Abstract
In Escherichia coli, the cold shock response is exerted upon a temperature change from 37°C to 15°C and is characterized by induction of several cold shock proteins, including polynucleotide phosphorylase (PNPase), during acclimation phase. In E. coli, PNPase is essential for growth at low temperatures; however, its exact role in this essential function has not been fully elucidated. PNPase is a 3′-to-5′ exoribonuclease and promotes the processive degradation of RNA. Our screening of an E. coli genomic library for an in vivo counterpart of PNPase that can compensate for its absence at low temperature revealed only one protein, another 3′-to-5′ exonuclease, RNase II. Here we show that the RNase PH domains 1 and 2 of PNPase are important for its cold shock function, suggesting that the RNase activity of PNPase is critical for its essential function at low temperature. We also show that its polymerization activity is dispensable in its cold shock function. Interestingly, the third 3′-to-5′ processing exoribonuclease, RNase R of E. coli, which is cold inducible, cannot complement the cold shock function of PNPase. We further show that this difference is due to the different targets of these enzymes and stabilization of some of the PNPase-sensitive mRNAs, like fis, in the Δpnp cells has consequences, such as accumulation of ribosomal subunits in the Δpnp cells, which may play a role in the cold sensitivity of this strain.
The cold shock response in Escherichia coli is elicited when cells which are exponentially growing at 37°C are shifted to a lower temperature, such as 15°C (for a review, see reference 38). Upon a temperature downshift, there is a transient arrest of cell growth, termed acclimation phase, during which there is a severe inhibition of general protein synthesis. However, during this phase a specific set of genes is induced which encode cold shock proteins. The highly induced cold shock proteins include CspA (20) and its homologues, such as CspB (26), CspG (36), and CspI (55), polynucleotide phosphorylase (16, 43, 44), RNA helicase CsdA (52), initiation factor IF2 (21), transcription factor NusA (17), RecA (54), histone-like protein H-NS (13), DNA gyrase (23), and ribosome-binding factor RbfA (11).
E. coli polynucleotide phosphorylase (PNPase) is encoded by the pnp gene, is induced at low temperature (24, 58), and is essential for growth at low temperatures (29, 40). However, the exact role of PNPase in this essential function has not been fully elucidated. PNPase is an exoribonuclease, promotes the processive degradation of RNA, and is also responsible for residual RNA tailing observed in E. coli mutants devoid of the main polyadenylating enzyme, PAP I (34, 35) (32). In vitro, PNPase catalyzes the phosphorolytic degradation of RNA, releasing nucleoside 5′-diphosphate from the 3′ end of the substrate, and the reverse reaction of nucleoside 5′-diphosphate polymerization with release of phosphate (28).
PNPase is a homotrimer of a 711-amino-acid polypeptide and contains two RNA binding domains, KH and S1, located at the C terminus, and two RNase PH domains linked by an alpha-helical domain, which are responsible for its catalytic and homotrimerization properties (19, 22, 42, 50). Systematic structure-function analysis of PNPase domains has revealed that in addition to the two core domains, the alpha-helical domain is involved in PNPase enzymatic activity (4). PNPase has three catalytic activities, phosphorolysis, polymerization, and phosphate exchange; all of these three activities reside in the core domain, consisting of the two RNase PH domains and the alpha-helical domain. Comparison of the two core domains showed that sequence conservation is indicated in the glycine and alanine residues involved in the close packing of the main elements within each core domain. The differences between the two may relate to the different functions in PNPase activity, in that the RNase PH-like phosphorolytic catalytic site may be conserved in the second domain while the first core domain is involved in the flexible binding of RNA substrate and the formation of the trimer channel structure (50). The KH and S1 domains are primarily involved in RNA binding, contribute to substrate recognition, and are dispensable. Deletion of the RNA binding domains does not abolish any of the three catalytic activities, indicating that they are contained in a domain independent of the catalytic center.
As mentioned above, during the acclimation phase, CspA homologues are highly induced. PNPase was shown to be required to repress production of CspA homologues by degrading them selectively at the end of the acclimation phase. In a pnp mutant, expression of CspA homologues was significantly prolonged upon cold shock (41, 56). PNPase also associates with the endonuclease RNase E and other proteins in the RNA degradosome (for a review, see reference 7).
To elucidate the cold shock function of PNPase, we screened for genes which when expressed from multicopy plasmids can complement the cold-sensitive phenotype of pnp deletion cells at 15°C. We observed that another 3′-to-5′ processing exoribonuclease, RNase II, can complement PNPase function at low temperature. This result suggested that the RNase activity of PNPase is critical for its cold shock function. This was further supported by the observation that RNase PH domains 1 and 2 of PNPase are important for its cold shock function. We also showed that the polymerization activity of PNPase is not essential for its cold shock function. Interestingly, the third 3′-to-5′ processing exoribonuclease, RNase R of E. coli, which is cold inducible (6), was not able to complement the cold shock function of PNPase. We further showed that this difference is due to the different targets of these enzymes and that stabilization of some of the PNPase-sensitive mRNAs, like fis, has consequences, such as accumulation of ribosomal subunits in the Δpnp cells, which may play a role in the cold sensitivity of this strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain JM83 [F− araΔ(lac-proAB) rpsL(Strr)] (57) was used as the wild-type strain. The Δpnp strain, in which the entire pnp gene was replaced with a kanamycin cassette, was constructed according to the method described by Datsenko and Wanner (12). The deletion was confirmed by PCR analysis. The rnb, rnr, and pcnB deletion strains used in this study were obtained from the Keio collection, Japan, and the Δpnp ΔpcnB and Δrnr Δrnb double deletion strains were created by P1 transduction after removing the kanamycin cassette from each strain by the method described by Datsenko and Wanner (12). The bacterial cultures were grown in Luria-Bertani broth (LB) (1% tryptone, 0.5% yeast extract, and 0.5% NaCl). Ampicillin (50 μg ml−1) was supplemented as required.
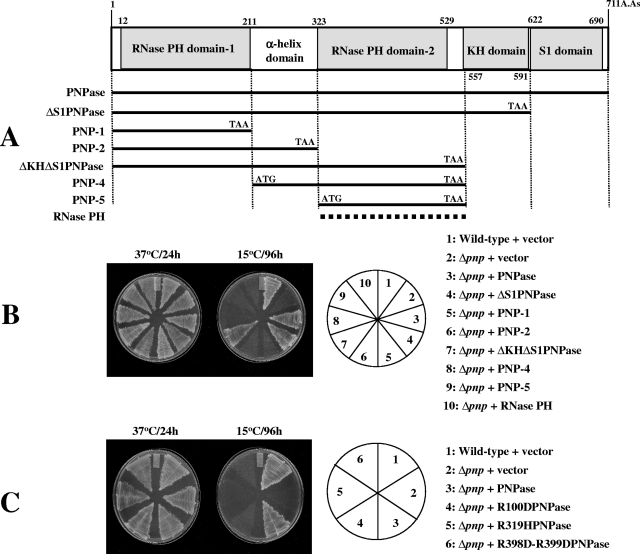
The plasmids pINIII-pnp, pINIII-rnb, and pINIII-rnr were constructed by cloning the regions corresponding to pnp, rnb. and rnr, respectively, using NdeI and BamHI. The pINIII plasmids containing different domains of PNPase were created using NdeI and BamHI as per the scheme shown in Fig. 2A.
FIG. 2.
RNase activity of PNPase is critical for its cold shock function. (A) Schematic representation of domains of PNPase and plasmids constructed. Different domains of PNPase were cloned in the pINIII vector using NdeI and BamHI sites, as shown. The RNase PH plasmid shown with a dashed line is the pINIII plasmid carrying rph, the gene encoding E. coli RNase PH. (B) E. coli wild-type cells were transformed with the pINIII plasmid as a control, and Δpnp cells were transformed with the pINIII plasmid alone or the pINIII plasmid containing either the entire PNPase, different domains of PNPase, or RNase PH. The plasmids carrying different domains of PNPase or RNase PH are designated as in panel A. The cells were streaked on LB plates containing ampicillin (50 μg ml−1) and incubated at 37°C or 15°C. Results for plates incubated at 37°C for 24 h or at 15°C for 96 h are presented. (C) E. coli wild-type cells were transformed with the pINIII plasmid as a control, and Δpnp cells were transformed with the pINIII plasmid alone or the pINIII plasmid containing either the wild-type PNPase or PNPase carrying the R100D, R319H, or R398D-R399D mutations. The cells were streaked on LB plates containing ampicillin (50 μg ml−1) and incubated at 37°C or 15°C. Results for plates incubated at 37°C for 24 h or at 15°C for 96 h are presented.
Site-directed mutagenesis.
Point mutations were introduced in pnp through site-directed mutagenesis using PCR by the methods described by Lerner et al. and Spee et al. (27, 48). The pINIII vector carrying the wild-type pnp gene was used as a template for PCR. The oligonucleotides used for PCR were as follows: for R100DPNPase, 5′-TTG ACC GCC CGA TTG ATC CGC TGT TCC CGG AA-3′ and 5′-TTC CGG GAA CAG CGG ATC AAT CGG GCG GTC AA-3′; for R319HPNPase, 5′-CGC GTA TCG ACG GTC ATG AAA AAG ATA TGA TC-3′ and 5′-GAT CAT ATC TTT TTC ATG ACC GTC GAT ACG CG-3′; and for R398D-R399DPNPase, 5′-GGT TCT CCG AAG GAT GAT GAA ATT GGT CAC G-3′ and 5′-CGT GAC CAA TTT CAT CAT CCT TCG GAG AAC C-3′. After the PCR, the template DNA was degraded by DpnI treatment. The resultant plasmids expressing R100DPNPase, R319HPNPase, or R398D-R399DPNPase were isolated and sequenced to confirm the mutant sequences.
RNA isolation.
E. coli cells grown overnight in LB medium at 37°C were diluted in fresh medium. The wild-type and Δpnp cells carrying pINIII vector alone and the Δpnp cells expressing the PNPase, RNase II, or RNase R proteins were grown at 37°C to exponential phase (optical density at 600 nm [OD600] of 0.5) and then were transferred to 15°C. Samples were removed after 1 and 5 h. For primer extension to detect the fis mRNA level, the RNAs were isolated at 37°C before shifting to 15°C and after 72 h at 15°C. The total RNA was extracted by the hot phenol method as previously described (47). It was quantified by measuring absorbance at 260 nm. The purity of RNA was confirmed by agarose gel electrophoresis.
Primer extension.
Primer extension was carried out by a method described previously (39). The deoxyoligonucleotide used for detection of yfiA was 5′-CTACTCTTCTTCAACTTCTTCGACGAAG-3′, and that for detection of fis was 5′-TTAGTTCATGCCGTATTTTTTC-3′ (25). The oligonucleotides were labeled with [γ32-P]ATP (DuPont-New England Nuclear) by using T4 polynucleotide kinase (Gibco BRL). Primer extension was carried out with 5 μg of RNA at 42°C for 1 h in a final reaction volume of 10 μl, with 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, 0.4 pmol of 32P-labeled primer, 0.5 mM (each) of the deoxynucleoside triphosphates, 10 U RNase inhibitor (Boehringer Mannheim), and 6.25 U of reverse transcriptase (Boehringer Mannheim). Equal amounts of RNA samples were used for primer extension, and the products were analyzed on a 6% polyacrylamide gel under denaturing conditions.
Isolation of polysomes and sucrose density gradient sedimentation.
Polysomes were prepared and resolved as described previously (46), with minor modifications. E. coli cells were grown at 37°C in LB medium supplemented to log phase. Upon reaching an appropriate culture density (OD600 of 0.5; 100-ml culture), polysomes were trapped by the addition of chloramphenicol to the culture to a final concentration of 0.1 mg ml−1. After an additional 4 min of incubation, cells were harvested by centrifugation. The cell pellet was resuspended in 1 ml of buffer BP (20 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 mM NH4Cl, and 5 mM β-mercaptoethanol). The cell suspension was placed in a Beckman ultracentrifuge tube. Cells were then lysed by immersing the tube into a liquid nitrogen bath for 1 min, followed by thawing them in a water bath at room temperature until no traces of ice remained. This freeze-thaw cycle was repeated two more times, and the lysate was subsequently subjected to centrifugation at 100,000 × g for 10 min in a Beckman TLA100.3 rotor. Polysomes were resolved by applying 0.2 ml of the supernatants to a 5 to 40% linear sucrose density gradient (10 ml) in buffer BP with subsequent ultracentrifugation at 4°C in a Beckman SW41 rotor for 2.5 h at 35,000 rpm.
The ribosome profile was analyzed at a 254-nm wavelength. rRNA was detected. Peaks corresponding to polysomes, and 70s, 50S, and 30S ribosomes are shown. The peak adjacent to the 30S ribosomal peak consists of small RNAs including small mRNAs, tRNAs, etc. For quantitation, the heights of individual peaks were measured from the base for each of the peaks in the profile carried out for the wild-type cells containing only the vector, and these values were considered to be 100%, and the base-to-peak values of corresponding peaks from other conditions were expressed as relative percentages.
RESULTS AND DISCUSSION
Genetic screening for protein counterparts of PNPase.
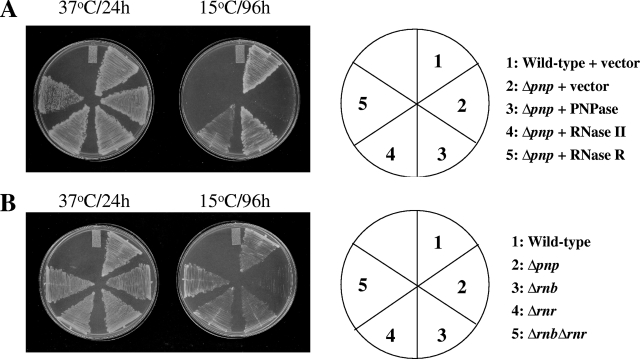
In order to elucidate the role of PNPase in acclimation of cells to cold, we explored genes which can complement the cold-sensitive phenotype of pnp deletion (Δpnp) cells at 15°C. A genomic library was created in the plasmid pUC19 by partial digestion of the E. coli chromosomal DNA with Sau3A1. The partially digested DNA fragments were cloned into the plasmid using BamHI. The Δpnp cells were transformed with the genomic library. Transformants were isolated for their ability to grow at 15°C. Colony PCR analysis of the transformants showed that 147 clones contained the entire pnp gene. The remaining seven clones showed the presence of the rnb gene. This gene encodes RNase II (RNase II) (53). We repeated the screening and in the second set obtained 53 colonies at 15°C. We observed that out of these, 32 candidates contained the entire pnp gene, 3 candidates had the pnp gene without the C-terminal region encoding the KH and S1 domains, and 1 candidate contained the pnp gene without the region encoding only the S1 domain region. The remaining 17 candidates showed the presence of the rnb gene. Thus, in addition to PNPase, RNase II was the only other candidate protein that emerged from these studies that is able to complement the PNPase function at low temperature. To confirm if this is indeed the case, the wild-type or Δpnp cells harboring the pINIII plasmid as controls and the Δpnp cells expressing plasmid-encoded PNPase or RNase II were streaked on LB-ampicillin plates and incubated at 37°C and 15°C. As expected, all the cells were able to grow at 37°C (Fig. 1A). The wild-type cells were able to grow at 15°C, while the Δpnp cells showed a cold-sensitive phenotype. The growth of Δpnp cells was restored at 15°C by the presence of either PNPase or RNase II on a plasmid (Fig. 1A). This result confirms that RNase II can complement PNPase function at low temperature.
FIG. 1.
(A) RNase II can complement the cold-sensitive phenotype of the Δpnp cells at 15°C. The E. coli wild-type cells were transformed with the pINIII plasmid as a control, and the Δpnp cells were transformed with the pINIII plasmid alone (control) or containing pnp, rnb, or rnr. The cells were streaked on LB plates containing ampicillin (50 μg ml−1) and incubated at 37°C and 15°C. Results with plates incubated at 37°C for 24 h or at 15°C for 96 h are presented. (B) RNase II or RNase R is not essential for cell growth at 15°C. E. coli wild-type, Δpnp, Δrnb, Δrnr, and Δrnb Δrnr cells were streaked on LB plates and grown at 37 or 15°C. Results with plates incubated at 37°C for 24 h or at 15°C for 96 h are presented.
E. coli has three major 3′-to-5′ processing exoribonucleases involved in mRNA turnover, namely, PNPase, RNase II, and RNase R. These enzymes are primarily involved in RNA metabolism in E. coli, and based on the observation that double deletion of PNPase and RNase II (16) or PNPase and RNase R is lethal at 37°C (9), these may have an overlapping role(s). In spite of this, these proteins exhibit several different properties. PNPase is a phosphorylase and requires phosphate for RNA degradation, while RNase II and RNase R are hydrolases. Degradation of RNAs by PNPase results in a release of XDPs, while that by RNase II and RNase R results in a release of XMPs. RNase II and RNase R belong to the RNR family; these exhibit approximately 60% similarity in their secondary structures. However, RNase R is larger than RNase II. Both PNPase and RNase R but not RNase II are induced by cold shock. Our genetic screen showed that only RNase II can complement the cold shock function of PNPase. However, we also tested if RNase R can complement the cold shock function of PNPase in our plate assay. As seen in Fig. 1A, consistent with the results of genetic screening, RNase R failed to support growth of Δpnp cells at 15°C. Interestingly, deletion of RNase II either singly or in combination with RNase R does not affect cell growth at 15°C (Fig. 1B).
Importance of RNase domains of PNPase in its cold shock function.
Our genetic screen revealed the importance of the RNase activity of PNPase. Therefore, we carried out domain analysis of PNPase to test if its RNase domains were sufficient for carrying out its cold shock function. As shown in Fig. 2, PNPase consists of two RNase PH domains linked together by an α-helix domain at the N-terminal end. Its C-terminal end consists of two RNA binding domains, the KH domain and the S1 domain. Constructs were created as depicted in Fig. 2A and cloned in the pINIII vector. The Δpnp cells were transformed with various plasmids as shown in Fig. 2B and grown on LB plates containing ampicillin at 37 and 15°C. Wild-type cells containing vector alone were also included as a control. As seen in Fig. 2B, cells carrying all the constructs were able to grow at 37°C. The wild-type cells containing vector alone or the Δpnp cells containing the entire pnp gene were able to grow at 15°C. The Δpnp cells carrying the vector alone were not able to grow at 15°C, as expected. The Δpnp cells carrying PNPase constructs in which the S1 domain was deleted either alone (no. 4; ΔS1PNPase) or together with the KH domain (no. 7; ΔKHΔS1PNPase) were able to grow at 15°C, indicating that these domains are dispensable for the function of PNPase at low temperature. This is also consistent with our observation that in our genetic screen described above, four clones, which were able to grow at 15°C, showed the presence of the pnp gene in which either the S1 domain alone or both the KH and S1 domains were absent. On the other hand, constructs in which either (or both) of the RNase PH domains was deleted (no. 5, 6, 8, and 9; PNP-1, PNP-2, PNP-4, and PNP-5, respectively) were not able to support growth of the Δpnp cells at 15°C.
In E. coli, the only 3′-5′ exoribonucleases that catalyze Pi-dependent degradation of RNA, leading to release of nucleoside diphosphates, are PNPase and RNase PH (14). The primary role of PNPase is in mRNA degradation (16), while that of RNase PH is suggested to be in tRNA processing (15). RNase PH shows similarity to RNase PH domain 2 of PNPase (50). Since our results emphasized the importance of the RNase PH domain of PNPase, we tested whether E. coli RNase PH (15) can complement the cold-sensitive phenotype of the Δpnp cells (no. 10). RNase PH is not essential for growth at low temperature (data not shown). As seen in Fig. 2, RNase PH was not able to support the growth of Δpnp cells at 15°C. This is consistent with our result that RNase PH did not come up in our genetic screen as a candidate to complement the cold-sensitive phenotype of Δpnp cells. Taken together, the results show that both RNase domains of PNPase are essential for its cold shock function, and RNase PH, which has only one RNase domain, or the PNPase construct devoid of either of the RNase domains is not adequate to carry out its cold shock function. The observation that the RNA binding domains of PNPase are dispensable for its essential activity at low temperature is also consistent with a previous report that a plasmid expressing a truncated PNPase devoid of the entire S1 domain was able to complement the low-temperature growth defect of a pnp nonsense mutant (58). Also, it was reported that the S1 or KH domain might enhance the processivity of PNPase but they are not required for activity against single-stranded substrates. PNPase lacking either the S1 or KH domain does indeed retain phosphorolytic activity against model substrates, suggesting that its catalytic activity resides within the core PH domain, also called the second core domain (49). However, this observation conflicts with a recent report by Matus-Ortega et al. (31), wherein the authors concluded that the RNA binding domains, particularly the KH domain, are vital at low temperature. One possible reason for this may be the different genetic backgrounds of the cells used. These authors used strain SK10019, in which both the pnp and rph genes are mutated or absent. It was shown that the Δpnp Δrph strain exhibits significantly slower growth (doubling time, 480 min) even at 31°C than the Δpnp (doubling time, 55 min) and Δrph (doubling time, 49 min) strains (59). In our studies, a Δpnp strain was used.
Previously it was reported that the mutations in RNase PH domains 1 and 2 abolish the catalytic activity of PNPase in vitro. Deletion of the KH and S1 domains, on the other hand, did not abolish the catalytic activity (22). Out of various mutations described in that study, we chose two mutations, R100D and R398D-R399D, present in the RNase PH1 and PH2 domains, respectively, to test if the mutant proteins lose their cold shock function. We also chose a mutation, R319H, described in that study, which resides in the α-helical domain and abolishes the enzymatic activity of PNPase in vitro. This is consistent with an observation by another group that the α-helical domain linking the two RNase PH domains of PNPase is involved in its enzymatic activity (4). The mutations were created by site-directed mutagenesis, and the pINIII plasmids carrying the mutant pnp constructs were transformed into Δpnp cells and grown on LB plates with ampicillin at 37 and 15°C. Wild-type cells and Δpnp cells carrying vector alone or expressing the wild-type PNPase were also included in the plate assay as controls. As seen from Fig. 2C, all cells showed growth at 37°C, while only the wild-type cells or Δpnp cells expressing the wild-type PNPase were able to grow at 15°C. The Δpnp cells did not show growth at 15°C, as expected. The cells expressing R100DPNPase, R319HPNPase, or R398D-R399DPNPase were not able to grow at 15°C. This clearly shows that the RNase activity of PNPase is essential for its cold shock function.
Polymerization activity of PNPase is not essential for its cold shock function.
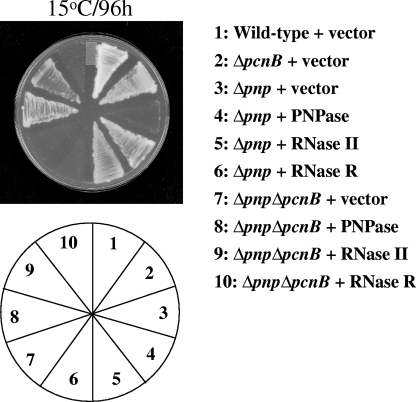
In addition to the phosphorolysis and phosphate exchange activity, PNPase also has polymerization activity (22). As described above, our genetic screen revealed that RNase II can complement the cold-sensitive phenotype of the Δpnp cells. However, RNase II does not have the polymerization activity of PNPase. This suggested the possibility that the polymerization activity of PNPase may not play a critical role in its cold shock function. We tested this possibility. E. coli has two enzymes which modify the 3′ ends of RNAs, poly(A) polymerase, which is encoded by the pcnB gene, and PNPase. They add a poly(A) and/or poly(X) tail at the 3′ end of RNA, which is important for their metabolism (33). The double deletion mutant strain devoid of these two enzymes (Δpnp ΔpcnB strain) lacks poly(A) or poly(X) tails on the 3′ ends of RNAs. As seen from Fig. 3, the wild-type or ΔpcnB strain with vector alone can grow at 15°C while the Δpnp strain with vector alone cannot grow at this temperature, as expected. This strain showed growth at 15°C when expressing PNPase or RNase II but not RNase R. The Δpnp ΔpcnB strain showed cold sensitivity, as expected, which can be complemented by expression of PNPase or RNase II but not RNase R. The complementation of the double deletion mutant by RNase II is interesting. Since RNase II lacks polymerization activity but can complement the cold-sensitive phenotype of the Δpnp ΔpcnB strain, it can be concluded that the cold-sensitive phenotype of the double deletion strain is not due to the absence of the two polymerization enzymes of E. coli and the polymerization activity of PNPase does not play a role in its cold shock function. The cold sensitivity of the Δpnp or ΔpnpΔpcnB strain is thus due to the absence of the RNase activity of PNPase.
FIG. 3.
The polymerization activity of PNPase is not essential for its cold shock function. E. coli wild-type, Δpnp, ΔpcnB, and Δpnp ΔpcnB cells were transformed with the pINIII plasmid alone as a control. The Δpnp and ΔpnpΔpcnB cells were also transformed with the pINIII plasmid containing PNPase, RNase II, or RNase R. The cells were streaked on LB plates containing ampicillin (50 μg ml−1) and incubated at 15°C. Results for plates incubated for 96 h are presented.
PNPase and RNase II have different substrate specificities from that of RNase R.
The observation that the double deletion mutant of PNPase and RNase R is not viable (9) suggests that they may have some overlapping functions. However, our studies showed that RNase R cannot complement the cold shock function of PNPase. Previous studies showed that RNase R may have a different role during cold shock adaptation from that of PNPase and RNase II, in that RNase R has a more important role in maturation of SsrA/tmRNA than the other two (6, 45). RNase II and RNase R share many catalytic properties; however, certain differences are present, such as the fact that RNase R acts poorly on DNA but can degrade rRNAs quite well. On the other hand, RNase II is essentially inactive against these (8). It was proposed that these enzymes are relatively nonspecific exoribonucleases; however, our observation that RNase II but not RNase R can complement PNPase function at low temperature suggests that they may have certain differences in their specificities. In order to test this possibility, we chose one gene that was shown to be a target of PNPase based on our DNA microarray data comparing the cold shock response (15°C for 1 and 5 h) of the wild-type strain and the Δpnp strain (not shown) and evaluated the effect of expression of all the three enzymes on the level of its mRNA. We chose the gene, yfiA, encoding protein Y, for this experiment. Protein Y is induced by cold shock (1, 2). The level of mRNA for this gene was elevated in the Δpnp strain compared to that in the wild-type strain after 5 h at 15°C. It inhibits translation initiation during cold shock but not at normal temperatures. It reduces translation initiation during stress and quickly releases ribosomes for renewed translation initiation.
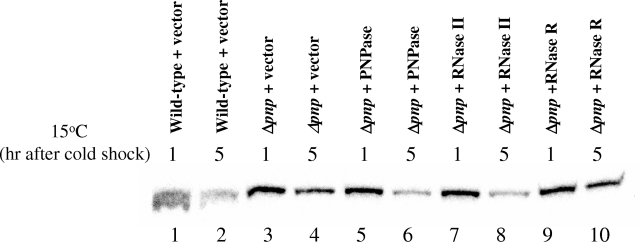
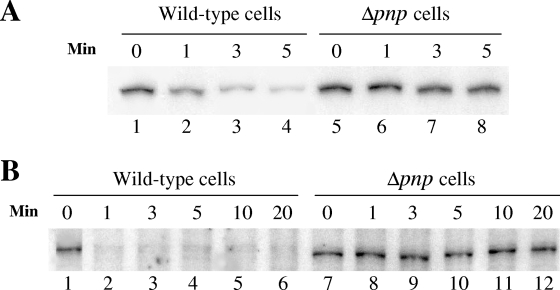
The wild-type and Δpnp cells carrying the pINIII vector alone and the Δpnp cells expressing the PNPase, RNase II, or RNase R protein were grown at 37°C to exponential phase (OD600 of 0.5) and then were transferred to a 15°C environment. Samples were removed after 1 and 5 h for RNA extraction. Primer extension was carried out to detect the yfiA mRNA level. Results of the primer extension are shown in Fig. 4. In the wild-type strain, the yfiA mRNA level goes down after 5 h at 15°C (compare lanes 1 and 2), while in the Δpnp strain, the yfiA mRNA level is significantly higher (lane 4). As shown later in Fig. 7, this high level of the yfiA transcript is due to its stabilization in Δpnp cells. This stabilization is lost in the Δpnp strain expressing plasmid-encoded PNPase (lane 6) and also in cells expressing plasmid-encoded RNase II (lane 8). However, in the cells expressing plasmid-encoded RNase R, the yfiA mRNA remains stabilized (lane 10). This result suggests that some of the targets of PNPase which are important for cold shock adaptation of cells are different from those of RNase R and thus RNase R cannot complement the cold shock function of PNPase.
FIG. 4.
PNPase has different targets from those of RNase R. Cells from which total RNAs were isolated by the hot phenol method are indicated above the respective lanes. The exponentially growing cells were cold shocked at 15°C for 1 h and 5 h, and samples were collected for RNA isolation. Equal amounts of RNA samples were used for primer extension with deoxyoligonucleotide corresponding to the yfiA gene. The products were analyzed by 6% polyacrylamide gel electrophoresis under denaturing conditions.
FIG. 7.
An increase in transcript levels of yfiA (A) and fis (B) in the Δpnp cells is due to stabilization of the respective mRNAs. Total RNAs were isolated from wild-type and Δpnp cells by the hot phenol method. The exponentially growing cells were cold shocked at 15°C for 1 h (for the yfiA gene) or 72 h (for the fis gene), the transcription was stopped by adding rifampin, and samples were collected for RNA isolation at the times indicated in the figure. Equal amounts of RNA samples were used for primer extension with deoxyoligonucleotides corresponding to the yfiA or fis gene. The products were analyzed by 6% polyacrylamide gel electrophoresis under denaturing conditions.
Deletion of PNPase causes 30S, 50S, and 70S ribosomal subunits to accumulate.
Next, we tested the possibility of the ribosome profile in the Δpnp cells being affected, since several mRNAs are stabilized by deletion of PNPase and also since deletion of PNPase renders the cells sensitive to low temperatures and it is well documented that the cold-sensitive mutants of E. coli often are defective in ribosome metabolism (5, 10, 18, 37, 51). A defective ribosomal profile would affect the translation, leading to growth retardation.
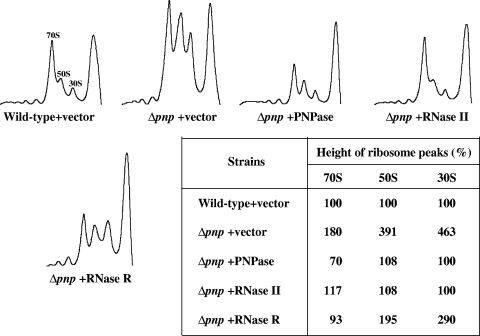
Wild-type and Δpnp cells containing pINIII vector alone and Δpnp cells expressing PNPase, RNase II, or RNase R were cold shocked at 15°C for 72 h, and samples were removed for polysome profile analysis. Cell lysates were subjected to 5 to 40% sucrose density gradient fractionation, as described in Materials and Methods. As shown in Fig. 5, in the Δpnp cells carrying the pINIII vector alone, the 30S (>4.5 times), 50S (∼4 times), and 70S (∼2 times) ribosomal subunits were accumulated. Plasmid-based expression of PNPase or RNase II was able to counteract this accumulation, restoring the normal ribosome profile. The table included in Fig. 5 shows quantitation of the ribosome profile values for easy comparison. In the cells expressing RNase R, the 50S ribosomal subunits (∼2 times) and the 30S ribosomal subunits (∼3 times) were still accumulated, although the accumulation was less than that in the Δpnp cells carrying the pINIII vector alone. The 70S ribosomal subunits were not accumulated in this strain.
FIG. 5.
Deletion of PNPase leads to accumulation of 30S, 50S, and 70S ribosomal subunits. The E. coli wild-type and Δpnp cells were transformed with the pINIII plasmid alone as a control. The Δpnp cells were also transformed with the pINIII plasmid containing PNPase, RNase II, or RNase R. The cells growing exponentially at 37°C were cold shocked at 15°C, and samples were collected 72 h after the cold shock. Polysomes were isolated and resolved as described in Materials and Methods.
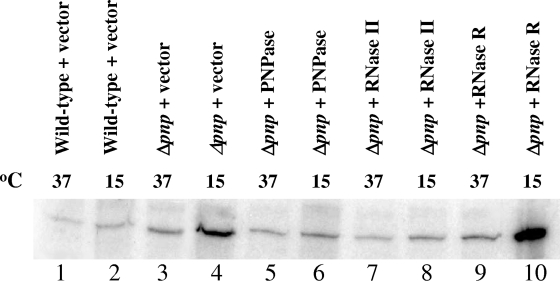
We next tested if this accumulation is a consequence of stabilization of fis in the Δpnp cells. Fis is a small, 11.2-kDa DNA binding protein that serves as a transcription regulator of many genes, including rRNA genes (3, 30). We carried out primer extension to detect fis mRNA levels in the cells from which the ribosome profile was isolated. Isolation of mRNA and primer extension was carried out as described in Materials and Methods. As shown in Fig. 6, a significantly higher level of fis mRNA was detected after 72 h at 15°C in the Δpnp cells than in the wild-type cells (compare lanes 2 and 4). As shown later in Fig. 7, this high level of fis transcript is due to its stabilization in Δpnp cells. This stabilization was lost in the Δpnp cells overexpressing PNPase (lane 6) or RNase II (lane 8) but not RNase R (lane 10). This result suggests that the unusual accumulation of ribosomal subunits seen in Fig. 5 may be a consequence of stabilization of fis mRNA in the Δpnp cells. It is interesting that the fis mRNA is significantly stabilized in the Δpnp cells expressing RNase R (lane 10), but the accumulation of ribosomal subunits in this strain is less than that in the Δpnp cells carrying vector alone. This may be due to the fact that RNase R but not RNase II can efficiently degrade rRNA (8). However, as seen in Fig. 5, this degradation is not complete, and the cells retain a significant amount of accumulated ribosomal subunits. It is possible that this accumulation results in the cold sensitivity of the Δpnp cells. One cannot exclude the possibility that other mRNAs which are stabilized in the Δpnp cells may also influence the ribosome profile in this strain. A time-dependent analysis of genes affected by PNPase deletion upon cold shock would shed light on this aspect.
FIG. 6.
fis mRNA is stabilized in the absence of PNPase. Cells from which total RNAs were isolated by the hot phenol method are indicated above the respective lanes. The cells growing exponentially at 37°C were cold shocked at 15°C, and samples were collected for RNA isolation before cold shock and 72 h after cold shock. Equal amounts of RNA samples were used for primer extension with deoxyoligonucleotide corresponding to the fis gene. The products were analyzed by 6% polyacrylamide gel electrophoresis under denaturing conditions.
In order to confirm that the high levels of yfiA and fis mRNAs seen in the Δpnp cells compared to those in the wild-type cells upon cold shock are due to their stabilization and not to their increased transcription, we analyzed stability of these mRNAs in these two strains. Cells were cold shocked as described above, rifampin was added to stop the transcription, and RNA samples were isolated at various time points. Primer extension was then carried out to detect the respective mRNAs. As seen in Fig. 7, both yfiA and fis mRNAs were stabilized in the Δpnp cells. In the case of yfiA mRNA, the half-life is less than 1 min (Fig. 7A, lane 2) in the wild-type cells while a significant amount of mRNA is detected even after 5 min in the Δpnp cells (lane 8). Similarly, in the wild-type cells, the fis mRNA rapidly disappears and is barely detectable even at 1 min (Fig. 7B, lane 2), while in the Δpnp cells, it is stable even after 20 min following rifampin addition (lane 12). This result proves that indeed these two genes are targets of the RNase activity of PNPase at low temperature and are stabilized in the Δpnp cells.
Conclusion.
The RNase activity of PNPase is critical for its cold shock function, while its polymerization activity is dispensable. Another 3′-5′ processing exonuclease, RNase II, can complement cold shock function of PNPase, but RNase R cannot. This is attributed to the difference in the substrate specificities of these enzymes. Further analysis of targets of PNPase which can be acted upon by RNase II but not by RNase R can shed light on the essential role of PNPase during cold shock adaptation of cells. Stabilization of some of the PNPase-sensitive mRNAs, such as fis, has consequences such as accumulation of ribosomal subunits in the Δpnp cells, which may play a role in the cold sensitivity of this strain.
Acknowledgments
We thank Hirotada Mori from the Keio collection, Japan, for the gift of the pcnB, rnb, and rnr deletion strains.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Agafonov, D. E., V. A. Kolb, I. V. Nazimov, and A. S. Spirin. 1999. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. USA 9612345-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agafonov, D. E., V. A. Kolb, and A. S. Spirin. 2001. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2399-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, L., L. Nilsson, E. Vijgenboom, and H. Verbeek. 1990. FIS-dependent trans-activation of tRNA and rRNA operons of Escherichia coli. Biochim. Biophys. Acta 1050293-301. [DOI] [PubMed] [Google Scholar]
- 4.Briani, F., M. Del Favero, R. Capizzuto, C. Consonni, S. Zangrossi, C. Greco, L. De Gioia, P. Tortora, and G. Deho. 2007. Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 89145-157. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, R. E., and P. S. Sypherd. 1974. Genetic analysis of cold-sensitive ribosome maturation mutants of Escherichia coli. J. Bacteriol. 1171082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 501349-1360. [DOI] [PubMed] [Google Scholar]
- 7.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 6171-87. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Z. F., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 27721624-21629. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 27314077-14080. [DOI] [PubMed] [Google Scholar]
- 10.Dammel, C. S., and H. F. Noller. 1993. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 7660-670. [DOI] [PubMed] [Google Scholar]
- 11.Dammel, C. S., and H. F. Noller. 1995. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9626-637. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245255-259. [DOI] [PubMed] [Google Scholar]
- 14.Deutscher, M. P. 1993. Promiscuous exoribonucleases of Escherichia coli. J. Bacteriol. 1754577-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deutscher, M. P., G. T. Marshall, and H. Cudny. 1988. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc. Natl. Acad. Sci. USA 854710-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, D. I., E. R. Olson, C. Georgopoulos, K. Tilly, I. Herskowitz, and F. Banuett. 1984. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol. Rev. 48299-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyl, D., A. Bock, and H. G. Wittmann. 1977. Cold-sensitive growth of a mutant of Escherichia coli with an altered ribosomal protein S8: analysis of revertants. Mol. Gen. Genet. 152331-336. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, T. J., J. D. Thompson, and J. Heringa. 1993. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324361-366. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 295881-5889. [DOI] [PubMed] [Google Scholar]
- 22.Jarrige, A., D. Brechemier-Baey, N. Mathy, O. Duche, and C. Portier. 2002. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J. Mol. Biol. 321397-409. [DOI] [PubMed] [Google Scholar]
- 23.Jones, P. G., R. Krah, S. R. Tafuri, and A. P. Wolffe. 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 1745798-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, P. G., R. A. VanBogelen, and F. C. Neidhardt. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1692092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch, C., and R. Kahmann. 1986. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J. Biol. Chem. 26115673-15678. [PubMed] [Google Scholar]
- 26.Lee, S. J., A. Xie, W. Jiang, J. P. Etchegaray, P. G. Jones, and M. Inouye. 1994. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol. Microbiol. 11833-839. [DOI] [PubMed] [Google Scholar]
- 27.Lerner, C. G., T. Kobayashi, and M. Inouye. 1990. Isolation of subtilisin pro-sequence mutations that affect formation of active protease by localized random polymerase chain reaction mutagenesis. J. Biol. Chem. 26520085-20086. [PubMed] [Google Scholar]
- 28.Littauer, U. Z. 2005. From polynucleotide phosphorylase to neurobiology. J. Biol. Chem. 28038889-38897. [DOI] [PubMed] [Google Scholar]
- 29.Luttinger, A., J. Hahn, and D. Dubnau. 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19343-356. [DOI] [PubMed] [Google Scholar]
- 30.Mallik, P., T. S. Pratt, M. B. Beach, M. D. Bradley, J. Undamatla, and R. Osuna. 2004. Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J. Bacteriol. 186122-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matus-Ortega, M. E., M. E. Regonesi, A. Pina-Escobedo, P. Tortora, G. Deho, and J. Garcia-Mena. 2007. The KH and S1 domains of Escherichia coli polynucleotide phosphorylase are necessary for autoregulation and growth at low temperature. Biochim. Biophys. Acta 1769194-203. [DOI] [PubMed] [Google Scholar]
- 32.Mohanty, B. K., and S. R. Kushner. 2003. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 50645-658. [DOI] [PubMed] [Google Scholar]
- 33.Mohanty, B. K., and S. R. Kushner. 2006. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 345695-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 9711966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 36982-994. [DOI] [PubMed] [Google Scholar]
- 36.Nakashima, K., K. Kanamaru, T. Mizuno, and K. Horikoshi. 1996. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J. Bacteriol. 1782994-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nashimoto, H., W. Held, E. Kaltschmidt, and M. Nomura. 1971. Structure and function of bacterial ribosomes. XII. Accumulation of 21S particles by some cold-sensitive mutants of Escherichia coli. J. Mol. Biol. 62121-138. [DOI] [PubMed] [Google Scholar]
- 38.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6125-136. [PubMed] [Google Scholar]
- 39.Phadtare, S., V. Tadigotla, W. H. Shin, K. Sengupta, and K. Severinov. 2006. Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J. Bacteriol. 1882521-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piazza, F., M. Zappone, M. Sana, F. Briani, and G. Deho. 1996. Polynucleotide phosphorylase of Escherichia coli is required for the establishment of bacteriophage P4 immunity. J. Bacteriol. 1785513-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154573-580. [DOI] [PubMed] [Google Scholar]
- 42.Regnier, P., M. Grunberg-Manago, and C. Portier. 1987. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J. Biol. Chem. 26263-68. [PubMed] [Google Scholar]
- 43.Reiner, A. M. 1969. Characterization of polynucleotide phosphorylase mutants of Escherichia coli. J. Bacteriol. 971437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiner, A. M. 1969. Isolation and mapping of polynucleotide phosphorylase mutants of Escherichia coli. J. Bacteriol. 971431-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards, J., P. Mehta, and A. W. Karzai. 2006. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 621700-1712. [DOI] [PubMed] [Google Scholar]
- 46.Ron, E. Z., R. E. Kohler, and B. D. Davis. 1966. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science 1531119-1120. [DOI] [PubMed] [Google Scholar]
- 47.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 321337-1346. [DOI] [PubMed] [Google Scholar]
- 48.Spee, J. H., W. M. de Vos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stickney, L. M., J. S. Hankins, X. Miao, and G. A. Mackie. 2005. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J. Bacteriol. 1877214-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symmons, M. F., G. H. Jones, and B. F. Luisi. 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 81215-1226. [DOI] [PubMed] [Google Scholar]
- 51.Tai, P. C., D. P. Kessler, and J. Ingraham. 1969. Cold-sensitive mutations in Salmonella typhimurium which affect ribosome synthesis. J. Bacteriol. 971298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toone, W. M., K. E. Rudd, and J. D. Friesen. 1991. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 1733291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wade, H. E. 1961. The autodegradation of ribonucleoprotein in Escherichia coli. Biochem. J. 78457-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 4860-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1811603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanaka, K., and M. Inouye. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 1832808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 58.Zangrossi, S., F. Briani, D. Ghisotti, M. E. Regonesi, P. Tortora, and G. Deho. 2000. Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli. Mol. Microbiol. 361470-1480. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, Z., and M. P. Deutscher. 1997. An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J. Bacteriol. 1794391-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]