Abstract
The rhizobacterium Stenotrophomonas rhizophila accumulates the compatible solutes glucosylglycerol (GG) and trehalose under salt stress conditions. The complete gene for the GG synthesis enzyme was cloned and sequenced. This enzyme from S. rhizophila represented a novel fusion protein composed of a putative C-terminal GG-phosphate synthase domain and an N-terminal putative GG-phosphate phosphatase domain, which was named GgpPS. A similar gene was cloned from Pseudomonas sp. strain OA146. The ggpPS gene was induced after a salt shock in S. rhizophila cells. After the salt-loaded cells reached stationary phase, the ggpPS mRNA content returned to the low level characteristic of the control cells, and GG was released into the medium. The complete ggpPS gene and a truncated version devoid of the phosphatase part were obtained as recombinant proteins. Enzyme activity tests revealed the expected abilities of the full-length protein to synthesize GG and the truncated GgpPS to synthesize GG-phosphate. However, dephosphorylation of GG-phosphate was detected only with the complete GgpPS protein. These enzyme activities were confirmed by complementation experiments using defined GG-defective mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Genes coding for proteins very similar to the newly identified fusion protein GgpPS for GG synthesis in S. rhizophila were found in genome sequences of related bacteria, where these genes are often linked to a gene coding for a transporter of the Mfs superfamily.
In their natural habitats, bacteria are confronted with seasonal as well as sudden changes in many environmental conditions. Therefore, they developed highly sophisticated acclimation strategies for coping with such situations. Members of the gram-negative bacterial genus Stenotrophomonas were found in a wide variety of environments and geographical regions, as well as associated with humans as a nosocomial pathogen (2, 6). A new species, S. rhizophila, was described, which exclusively comprises strains isolated from the rhizosphere of plants (30). Bacteria living in the rhizosphere are often confronted by changes in the osmotic potential, which are balanced by the accumulation of compatible solutes (22). Accordingly, S. rhizophila accumulates glucosylglycerol (GG) and trehalose under salt stress conditions (27, 30).
GG and trehalose are compatible solutes that allow bacteria, yeast, and plants to acclimate to enhanced salinities, desiccation, and cold stress. These compounds are accumulated in very high amounts without disturbing cellular metabolism. The salt-proportional accumulation results in a decrease in the intracellular osmotic potential necessary for osmotic water uptake and a rise of turgor and growth. Trehalose has been detected in many gram-negative bacteria, including Escherichia coli, as a naturally compatible solute (8). In contrast, GG was initially thought to be a characteristic of moderately halotolerant cyanobacteria (see reference 11). Later, it was also found in heterotrophic bacteria such as different species of the genus Pseudomonas (21, 24). Results of nuclear magnetic resonance testing showed that in all cases, α-2-d-glucosylglycerol was used as a compatible solute (21). Corresponding to its structure, composed of a carbohydrate and a polyol part, GG showed intermediate protective features when it was investigated in vitro (12).
GG and trehalose are synthesized by a similar two-step biosynthetic pathway. Nucleotide sugars are used in a glucosyltransferase reaction catalyzed by GG-phosphate synthase (GgpS) or trehalose-phosphate synthase (Tps), leading to a phosphorylated intermediate, which is subsequently dephosphorylated by a specific phosphatase (11). During the last few years, genes encoding enzymes for trehalose synthesis have been cloned from many heterotrophic bacteria (e.g., E. coli [8]), while genes for GG synthesis were identified in two cyanobacterial strains (7, 19). The Tps proteins from heterotrophic bacteria and the GgpS proteins from cyanobacteria share a similarity of around 40%, implying a common ancestor, which allows their annotation in many newly appearing genome sequences from different bacterial strains.
The aim of the present study was to analyze the molecular basis for the compatible solute biosynthesis in the newly described species S. rhizophila. Moreover, until now, no enzyme involved in GG biosynthesis from heterotrophic bacteria was functionally characterized. The gene for the GG synthesizing enzyme in S. rhizophila was sequenced, and the postulated enzyme activities were experimentally verified. This gene was named ggpPS, since it codes for a new class of enzymes for GG synthesis, which are composed of an N-terminal GG-phosphate phosphatase (GgpP) domain fused with a C-terminal GG-phosphate synthase (GgpS) domain. Very similar putative GgpPS proteins are encoded in genomes of many heterotrophic bacteria, while all cyanobacteria harbor separate ggpS and ggpP genes.
MATERIALS AND METHODS
Strains and culture conditions.
The type strain DSM 14405T of the species S. rhizophila (30), which was isolated from the rhizosphere of the oilseed rape plant, was used. Moreover, Pseudomonas sp. strain OA146, isolated from coastal waters of the Southern Baltic Sea and accumulating only GG, was analyzed (21). For physiological experiments, Stenotrophomonas was cultivated in mineral medium (as described in reference 23) containing malic acid as the carbon source, supplemented with d,l-methionine (40 mg liter−1). In salt shock experiments, crystalline NaCl (3%, final concentration) was added directly to the cultures at the beginning of the experiments. Cells were cultivated in Erlenmeyer flasks (100 to 2,000 ml) shaken at 180 rpm and at 30°C. Samples were taken from freshly diluted cultures showing similar optical densities at 600 nm (A600). E. coli strain TG1 was used for DNA cloning, and strain LMG194 was used for protein expression. E. coli was cultivated in LB medium at 37°C. Synechocystis sp. strain 6803 was grown photoautotrophically as described previously (10).
DNA manipulations.
Total DNA from Stenotrophomonas and Pseudomonas spp. was isolated from lysozyme-treated cells and purified by using the detergent cetyltrimethylammonium bromide (10). All other techniques, such as transformation of E. coli, ligation, and restriction analysis (restriction enzymes were obtained from New England Biolabs), were standard methods (28). Plasmid DNA was isolated by using a GFX Micro Plasmid Prep kit (Amersham Biosciences). Sequencing was performed with a CEQ dye terminator cycle sequencing Quick Start kit (Beckman Coulter). Computer analyses of DNA and protein sequences were performed with BLAST (1), ClustalX (version 1.64b), and Clone-Manager, version 4.1 (Sci-Ed Software) software.
Cloning and sequencing of ggpPS and tps.
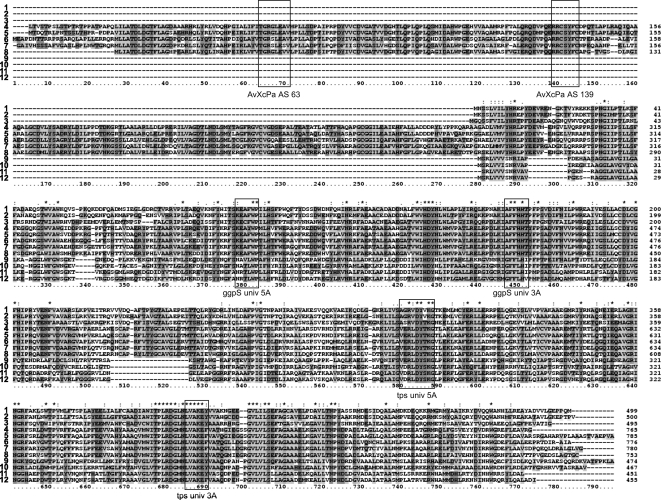
A PCR strategy using degenerated primers was applied. For primer design, the amino acid sequences of the previously characterized GgpS proteins from the cyanobacterium Synechocystis sp. strain 6803 (accession no. NP_441672; sll1566 [19]) and Synechococcus sp. strain PCC 7002 (accession no. CAA06963 [7]) were compared to the Tps proteins from bacteria and yeast to define well-conserved parts. Several boxes appeared (Fig. 1), which were used for primer development (all primers are listed in Table 1). The primer combinations applied to clone the GG synthesis gene were as follows: ggpSuniv5A and ggpSuniv3A, producing a fragment of about 220 bp; and AvXcPa AS 63, AvXcPa AS 111, and AvXcPa AS 139 together with ggpSD5o, producing fragment sizes of about 1 kb, 900 bp, and 800 bp, respectively. Additionally, the strategy of inverse PCR was used, for which the NcoI-cut chromosomal DNA of S. rhizophila was ligated with the NcoI-cut vector pUCBM21, followed by PCR with the primer M13fw or M13rev and the specific ggpS3i primer. This strategy produced the 3′ end of the ggpPS gene. The upstream sequence of ggpPS from S. rhizophila harboring the ycaD gene was sequenced directly from chromosomal DNA of S. rhizophila by a commercial enterprise (Firma Meixner, DLMBC, Berlin, Germany). The sequence of the entire operon composed of the ycaD and ggpPS genes of S. rhizophila can be found under the updated EMBL accession no. AJ878740.2. Using the same strategy, we obtained the sequence of the complete ggpPS gene from Pseudomonas sp. strain OA146 (EMBL accession no. AJ318784).
FIG. 1.
Amino acid sequence alignment of GgpS (1-8) and selected Tps sequences (9-12) with the corresponding proteins from S. rhizophila DSM 14405 showing a high degree of similarity and also some obvious differences between these two related enzyme groups. Highly conserved boxes used for the primer design are marked. The designations of most primers used in this study for gene cloning are indicated below the boxes (Table 1). 1, Synechocystis sp. strain PCC 6803 (accession no. P74258); 2, Synechococcus sp. strain PCC 7002 (accession no. O65979); 3, Synechococcus sp. strain WH 8102 (accession no. CAE07796); 4, Stenotrophomonas rhizophila DSM 14405 (accession no. AJ878740.2); 5, Xanthomonas campestris pv. campestris ATCC 33913 (accession no. NP_638382); 6, Azotobacter vinelandii AvOP (accession no. ZP_00416807); 7, Pseudomonas sp. strain OA146 (accession no. AJ318784); 8, Azoarcus sp. strain EbN1 (accession no. YP_157460); 9, E. coli strain K-12 (accession no. BAA15717); 10, Azotobacter vinelandii AvOP (accession no. ZP_00419964); 11, Stenotrophomonas maltophilia strain R551-3 (accession no. ZP-01646117); 12, Xanthomonas campestris pv. campestris ATCC 33913 (accession no. NP_638429).
TABLE 1.
Oligonucleotides used for amplification and cloning of the corresponding genes
| Primer | Sequencea | Restriction site | Application |
|---|---|---|---|
| ggpS univ 5A | AARGAAGCCTTYTGGCC | Cloning of internal ggpPS fragments | |
| ggpS univ 3A | GTGTGGTGGAARAAGGC | ||
| AvXcPa AS 63 | ACCGGCCGCAGCCTGGAA | ||
| AvXcPa AS 111 | CGCCGCTGCTCCTAYTTCTGC | ||
| AvXcPa AS 139 | CGCCGCTGCTCCTAYTTCTGC | ||
| ggpSD5o | TGCGGTTGACCTTCAGGA | ||
| ggpS3i | TTTCAGGTCCGGGCGCAGTT | Inverse PCR of 3′ end of ggpPS | |
| tps univ 5A | TTTTTTYTGCACATYCC | Cloning of the partial tps gene | |
| tps univ 3A | ATGAACYTGGTGGCCAA | ||
| ggp5C | TTYCTGCACACCCCSTTYCC | ||
| ggpS5i | ATGATGCATGTGTTCTGGGA | Labeling of the ggpPS probe | |
| GgpS5′Xho | CTCGAGTTGACTGTATCGACA | XhoI | Cloning of the complete ggpPS gene into expression vector |
| ggpS3′Hind | AAGCTTTCAACCGCGCACCGC | HindIII | |
| ggpS5′AS284 | CTCGAGGCCGGCGCTGACCTG | XhoI | Cloning of the truncated ggpPS gene |
| Syn_compl.5′ | GGGTACCATGAATTCATCCCTTGTGAT | KpnI | Amplification of ggpS from Synechocystis sp. strain PCC 6803 |
| Syn_compl.3′ | GTCTAGACTACATTTGGGGGGGCTCTC | XbaI | |
| SF_compl.5′ | GGGTACCATGACTGTATCGACACCCTCT | KpnI | Amplification of full-length ggpPS and its truncated version from S. rhizophila for complementation |
| Sk_compl.5′ | GGGTACCATGGGCGCTGACCTGGTGATG | KpnI | |
| SF_compl.3′ | GGAGCTCTCAACCGCGCACCGCCTCCA | SacI | |
| proggpS-5′ | GGGGTACCTCCAGGCATATTAGTTCA | KpnI | Amplification of the ggpS (strain PCC 6803) promoter region from nucleotide −193 to +495 (20) |
| proggpS-3′ (1027-KpnI) | GGGTACCATTCATAAAATCAGCGG | KpnI |
Added restriction sites are underlined.
A partial tps gene of S. rhizophila was cloned after PCR amplification, using the primer combinations, as follows: tpsuniv5A and tpsuniv3A, producing a fragment of about 330 bp; and ggp5C and tpsuniv3A, producing a fragment of about 550 bp.
Gene expression and compatible solute accumulation after salt shock.
Northern blot hybridization experiments were performed to estimate the steady-state level and size of ggpS mRNA. Total RNA was isolated from cells (10 ml), using a High Pure RNA isolation kit (Roche). Methods used for the separation of RNA, blotting, and hybridization were as described in detail previously (10). A specific DNA hybridization probe was obtained after PCR amplification of the partial ggpPS gene (using the primer combinations ggpS5i and ggpS3i). The DNA was labeled by [α-32P]dATP (Amersham Pharmacia Biotech) using a HexaLabel DNA labeling kit (MBI Fermentas). Hybridization signals were recorded and quantified by using a phosphorimager (model BAS1000; Fuji). To correct the quantitative data for variations in RNA loading, we made all calculations of the relative intensities of hybridization signals obtained after applying a radiolabeled 16S-rRNA gene probe from S. rhizophila, which was generated by PCR with 16S-rRNA gene primers (15), to the same filters.
GG was extracted from frozen cell pellets or purified from the supernatant and analyzed by high-performance liquid chromatography (29). The content of the compatible solute was related to the total protein content estimated in the lysates after the sonication of 1 ml of cells (according to the method described in reference 17).
Overexpression of the complete ggpPS gene and a truncated version in E. coli.
Fragments carrying the complete ggpPS gene and a truncated version lacking the 5′ end were cloned into the pBAD/HisA vector system (Invitrogen), fused with an N-terminal His-tag coding sequence. For PCR amplification of the complete ggpPS gene (2.3 kb), the primers ggpS5′Xho and ggpS3′Hind (Table 1) were used, while the primers ggpS5′AS284 and ggpS3′Hind were applied to obtain the truncated ggpS 1.4-kb fragment. For overexpression, the resulting plasmids pB1.4kb and pB2.3kb were transferred into E. coli strain LMG194. The cells were grown in LB medium at 20°C overnight. Production of recombinant proteins was induced by adding arabinose (0.002%). Proteins were extracted by sonication in 33 mM Tris-maleate buffer (pH 7.8) containing 2 mM Na2-EDTA, 3 mM β-mercaptoethanol, and 5 mg ml−1 bovine serum albumin (BSA). Proteins were purified by affinity chromatography on Ni-nitrilotriacetic acid columns (Pro Bond resin; Invitrogen) and eluted by increasing imidazole concentrations.
Estimation of GG-phosphate synthase and GG-phosphate phosphatase enzyme activities.
Proteins were immediately used to measure the GgpS activity, using a coupled enzyme assay, where the release of ADP and UDP from ADP-glucose and UDP-glucose, respectively, was followed photometrically at 340 nm (Cary 50 model UV/Vis spectrophotometer; Varian). The standard GgpS enzyme assay contained 710 μl Tricine buffer (140 mM [pH 7.8]) with MgCl2 (8.8 mM), KCl (85 mM), phosphoenolpyruvate (3.5 mM), lactate dehydrogenase (5 U), and pyruvate kinase (5 U); 75 μl NADH (4 mM); 75 μl NaCl (1 M); 50 μl ADP-glucose or UDP-glucose (100 mM). To the standard assay mixture, we added 50 μl containing about 10 μg of recombinant protein. After the mixture equilibrated for about 3 min, the measurement was started by the addition of 50 μl of glycerol-3-phosphate (100 mM), and NADH oxidation was recorded for 5 to 10 min at 340 nm and 30°C.
To verify GG generation, samples were analyzed by gas chromatography (GC), using a Focus GC (Thermo Scientific) unit equipped with a TR-5MS column (30 m by 0.25 mm by 0.25 μm; Thermo Scientific) and an AS 3000 autosampler. After protein was precipitated with acetone and 50 μg sorbitol was added as an internal standard, the extracts were dried in a vacuum centrifuge. Trimethylsilyl derivatives of sugars were obtained by incubation with 65 μl of pyridine/methoxylamine (20 mg ml−1; 90 min at 30°C) and 35 μl of N,O-bis(trimethylsilyl)-trifluoroacetamide (60 min at 60°C; Sigma). Splitless injection of a 1-μl sample was performed at 300°C. The initial column temperature was set at 160°C and increased at a rate of 5°C min−1, to a final temperature of 280°C that was held for 10 min at the end of the cycle. Nitrogen was used as the carrier gas. Data were analyzed with ChromQuest version 4.2.34 software (Thermo Electron).
GgpP activity was measured by the release of GG from laboratory-made GG-phosphate, which was purified from salt-treated cells of a Synechocystis sp. strain PCC 6803 mutant defective in GG-phosphate phosphatase activity (10). The assay mixture contained 15 μl of GG-phosphate (20 μg), 50 μl of HEPES buffer (100 mM [pH 7.5]) containing 5 mg ml−1 BSA) with 7.5 μl NaCl (1 M), 14 μl MgCl2 (120 mM), 3.5 μl distilled water, and 10 μl protein extract containing about 2 μg of recombinant protein. The mixture was incubated at 30°C overnight. The amount of dephosphorylated GG was measured by GC. Protein content was estimated according to the method described in reference 4.
Complementation of the Synechocystis mutants.
To verify the proposed enzyme activities of the newly identified GG synthesis enzyme, complementation experiments were performed using mutants of the cyanobacterium Synechocystis sp. strain PCC 6803 with defined defects in the open reading frame (ORF) sll1566 (ΔggpS) or slr0746 (ΔggpP or ΔstpA) as described previously (10, 19). The full-length and the truncated versions of the ggpPS gene from S. rhizophila were amplified from chromosomal DNA and cloned into the KpnI/SacI cut vector pIGA (14) (Table 1 lists primers). As positive and negative controls, we used the ggpS gene from Synechocystis sp. strain PCC 6803 and the gfp gene, respectively. To ensure salt-dependent expression, the ggpS promoter region (from nucleotide −193 to +495, relative to the determined transcription start site) (20) was fused to the coding sequences of the complete ggpPS gene or its truncated version. Transformation of the Synechocystis mutants with the pIGA constructs was achieved as described previously (14). For the experiments, axenic cultures of the cyanobacteria were grown photoautotrophically under continuous illumination of 30 μmol photons s−1 m−2 at 30°C. The changed salt resistance was tested on agar plates, where cells were spotted onto BG11 medium (26) (pH 8), containing 0.9% Kobe agar and various NaCl concentrations. Plates were incubated for at least 10 days. The accumulation of GG was measured in ethanolic extracts by GC as described above.
RESULTS
Cloning of the ggpPS and tps genes.
The GG biosynthesis gene of S. rhizophila type strain DSM 14405 was searched using a PCR strategy with degenerated primers, which were deduced from well-conserved regions in known proteins (Fig. 1). Fragments of expected sizes were obtained. Their nature as internal ggpPS fragments was verified by sequencing. Finally, the complete gene was obtained (EMBL accession no. AJ878740.2). The amino acid sequence of this first enzyme for GG synthesis from a heterotrophic bacterium showed an N-terminal extension (Fig. 1) that was long compared to that of previously known GgpS sequences from cyanobacteria (7, 19). This unusual structure was confirmed by cloning a similar gene from the GG-accumulating heterotrophic bacterium Pseudomonas sp. strain OA146 (EMBL accession no. AJ318784). Additionally, proteins with significant similarities to GgpS from cyanobacteria and a 240-amino-acid residue N-terminal addition are carried in the complete genome sequences of the following heterotrophic bacteria: Xanthomonas campestris strain ATCC 33913 (accession no. AAM42306 [5]), Azotobacter vinelandii strain AvOP (accession no. ZP_00416807), Azoarcus sp. strain EbN1 (accession no. YP_157460 [25]), Pseudomonas mendocina strain YMP (accession no. YP_001188605), and Marinobacter sp. strain ELB17 (accession no. ZP_01735427).
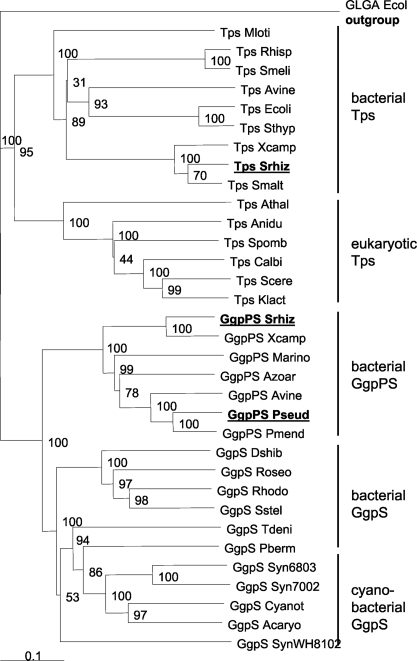
The C-terminal parts of all these proteins were very similar and harbored a so-called OtsA domain (COG0380), which belongs to the large group of the glycosyltransferase family 20 (PFAM 00982). This domain of the putative GG synthesis proteins shares a range of sequence identity of 60% to more than 80%, while its identity to the functionally characterized GgpS proteins from cyanobacteria was around 45% (Table 2). As indicated by the OtsA domain, these proteins are also similar to those of the Tps sequences, where the average identity was 38% or less. Both enzyme groups involved in GG or trehalose synthesis can be clearly distinguished by the insertion of about 20 amino acid residues around position 515, which is present in all GG synthesis enzymes, regardless of whether they originate from cyanobacteria or heterotrophic bacteria, but this sequence is missing from all Tps sequences (Fig. 1). Other parts of the overall C-terminal sequence are quite similar among GgpS and Tps proteins from different organisms, particularly at the well-conserved regions used to design the primers. When the highly conserved C-terminal part was used for a phylogenetic comparison, a separation between the newly discovered putative GG synthesis enzymes (Fig. 2, branch GgpPS) and the cyanobacterial GgpS proteins became visible; however, both cluster together and are clearly separated from the two clusters of Tps proteins from prokaryotes or eukaryotes (Fig. 2).
TABLE 2.
Comparison of the GgpPS protein of S. rhizophilaa
| Protein | Size (aa) | Identity/similarity (%)b | Similar protein/size (aa) | Organism | Accession no. |
|---|---|---|---|---|---|
| GgpPS | 765 | 84/90 | GgpPS/785 | Xanthomonas campestris ATCC 33913 | NP_638382 |
| 765 | 63/75 | GgpPS/750 | Azotobacter vinelandii AvOP | ZP_00416807 | |
| 765 | 63/70 | GgpPS/755 | Pseudomonas sp. strain OA146 | AJ318784 | |
| 765c | 43/61 | GgpS/500 | Synechococcus sp. strain PCC 7002 | O65979 | |
| 765c | 46/64 | GgpS/499 | Synechocystis sp. strain PCC 6803 | P74258 | |
| 765c | 45/63 | GgpS/511 | Parvularcula bermudensis strain HTCC2503 | ZP_01018536 | |
| 765c | 44/60 | GgpS/518 | Thiobacillus denitrificans strain ATCC 25259 | YP_316384 | |
| 765d | 29/46 | Spp/244 | Synechocystis sp. strain PCC 6803 | AAG31136 | |
| 765d | 43/54 | Tps/278e | Thiobacillus denitrificans strain ATCC 25259 | YP_316385 | |
| 765d | 41/54 | Tps/511e | Parvularcula bermudensis strain HTCC2503 | ZP_01018537 | |
| 765d | None | GgpP (StpA) | Synechocystis sp. strain PCC 6803 | AAB41279 |
For an amino acid (aa) sequence comparison, see Fig. 1; data are taken from S. rhizophila DSM 14405 with similar proteins (using BLAST [1]).
Similarities/identities are given for the overlapping region.
Similarity is restricted to the C-terminal part of GgpPS from S. rhizophila.
Similarity is restricted to the N-terminal part of GgpPS from S. rhizophila.
Tps, probably the wrong automatic annotation was used.
FIG. 2.
Phylogenetic comparison of putative GG synthesis protein and selected Tps protein sequences from heterotrophic bacteria, including the GgpPS and partial Tps sequences from S. rhizophila DSM 14405, cyanobacteria, yeast, and plants (protein sequences were extracted from GenBank). Sequences obtained in this study are shown in bold. The comparison was made using ClustalX. Bootstrap values are included. The glycogen synthase (GlgA) protein from E. coli was used as the outgroup. Acaryo, Acaryochloris marina MBIC11017 (accession no. YP_001517954); Anidu, Aspergillus nidulans (accession no. AAB05869); Athal, Arabidopsis thaliana (accession no. AC007260); Avine, Azotobacter vinelandii AvOP (accession nos. ZP_00419964, ZP_00416807); Azoar, Azoarcus sp. strain EbN1 (accession no. YP_157460; Calbi, Candida albicans (accession no. CAA69223); Cyanot, Cyanothece sp. strain CCY0110 (accession no. ZP_01726261); Dshib, Dinoroseobacter shibae DFL12 (accession no. ZP_0158649); Ecoli, E. coli BAA15717; Klact, Kluyveromyces lactis (accession no. CAA51164); Marino, Marinobacter sp. strain ELB17 (accession no. ZP_01734527); Mloti, Mesorhizobium loti MAFF303099 (accession no. NP_109451); Pberm, Parvularcula bermudensis HTCC2503 (accession no. ZP_01018536); Pseud, Pseudomonas sp. strain OA146 (accession no. Q93JY3); Pmend, Pseudomonas mendocina YMP (accession no. YP_001188605); Rhisp, Rhizobium sp. strain NGR234 (accession no. AAB91813); Roseo, Roseovarius HTCC2601 (accession no. ZP_01444455); Rhodo, Rhodobacterales bacterium HTCC2654 (accession no. ZP_01013028); Sthyp, Salmonella enterica serovar Typhimurium LT2 (accession no. AAL20844); Scere, Saccharomyces cerevisiae (accession no. CAA85083); Smalt, Stenotrophomonas maltophilia strain K279a (accession no. ZP_01646117); Smeli, Sinorhizobium meliloti 1021 (accession no. NP_435371); Spomb, Schizosaccharomyces pombe (accession no. CAA82861); Srhiz, Stenotrophomonas rhizophila DSM 14405 (accession no. AJ878740.2); Sstel, Sagittula stellata E-37 (accession no. ZP_01747210); Syn6803, Synechocystis sp. strain PCC 6803 (accession no. P74258); Syn7002, Synechococcus sp. strain PCC 7002 (accession no. O65979); SynWH8102, Synechococcus sp. strain WH 8102 (accession no. CAE07796); Thiobacillus denitrificans ATCC 25259 (accession no. ZP_00334532); Xcamp, Xanthomonas campestris pv. campestris ATCC 33913 (accession nos. NP_638429, NP_638382).
The additional N-terminal part in the new GG synthesis enzyme from S. rhizophila was used for separate similarity searches using the BLAST algorithm (1). This protein part contains an S6PP domain (PFAM 05116), which belongs to the large group of phosphohydrolases of the “HAD-protein-superfamily” (COG0561). Again, very high identities of 60 to 80% were found for this domain among the putative GG synthesis proteins of related bacteria such as X. campestris (Table 2). Besides finding many similar protein sequences from genome sequencing projects, we found that the highest similarities to a functionally characterized protein were displayed by sucrose-phosphate phosphatases (S6PP), e.g., from Synechocystis sp. strain PCC 6803 (Table 2). Obviously, the putative GG synthesis proteins from the heterotrophic bacteria seem to present a fusion capable of performing the two enzymatic reactions necessary to synthesize GG. The N-terminal, putative phosphatase domain could be involved in the dephosphorylation of the intermediate GG-phosphate, which is probably produced by the C-terminal glucosyltransferase/synthase domain. Therefore, the corresponding genes encode a new class of composite GG synthesis enzymes and were named ggpPS.
It should be mentioned that our similarity searches detected many additional putative GgpS sequences in genomes from cyanobacteria and from diverse heterotrophic bacteria that exhibit only the synthase domain, like the GgpS enzyme from Synechocystis sp. strain PCC 6803. Those putative GgpS sequences from heterotrophic bacteria, e.g., Thiobacillus denitrificans, cluster together with the cyanobacterial proteins (Fig. 2). Interestingly, separate ORFs displaying significant similarities to the N-terminal part of GgpPS from S. rhizophila were found up- or downstream from these putative GgpS-encoding genes in the genomes of the same heterotrophic bacteria (Table 2). In contrast, all the cyanobacterial genomes harbor another class of GG-phosphate phosphatases (GgpP or StpA), which is specific for cyanobacteria and was functionally characterized in Synechocystis sp. strain PCC 6803 (10). This class of cyanobacterial GgpP proteins did not show any similarities to the N-terminal extension of the newly discovered GgpPS proteins nor to the HAD superfamily proteins from heterotrophic bacteria harboring a cyanobacterial-type GgpS without a fused phosphatase domain (Table 2).
In further experiments, the upstream sequence of the ggpPS gene from S. rhizophila was sequenced (EMBL accession no. AJ878740.2). An ORF was identified immediately upstream of the ggpPS coding sequence that codes for a putative transporter. According to the corresponding protein from E. coli, it was named ycaD (3). The corresponding protein sequence harbors a conserved domain, PRK03633, which is characteristic for transporter proteins of the “major facilitator superfamily” (Mfs). Proteins with the highest similarity (60 to 80% identical amino acid residues) were detected in the genomes of related bacteria such as X. campestris strain ATCC 33913, A. vinelandii strain AvOP, P. mendocina strain YMP, and Marinobacter sp. strain ELB17. The same bacterial strains contain genes coding for GgpPS proteins. In each case, these genes are closely linked and could form an operon.
Because of its ability to accumulate trehalose in addition to GG, a tps gene was searched for in the genome of S. rhizophila. An internal fragment covering about 20% of the entire tps sequence was obtained. A complete tps gene was extracted from the ongoing genome sequencing project of a closely related bacterium, S. maltophilia strain R551-3 (accession no. ZP_01646117). The deduced amino acid sequences show a high degree of similarity to Tps proteins from related gram-negative bacteria such as Xanthomonas (Fig. 1), while Tps from eukaryotes (yeast and plants) and all sequences for GG synthesis enzymes cluster in separate branches (Fig. 2).
Expression of ggpPS after salt stress.
To make a functional analysis of the newly discovered gene for GG biosynthesis, we analyzed its expression after it was subjected to salt shock. Expression of the ggpPS gene was first characterized in Northern blotting experiments using a radiolabeled probe. A smear was obtained with RNA from salt-treated cells, while only very faint signals were detected from RNA of cells grown under control conditions. The maximal transcript sizes corresponding to the largest signals were found at about 3.8 kb (not shown). This size corresponded well to an expression of the ggpPS together with the putative ycaD transporter gene situated upstream.
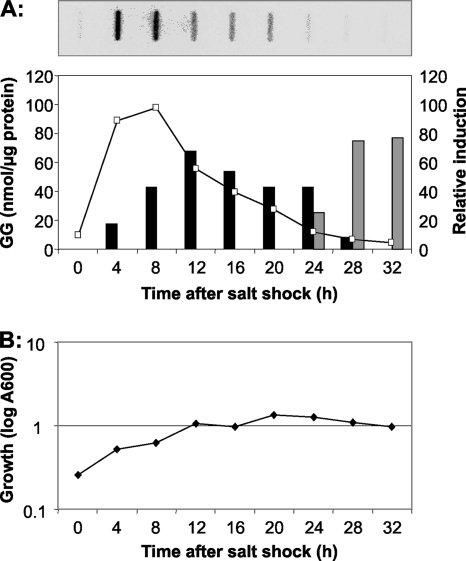
In a time course experiment, the ggpPS gene expression and the GG accumulation were compared. With RNA extracted from control cells grown under low-salt conditions, almost no signal was obtained independently of the growth phase. However, within only 30 min after the addition of NaCl, a strong induction was detected for ggpPS. These mRNA levels further increased and showed a maximum (10-fold increase) level at around 6 h after salt shock was induced (Fig. 3). Thereafter, the mRNA level became diminished, reaching a twofold lower level at the late exponential phase. The transcript level of the ggpPS in stationary-phase cells returned to very low levels, comparable to those of control cells, where almost no hybridization signal was obtained (Fig. 3). The accumulation kinetics of the compatible solute GG followed the gene expression pattern. Control cells grown in low-salt medium did not contain GG at any growth phase. Immediately after they were exposed to salt shock, Stenotrophomonas cells started GG accumulation, without a significant lag phase. However, GG was detected only in salt-shocked cells during exponential growth (Fig. 3). The decrease of cellular GG after the transition of cells into stationary phase is obviously based on a selective release of this compatible solute into the medium, since in parallel to the decrease of cellular GG, the amount of GG started to increase in the medium (Fig. 3A). Nearly the total amount of GG was detected in the medium; therefore, a controlled degradation can be ruled out.
FIG. 3.
Alteration of the ggpPS-specific mRNA steady-state levels in S. rhizophila DSM 14405 after a salt shock of 3% NaCl was applied (A). During the complete growth phase, samples were taken at the given time points, and total RNA was isolated and analyzed by hybridization with a 32P-labeled probe. The signals were quantified using a phosphorimager and were related to a hybridization signal of a 16S-rRNA gene probe applied to the same filters. The mRNA levels obtained at time point 0 were set to the level 10, while all other mRNA levels represent induction ratios relative to this time point. For comparison, the amount of GG in cells (panel A, black columns) and in the medium (panel A, gray columns) was estimated. (B) The growth curve is displayed for S. rhizophila cells shocked with 3% NaCl at time point 0 h.
Experimental verification of the enzyme activity of ggpPS.
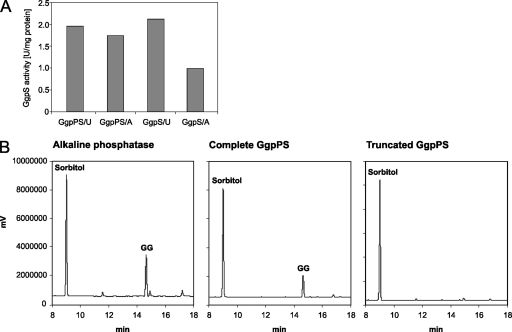
To have direct evidence for the function of the putative new GG synthesis protein from heterotrophic bacteria, we initially undertook attempts to specifically mutate the gene in S. rhizophila. Unfortunately, we could not establish a directed mutation protocol for this bacterium. Alternatively, we measured the predicted enzymatic activities of the gene products. For this purpose, the ggpPS gene was expressed after it was fused to a His tag coding sequence in E. coli. Besides the complete gene, a truncated form coding only for the C-terminal GgpS part was cloned into the same vector system. Recombinant proteins of the expected sizes of 89.5 and 60.1 kDa, from the full-length and truncated gene forms, respectively, were purified (not shown). Unfortunately, these enzymes seemed to be rather unstable. They did not show enzymatic activities in the GgpS enzyme assay developed for the cyanobacterial GgpS protein (20). Alternatively, we used a coupled enzyme assay, which allows for the direct monitoring of glucosyltransferase activities in a cuvette of a photometer. This test worked reproducibly and accurately with the recombinant GgpS from Synechocystis sp. strain PCC 6803 (not shown). In contrast, only in a few cases were we able to detect low GgpS activities with the newly identified GG synthesis enzymes of S. rhizophila. These measurements indicated GgpS activity with the precursor ADP-glucose, as well as that of UDP-glucose in combination with glycerol-3-phosphate, using the full-length GgpPS protein and the truncated protein representing the C-terminal GgpS domain as well (Fig. 4A). The de novo synthesis of GG shown in the successful enzyme measurements was verified by the detection of the newly produced GG by GC.
FIG. 4.
Verification of the putative enzyme activities of GgpPS from S. rhizophila DSM 14405, using recombinant proteins obtained after expression of the complete and a truncated ggpPS gene in E. coli. (A) Measurements of enzyme activities with purified complete GgpPS and the truncated GgpS. Enzyme activities were measured in the presence of the precursor UDP-glucose (U) or ADP-glucose (A). (B) Detection of GG-phosphate phosphatase activities after equal amounts of GG-phosphate were incubated with alkaline phosphatase, the purified complete GgpPS, and the purified truncated GgpS. The GG released was detected using gas-liquid chromatography, where 50 μg of sorbitol (Sorbitol) was added as the internal standard.
The second enzyme activity, GG-phosphate phosphatase, could be detected only with the full-length protein. Incubation of GG-phosphate with the purified intact 89.5-kDa GgpPS protein led to almost complete dephosphorylation of GG-phosphate, comparable to that of the control reaction with alkaline phosphatase (Fig. 4B). In contrast, incubation of GG-phosphate with the truncated protein devoid of the N-terminal phosphatase domain did not result in the release of GG, which fully supports the prediction of protein function.
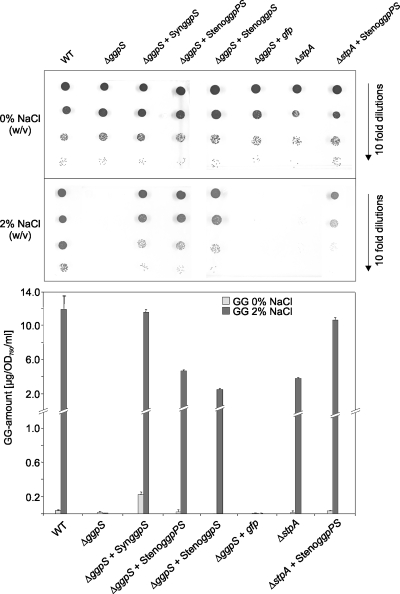
Because of the problems involved with developing a stable in vitro enzyme assay for these proteins, we used complementation of Synechocystis mutants as an additional strategy to confirm the combined activity of GG-phosphate synthase, as well as phosphatase, by the newly discovered GgpPS protein. Salt-sensitive mutants of Synechocystis sp. strain PCC 6803 defective in GG synthesis, either via the knockout of ggpS (the synthase/glucosyltransferase step [19]) or ggpP (the StpA, phosphatase step [10]), were used as hosts for the expression of the S. rhizophila gene under the control of the ggpS promoter from Synechocystis sp. strain PCC 6803. Expression of the ggpS gene of Synechocystis (control) and both the complete ggpPS gene and its truncated version of S. rhizophila in the ggpS defective mutant led to improved salt tolerance and a significant accumulation of GG (Fig. 5). Expression of gfp as the control for a gene that is not involved in compatible solute synthesis did not result in GG accumulation or in improved salt tolerance in comparison to that of the ggpS mutant as expected. Moreover, expression of the full-length ggpPS gene led to complementation of the salt-sensitive phenotype of the mutant defective in the phosphatase step (ΔstpA), which shows a severe drop in salt resistance because of the toxic effect of GG-phosphate accumulation (10). Obviously, GG-phosphate is dephosphorylated in cells of the complemented strain, resulting in much higher salt tolerance than in nontransformed mutant cells, which confirms the GG-phosphate phosphatase activity of the full-length GgpPS protein from S. rhizophila (Fig. 5).
FIG. 5.
Complementation of the defined Synechocystis sp. strain PCC 6803 mutants defective in GG synthesis by the ggpPS gene from S. rhizophila. The salt resistance and accumulation of GG were analyzed in cells of the Synechocystis wild type (WT), the salt-sensitive mutants of Synechocystis defective in GgpS (ΔggpS) or GgpP (ΔstpA), and the mutant strains expressing the Synechocystis ggpS gene (ΔggpS + SynggpS), the full-length ggpPS gene from S. rhizophila (ΔggpS + StenoggpPS and ΔstpA + StenoggpPS, respectively), the truncated version of the ggpPS gene from S. rhizophila (ΔggpS + StenoggpS), and the gfp gene (ΔggpS + gfp), respectively. (A) Effect of NaCl on the viability of cells grown for 10 days on BG11 medium supplemented with 0 or 2% NaCl (pH 8). (B) GG accumulation in cells of the strains grown in liquid BG11 medium supplemented with 0 or 2% NaCl for 1 day.
DISCUSSION
The complete gene encoding a new enzyme for GG synthesis was identified and cloned from S. rhizophila. A very similar gene for GG synthesis was obtained from Pseudomonas sp. strain OA146. The corresponding protein sequences revealed very high similarities over their whole lengths to GgpS proteins that have already been characterized, which made their proposed nature very likely (Fig. 1, 2). The newly obtained GgpPS proteins of S. rhizophila and those from Pseudomonas sp. strain OA146 grouped clearly into the corresponding cluster, which is closely linked to the cluster of separate GgpS proteins dominated by sequences from different cyanobacteria (Fig. 2). The newly discovered GG synthesis enzymes, called GgpPS, are a composite of an N-terminal GG-phosphate phosphatase and a C-terminal GG-phosphate synthase domain. Such composite enzymes displaying the synthase and phosphatase reactions were also found for the Tps2 protein in yeast (13) and for the sucrose phosphate synthases (SPS) proteins in plants (18). In the case of the SPS protein, the N-terminal domain contained all features for synthase activity, and a 20-kDa C-terminal domain displayed phosphatase activity. Despite the structural similarity to the combined SPS/S6PP proteins from plants, the SPS proteins from Synechocystis sp. strain PCC 6803 showed only SPS activity, since a few amino acid residues known to be essential for S6PP activity are missing in the putative C-terminal S6PP part of this gene product. The necessary S6PP activity is performed by a separate protein (18), which showed high sequence identities to the N-terminal part of the newly discovered GgpPS proteins.
The putative function of the GgpPS from S. rhizophila was verified using three different lines of evidence. First, the expression of the gene correlated very closely with the accumulation of GG in S. rhizophila. Control cells, regardless of the growth phase, did not show significant ggpPS expression and were virtually free of GG. Both processes were activated after salt shock, as was shown for many other bacteria (20). In the case of the GG accumulation, it seems to be regulated mainly by the transcriptional activity of the ggpPS gene. This is also obvious from the behavior of stationary-phase salt-treated cells, which switched off the ggpPS gene expression and released GG into the medium. How far the posttranslational regulation of enzyme activities is involved in the downregulation of GG synthesis, in addition to the transcriptional regulation, cannot be determined from the present data. It has been shown for trehalose synthesis in salt-stressed E. coli cells, as well as for GG synthesis in salt-stressed cyanobacterial cells, that transcriptional and protein activity regulation cooperate to ensure a stress-proportional accumulation of compatible solutes (8, 20).
Second, the enzyme activity measurements fully support the assumption that the proteins made from the cloned ggpPS genes from S. rhizophila are sufficient to synthesize the compatible solute GG in these heterotrophic bacteria. The full-length and the truncated C-terminal domains were able to produce GG-phosphate, while only the complete GgpPS protein with the N-terminal phosphatase domain was able to split GG-phosphate. Contrary to the cyanobacterial GgpS, which was found to be strictly dependent on ADP-glucose (9), the newly discovered GgpPS enzymes were able to use UDP-glucose and ADP-glucose as the glucosyl donors. The problems with the enzyme assays could not be solved in this study, which made a more detailed biochemical characterization impossible.
Therefore, as a third line of evidence, we showed the successful complementation of Synechocystis mutants. Again, the complete ggpPS gene and the truncated version expressing the C-terminal glucosyltransferase domain complemented the salt-sensitive phenotype of a Synechocystis ggpS defective mutant by the regeneration of GG accumulation (Fig. 5). The toxic accumulation of the intermediate GG-phosphate in the corresponding ΔggpP (ΔstpA) strain was also successfully diminished by the expression of the complete ggpPS from S. rhizophila.
The ggpPS gene in S. rhizophila is linked to a gene for a transporter of the Mfs superfamily. Both genes seem to be cotranscribed. Among other characteristics, it shows high similarities to an mfs transporter gene from E. coli called ycaD (3). Interestingly, proteins very similar to YcaD from S. rhizophila are also encoded upstream from the putative ggpPS genes in, e.g., A. vinelandii, which was proven to accumulate GG after salt shock (data not shown here), or X. campestris. The substrates for the YcaD protein from S. rhizophila and the most closely related ones are unknown; however, those transport proteins have been shown to be involved in sugar transport into or out of the cells. There are many examples showing that Mfs transporters are involved in the specific efflux of drugs or other substances from bacterial cells (16). The colocalization of ycaD and ggpPS in S. rhizophila and many related bacterial strains makes it tempting to speculate that this cotranscribed putative transporter may be involved in GG transport. Whether it is used for GG uptake or the observed selective release of GG from stationary phase cells needs further experimentation.
Acknowledgments
The work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG). The financial support of the Fond der Chemischen Industrie to M.H. is acknowledged.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, G., N. Roskot, and K. Smalla. 1999. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 373594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, M. J. Collado-Vides, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417459-463. [DOI] [PubMed] [Google Scholar]
- 6.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infections associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 117-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelbrecht, F., K. Marin, and M. Hagemann. 1999. Expression of the ggpS gene, involved in osmolyte synthesis in the marine cyanobacterium Synechococcus sp. strain PCC 7002, revealed regulatory differences to the freshwater strain Synechocystis sp. PCC 6803. Appl. Environ. Microbiol. 654822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giaever, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strom. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 1702841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagemann, M., and N. Erdmann. 1994. Activation and pathway of glucosylglycerol biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 1401427-1431. [Google Scholar]
- 10.Hagemann, M., A. Schoor, R. Jeanjean, E. Zuther, and F. Joset. 1997. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J. Bacteriol. 1791727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagemann, M., A. Schoor, S. Mikkat, U. Effmert, E. Zuther, K. Marin, S. Fulda, J. Vinnemeier, A. Kunert, C. Milkowski, C. Probst, and N. Erdmann. 1999. The biochemistry and genetics of the synthesis of osmoprotective compounds in cyanobacteria, p. 177-186. In A. Oren (ed.), Microbiology and biogeochemistry of hypersaline environments. CRC Press LLC, Boca Raton, FL.
- 12.Hincha, D., and M. Hagemann. 2004. Stabilization of model membranes during drying by compatible solutes involved in the stress tolerance of plants and microorganisms. Biochem. J. 383277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaasen, I., J. McDougall, and A. R. Strøm. 1994. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene 1459-15. [DOI] [PubMed] [Google Scholar]
- 14.Kunert, A., M. Hagemann, and N. Erdmann. 2000. Construction of promoter probe vectors for Synechocystis sp. PCC 6803 applying the light-emitting reporter systems Gfp and LuxAB. J. Microbiol. Methods 41184-194. [DOI] [PubMed] [Google Scholar]
- 15.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, West Sussex, United Kingdom.
- 16.Lewinson, O., J. Adler, N. Sigal, and E. Bibi. 2006. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol. Microbiol. 61277-284. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosenbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 18.Lunn, J. E. 2002. Evolution of sucrose synthesis. Plant Physiol. 1281490-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marin, K., E. Zuther, T. Kerstan, A. Kunert, and M. Hagemann. 1998. The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosylglycerol-phosphate synthase is involved in osmolyte synthesis. J. Bacteriol. 1804843-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin, K., J. Huckauf, S. Fulda, and M. Hagemann. 2002. Salt-dependent expression of glucosylglycerol-phosphate synthase involved in osmolyte synthesis of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1842870-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkat, S., E. Galinski, A. Minkwitz, G. Berg, and A. Schoor. 2000. Salt adaptation in moderately halotolerant bacteria: characterization of glycosylglycerol-synthesizing isolates from brackish coastal waters and the rhizosphere. Syst. Appl. Microbiol. 2331-41. [DOI] [PubMed] [Google Scholar]
- 22.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50101-136. [DOI] [PubMed] [Google Scholar]
- 23.Palleroni, N. J., and M. Doudoroff. 1972. Some properties and taxonomic subdivisions of the genus Pseudomonas. Annu. Rev. Phytopathol. 1073-100. [Google Scholar]
- 24.Pocard, J. A., L. T. Smith, G. M. Smith, and D. LeRudulier. 1994. A prominent role for glucosylglycerol in the adaptation of Pseudomonas mendocina SKB70 to osmotic stress. J. Bacteriol. 1766877-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 18327-36. [DOI] [PubMed] [Google Scholar]
- 26.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1111-16. [Google Scholar]
- 27.Roder, A., E. Hoffmann, M. Hagemann, and G. Berg. 2005. Synthesis of the compatible solutes glucosylglycerol and trehalose by salt-stressed cells of Stenotrophomonas strains. FEMS Microbiol. Lett. 243219-226. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schoor, A., N. Erdmann, U. Effmert, and S. Mikkat. 1995. Determination of the cyanobacterial osmolyte glucosylglycerol by high-performance liquid chromatography. J. Chromatogr. A 70489-97. [Google Scholar]
- 30.Wolf, A., A. Fritze, M. Hagemann, and G. Berg. 2002. Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 521937-1944. [DOI] [PubMed] [Google Scholar]