitochondriate cell, respectively, and were retained in plants to facilitate plastid function (3). A rickettsial origin of ATP/ADP translocases, however, would not explain the distinct phylogenetic relationship between chlamydial and plastidic ATP/ADP translocases.
The alternative scenario therefore assumes that ATP/ADP translocases originated in a chlamydial ancestor and were then transferred to plants and rickettsiae, respectively (15, 45). This hypothesis is consistent with the presence of sequences encoding (putative) ATP/ADP translocases in representatives of all major evolutionary lineages within the chlamydiae, including Chlamydiaceae, Parachlamydiaceae, Waddliaceae (GenBank accession number AAX45329), Simkaniaceae (AAX45330), and Criblamydiaceae (8, 15, 19, 45, 47-49, 55). A chlamydial origin of ATP/ADP translocases also gained recent support from two independent phylogenomic studies which, in addition, suggested a contribution of chlamydiae to the origin of the primary photosynthetic eukaryote (20, 54).
Our analysis comprising a larger data set of ATP/ADP translocases than previous studies, including novel chlamydial, plant and algal sequences and the first deltaproteobacterial ATP/ADP translocase, provides strong evidence for a chlamydial origin of ATP/ADP translocases. The tree topology obtained, which is well supported and in general agreement with previously published results (15, 45), can be best explained by ancient horizontal transfer events from a chlamydial ancestor to plants and rickettsiae and a third transfer from a rickettsia-like organism to deltaproteobacterial L. intracellularis.
The genome of L. intracellularis, however, does not show clear signs of recent lateral acquisition of foreign DNA. G+C skew analysis indicated no obvious regions with unusual G+C content indicative of genomic islands (data not shown). Consistently, the G+C content of the gene encoding NTT1Li is, at 33%, not different from the overall G+C content of the L. intracellularis genome, which contains only a few transposases (n = <10) and phage-like genes (n = 5). This indicates either that signposts of lateral gene transfer of NTT1Li were blurred due to amelioration (28), i.e., that the gene transfer event occurred a long time ago, or that the donor genome had a G+C content similar to that of the L. intracellularis genome. Interestingly, Rickettsia species show a genomic G+C content of between 29 and 32.5%, while chlamydiae have a generally higher genomic G+C content (34.7 to 41.3%). Taken together, these findings and the well-supported grouping of NTT1Li with rickettsial ATP/ADP translocases in our phylogenetic analysis (Fig. 1) point to rickettsiae as putative donors of the L. intracellularis ATP/ADP translocase.
In contrast to chlamydiae and rickettsiae, L. intracellularis is not related to a deeply branching evolutionary lineage of exclusively intracellular bacteria but belongs to the Desulfovibrionaceae (37, 46), most of which are free-living, sulfate-reducing microorganisms (11); its closest relatives are Bilophila wadsworthia and Desulfovibrio desulfuricans, from which L. intracellularis split about 200 million years ago (if a divergence rate for the 16S rRNA gene of 1%/50 million years is assumed) (40). Compared to rickettsiae and chlamydiae, which form deep branches in the Alphaproteobacteria and the Bacteria, respectively, and which evolved far more than a billion years ago (7, 13), L. intracellularis has thus only relatively recently adapted to an obligate intracellular life style. The acquisition of a gene encoding an ATP/ADP translocase (from a rickettsia-like donor) might have facilitated this process.
Conclusion.
In this study, we have analyzed the first ATP/ADP translocase from a deltaproteobacterial pathogen and could show that this ancient and important mechanism for host cell interaction is conserved among major human and animal pathogens. Biochemical characterization of this ATP/ADP translocase allowed first insights into the molecular basis of the intracellular life style of L. intracellularis.
Acknowledgments
Sequencing of the L. intracellularis genome was achieved by Connie J. Gebhart and Vivek Kapur (University of Minnesota) and funded by the United States Department of Agriculture. Steven McOrist is gratefully acknowledged for providing L. intracellularis samples. We are grateful to Sebastian Lücker for purification of DNA from L. intracellularis.
Work in the laboratory of Matthias Horn and Michael Wagner was funded by grants from the Austrian Science Fund FWF (Y277-B03), the GEN-AU (Genome Research in Austria) program (GZ 200.100/1-VI/1/2004), and the University of Vienna (Research Focus Project FS573001). Stephan Schmitz-Esser is supported by an FWF grant (P19252-B17). The work of Ilka Haferkamp and Michelle Ast was supported by the Deutsche Forschungsgemeinschaft (Graduate Research School 845 and GZ: NE418/9-2).
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Alexeyev, M. F., and H. H. Winkler. 1999. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 2851503-1513. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amiri, H., O. Karlberg, and S. E. Andersson. 2003. Deep origin of plastid/parasite ATP/ADP translocases. J. Mol. Evol. 56137-150. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396133-140. [DOI] [PubMed] [Google Scholar]
- 5.Arai, M., H. Mitsuke, M. Ikeda, J. X. Xia, T. Kikuchi, M. Satake, and T. Shimizu. 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32W390-W393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audia, J. P., and H. H. Winkler. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 1886261-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battistuzzi, F. U., A. Feijao, and S. B. Hedges. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casson, N., R. Michel, R. D. Mueller, and G. Greub. 2008. Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg. Infect. Dis. 14168-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collingro, A., E. R. Toenshoff, M. W. Taylor, T. R. Fritsche, M. Wagner, and M. Horn. 2005. ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 551863-1866. [DOI] [PubMed] [Google Scholar]
- 10.Daugherty, R. M., N. Linka, J. P. Audia, C. Urbany, H. E. Neuhaus, and H. H. Winkler. 2004. The nucleotide transporter of Caedibacter caryophilus exhibits an extended substrate spectrum compared to the analogous ATP/ADP translocase of Rickettsia prowazekii. J. Bacteriol. 1863262-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux, R., S. H. He, C. L. Doyle, S. Orkland, D. A. Stahl, J. LeGall, and W. B. Whitman. 1990. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J. Bacteriol. 1723609-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 13.Feng, D. F., G. Cho, and R. F. Doolittle. 1997. Determining divergence times with a protein clock: update and reevaluation. Proc. Natl. Acad. Sci. USA 9413028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuxelius, H. H., A. Darby, C. K. Min, N. H. Cho, and S. G. Andersson. 2007. The genomic and metabolic diversity of Rickettsia. Res. Microbiol. 158745-753. [DOI] [PubMed] [Google Scholar]
- 15.Greub, G., and D. Raoult. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl. Environ. Microbiol. 695530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haferkamp, I., S. Schmitz-Esser, N. Linka, C. Urbany, A. Collingro, M. Wagner, M. Horn, and H. E. Neuhaus. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432622-625. [DOI] [PubMed] [Google Scholar]
- 17.Haferkamp, I., S. Schmitz-Esser, M. Wagner, N. Neigel, M. Horn, and H. E. Neuhaus. 2006. Tapping the nucleotide pool of the host: novel nucleotide carrier proteins of Protochlamydia amoebophila. Mol. Microbiol. 601534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn, M. 12 May 2008, posting date. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed]
- 19.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304728-730. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J., and J. P. Gogarten. 2007. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iliffe-Lee, E. R., and G. McClarty. 1999. Glucose metabolism in Chlamydia trachomatis: the ‘energy parasite’ hypothesis revisited. Mol. Microbiol. 33177-187. [DOI] [PubMed] [Google Scholar]
- 22.Joseph, B., and W. Goebel. 2007. Life of Listeria monocytogenes in the host cells' cytosol. Microbes Infect. 91188-1195. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa, M., S. Goto, S. Kawashima, Y. Okuno, and M. Hattori. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32D277-D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingenberg, M. 1989. Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch. Biochem. Biophys. 2701-14. [DOI] [PubMed] [Google Scholar]
- 26.Krause, D. C., H. H. Winkler, and D. O. Wood. 1985. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc. Natl. Acad. Sci. USA 823015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroll, J. J., M. B. Roof, L. J. Hoffman, J. S. Dickson, and D. L. Harris. 2005. Proliferative enteropathy: a global enteric disease of pigs caused by Lawsonia intracellularis. Anim. Health Res. Rev. 6173-197. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44383-397. [DOI] [PubMed] [Google Scholar]
- 29.Lawson, G. H., and C. J. Gebhart. 2000. Proliferative enteropathy. J. Comp. Pathol. 12277-100. [DOI] [PubMed] [Google Scholar]
- 30.Linka, N., H. Hurka, B. F. Lang, G. Burger, H. H. Winkler, C. Stamme, C. Urbany, I. Seil, J. Kusch, and H. E. Neuhaus. 2003. Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 30627-35. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 321363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangold, H. K. 1967. Dünnschicht Chromatographie. Ein Laboratoriumshandbuch. Springer, Heidelberg, Germany.
- 33.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauch, C.-P., and G. Bilkei. 2005. Reproductive performance of gilts following an outbreak of acute proliferative enteropathy due to Lawsonia intracellularis. Vet. J. 170128-131. [DOI] [PubMed] [Google Scholar]
- 35.McClarty, G. 1999. Chlamydial metabolism as inferred from the complete genome sequence, p. 69-100. In R. S. Stephens (ed.), Chlamydia. American Society for Microbiology, Washington, DC.
- 36.McOrist, S. 2005. Defining the full costs of endemic porcine proliferative enteropathy. Vet. J. 1708-9. [DOI] [PubMed] [Google Scholar]
- 37.McOrist, S., C. J. Gebhart, R. Boid, and S. M. Barns. 1995. Characterization of Lawsonia intracellularis gen. nov., sp. nov., the obligately intracellular bacterium of porcine proliferative enteropathy. Int. J. Syst. Bacteriol. 45820-825. [DOI] [PubMed] [Google Scholar]
- 38.Mewes, H. W., S. Dietmann, D. Frishman, R. Gregory, G. Mannhaupt, K. F. Mayer, M. Munsterkotter, A. Ruepp, M. Spannagl, V. Stumpflen, and T. Rattei. 2008. MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res. 36D196-D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhaus, H. E., E. Thom, T. Mohlmann, M. Steup, and K. Kampfenkel. 1997. Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 1173-82. [DOI] [PubMed] [Google Scholar]
- 40.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 9612638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmieri, L., H. Rottensteiner, W. Girzalsky, P. Scarcia, F. Palmieri, and R. Erdmann. 2001. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 205049-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren, Q., K. H. Kang, and I. T. Paulsen. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32(Database issue)D284-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz-Esser, S., N. Linka, A. Collingro, C. L. Beier, H. E. Neuhaus, M. Wagner, and M. Horn. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J. Bacteriol. 186683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, D. G., and G. H. Lawson. 2001. Lawsonia intracellularis: getting inside the pathogenesis of proliferative enteropathy. Vet. Microbiol. 82331-345. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, V., N. Casson, and G. Greub. 2006. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture. Environ. Microbiol. 82125-2135. [DOI] [PubMed] [Google Scholar]
- 48.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Connor, I. Goodhead, H. Norbertzcak, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeno-Tarraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjaden, J., T. Mohlmann, K. Kampfenkel, G. Henrichs, and H. E. Neuhaus. 1998. Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16531-540. [Google Scholar]
- 51.Tjaden, J., H. H. Winkler, C. Schwoppe, M. Van Der Laan, T. Mohlmann, and H. E. Neuhaus. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 1811196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trentmann, O., C. Decker, H. H. Winkler, and H. E. Neuhaus. 2000. Charged amino-acid residues in transmembrane domains of the plastidic ATP/ADP transporter from arabidopsis are important for transport efficiency, substrate specificity, and counter exchange properties. Eur. J. Biochem. 2674098-4105. [DOI] [PubMed] [Google Scholar]
- 53.Trentmann, O., M. Horn, A. C. van Scheltinga, H. E. Neuhaus, and I. Haferkamp. 2007. Enlightening energy parasitism by analysis of an ATP/ADP transporter from chlamydiae. PLoS Biol. 5e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyra, H. M., M. Linka, A. P. Weber, and D. Bhattacharya. 2007. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8R212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandahl, B. B., S. Birkelund, and G. Christiansen. 2004. Genome and proteome analysis of Chlamydia. Proteomics 42831-2842. [DOI] [PubMed] [Google Scholar]
- 56.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, O. Khovayko, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, J. Ostell, K. D. Pruitt, G. D. Schuler, M. Shumway, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, R. L. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2008. Database resources of the National Center for Biotechnology Information, vol. 36, p.D13-D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler, H. H., and H. E. Neuhaus. 1999. Non-mitochondrial ATP transport. Trends Biochem. Sci. 2464-68. [DOI] [PubMed] [Google Scholar]
- 58.Wolf, Y. I., L. Aravind, and E. V. Koonin. 1999. Rickettsiae and chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet. 15173-175. [DOI] [PubMed] [Google Scholar]