Abstract
OhrR proteins can be divided into two groups based on their inactivation mechanism: 1-Cys (represented by Bacillus subtilis OhrR) and 2-Cys (represented by Xanthomonas campestris OhrR). A conserved cysteine residue near the amino terminus is present in both groups of proteins and is initially oxidized to the sulfenic acid. The B. subtilis 1-Cys OhrR protein is subsequently inactivated by formation of a mixed-disulfide bond with low-molecular-weight thiols or by cysteine overoxidation to sulfinic and sulfonic acids. In contrast, the X. campestris 2-Cys OhrR is inactivated when the initially oxidized cysteine sulfenate forms an intersubunit disulfide bond with a second Cys residue from the other subunit of the protein dimer. Here, we demonstrate that the 1-Cys B. subtilis OhrR can be converted into a 2-Cys OhrR by introducing another cysteine residue in either position 120 or position 124. Like the X. campestris OhrR protein, these mutants (G120C and Q124C) are inactivated by intermolecular disulfide bond formation. Analysis of oxidized 2-Cys variants both in vivo and in vitro indicates that intersubunit disulfide bond formation can occur simultaneously at both active sites in the protein dimer. Rapid formation of intersubunit disulfide bonds protects OhrR against irreversible overoxidation in the presence of strong oxidants much more efficiently than do the endogenous low-molecular-weight thiols.
Reactive oxygen species (ROS), including superoxide anion (O2−·), hydrogen peroxide (H2O2), hydroxyl radical (OH·), and organic hydroperoxide (ROOH), can damage many cell components and cause cell death (18). Commonly, cells will defend against ROS by employing ROS-sensing transcription factors that regulate the expression of detoxifying enzymes and DNA and protein repair systems (18). OxyR from Escherichia coli is one of the best-studied peroxide-sensing transcription factors. Reduced OxyR binds DNA upstream of the genes involved in oxidative stress defense but cannot activate the transcription of these genes. In the presence of H2O2, OxyR is oxidized and subsequently activates transcription by directly interacting with RNA polymerase (13). Only one cysteine, C199, is absolutely critical for redox sensing of OxyR (8), and this cysteine is oxidized by H2O2 to form a sulfenic acid. Subsequent to its initial oxidation, C199 forms a disulfide bond with C208 to activate OxyR (7, 9). However, different products of OxyR oxidation, including the C199 sulfenate and S-nitrosylated and S-glutathionylated derivatives, have also been proposed to be physiologically relevant (7, 9).
OhrR (organic hydroperoxide resistance regulator) is a peroxide-sensing transcription factor that regulates the expression of ohr genes, which encode thiol-dependent peroxidases that can reduce organic hydroperoxides to the corresponding alcohols (2, 19). The OhrR proteins from Bacillus subtilis and Xanthomonas campestris have been studied in the most detail, and structural studies have allowed visualization of both the reduced and DNA-bound forms of B. subtilis OhrR (6) and the reduced and oxidized forms of X. campestris OhrR (11). Although these two proteins share 40% amino acid identity and are structurally quite similar, they appear to differ significantly in their peroxide-sensing mechanisms. B. subtilis OhrR contains only a single Cys residue in each protein chain and functions by oxidation of this single residue (a 1-Cys mechanism). In contrast, X. campestris OhrR is representative of a large subgroup of OhrR proteins that contain two conserved Cys residues, and in this case protein oxidation leads to an intersubunit disulfide (a 2-Cys mechanism) (14).
The peroxide-sensing mechanism of Bacillus subtilis OhrR has been defined by biochemical comparisons of reduced and oxidized protein both in vitro and as recovered from cells (10, 17). The sole cysteine residue of B. subtilis OhrR, C15, is required for sensing of organic peroxides (4). This reactive C15 is initially oxidized by cumene hydroperoxide (CHP), a model organic peroxide, to the sulfenic acid derivative, as monitored in vitro (4). However, further studies have revealed that sulfenic acid formation is not sufficient to inactivate OhrR (10). The fate of the initially formed sulfenate depends on whether or not low-molecular-weight thiols are present and on the nature and concentration of the oxidant. In the absence of thiols, the sulfenate either can be irreversibly overoxidized to the sulfinic and sulfonic acids or can slowly cyclize to generate a protein sulfenamide (in which the initial sulfenate condenses with a neighboring backbone amide) (10). In the presence of low-molecular-weight thiols such as cysteine, the sulfenate can rapidly form a mixed disulfide (S cysteinylation). Unlike many organisms, B. subtilis lacks glutathione, and in cells treated with CHP the OhrR protein is oxidized to yield several different S-thiolated derivatives (including the mixed disulfides with cysteine, coenzyme A, and a structurally uncharacterized 398-Da thiol) (10). At high oxidant concentrations, or in the presence of linoleic acid hydroperoxide (LHP), significant amounts of overoxidation occur even in the presence of low-molecular-weight thiols, suggesting that under these conditions oxidation may be largely irreversible (17).
In contrast with B. subtilis OhrR, orthologs from other species often contain one or more Cys residues in their C-terminal domains. In the case of X. campestris OhrR, oxidation leads to an intersubunit disulfide bond between C22 and C127, resulting in the inactivation of the protein (14). Recent structural studies demonstrate that this oxidation event leads to a large-scale reorientation of the winged helix-turn-helix DNA-binding domains, thereby leading to derepression (11). Like the S thiolation of B. subtilis OhrR, protein disulfide bond formation protects the peroxidatic thiolate against overoxidation and provides a pathway by which active OhrR can be regenerated.
Here we demonstrate that the introduction of Cys residues into B. subtilis OhrR, at either of two positions, converts this protein into a 2-Cys OhrR in which derepression is accompanied by the formation of one or more intersubunit disulfide bonds. Thus, there is no fundamental difference between 1-Cys and 2-Cys OhrR family members, apart from the lack of a second appropriately positioned cysteine in the former class of proteins. These results provide insights into the evolutionary advantages of providing a second, protective thiol group as part of the protein (as in the 2-Cys family members) rather than relying on low-molecular-weight thiols.
MATERIALS AND METHODS
Site-directed mutagenesis and purification of OhrR.
The OhrR-FLAG G120C, Q124C, and GQ double mutants were generated using megaprimer PCR mutagenesis (15) using B. subtilis chromosomal DNA as the template as described previously (16). Mutant OhrR proteins were expressed ectopically using the xylose-inducible expression plasmid pXT as described previously (3). The OhrR-FLAG variants were expressed from their native promoter element after integration at the amyE locus in an ohrR mutant containing a PohrA-cat-lacZ operon fusion [HB2012; CU1065 ohrR::Kan SPβc2Δ2::Tn917::φ(ohrA-cat-lacZ) (4)]. The strains were checked for proper integration by PCR and the correct substitutions were confirmed by DNA sequencing of the PCR product.
For biochemical analyses, OhrR proteins were expressed using pET16b (Novagen)-based overexpression plasmids in Escherichia coli BL21(DE3)(pLysS). Proteins were purified following overexpression using heparin-Sepharose and Superdex-75 column chromatography as previously described (4). The molecular weight of each protein was checked by electrospray ionization-mass spectrometry (MS) and showed agreement with the calculated values. OhrR concentrations were determined using the calculated value of ɛ280 (14,440 M−1cm−1 for the wild type and corrected values for each of the mutant proteins) and aliquots were stored at −70°C in buffer A (20 mM Tris [pH 8.0], 100 mM NaCl, 5% [vol/vol] glycerol) containing 1 mM dithiothreitol (DTT) and 1 mM EDTA. For in vitro oxidation experiments, DTT and EDTA were removed using Bio-Spin 6 chromatography columns immediately prior to use. Protein structure figures were created using DeepView Swiss-Pdb Viewer 3.7 (GlaxoSmithKline R&D and the Swiss Institute of Bioinformatics) and Pov-Ray 3.6 (Persistence of Vision Raytracer Pty. Ltd.).
Analysis of OhrR oxidation in vitro by SDS-PAGE and MALDI-TOF MS.
Disulfide bond formation was monitored by nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples of OhrR variants (300 nM, monomer) either were left untreated or were treated with 3 μM CHP for 10 min in 1 ml of buffer A in the absence or presence of 1 mM cysteine. Proteins were precipitated with 110 μl of 100% trichloroacetic acid and recovered by centrifugation for 10 min at 16,100 × g. The protein precipitate was resuspended with iodoacetamide (IA) buffer (0.5 M Tris [pH 7.5], 5% [vol/vol] glycerol, 1 mM EDTA, 2% [wt/vol] SDS) containing 20 mM IA and further incubated for 1 h in the dark. Samples were resolved by nonreducing SDS-PAGE using a Tris-Tricine buffer system and photographed after staining with Coomassie brilliant blue R250 and subsequent destaining. The stained bands were cut and washed three times with 50 mM ammonium bicarbonate containing 50% (vol/vol) acetonitrile for 15 min at 37°C. The washed gel pieces were dried using a SpeedVac, rehydrated with 10 μl of trypsin solution (20 ng/μl trypsin in 40 mM ammonium bicarbonate containing 9% [vol/vol] acetonitrile), and incubated overnight at 37°C. The resulting tryptic digests (0.5 μl) were mixed with equal volumes of matrix and analyzed using an Applied Biosystems 4700 matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer.
Immunoblot analyses.
Cells were harvested and resuspended in Tris-borate-saline (TBS) buffer containing lysozyme (1 mg/ml), 100 mM IA, and protease inhibitor cocktail (Roche) and disrupted by sonication. OhrR-FLAG derivatives were immunoprecipitated using anti-FLAG beads (Sigma), resolved on a 15% Tris-Tricine SDS-PAGE gel, and electrophoretically transferred to Immunblot polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked in TBS with 0.5% Tween 20 (TBST) supplemented with 5% skim milk for 30 min at room temperature. The membrane was then incubated with anti-FLAG rabbit antibody (Sigma) overnight at room temperature. The membrane was washed three times for 10 min each in TBST, followed by incubation with alkaline phosphatase-coupled secondary goat anti-rabbit antibody for 1 h (Sigma). After the membrane was thoroughly washed, OhrR was visualized by adding the BCIP (5-bromo-4-chloro-3-indolylphosphate/Nitro Blue Tetrazolium substrate (Bio-Rad).
Analysis of OhrR oxidation in vivo.
To monitor protein oxidation in vivo, OhrR-FLAG was recovered from cells in the presence of 100 mM IA, isolated by SDS-PAGE, and then digested with trypsin as described previously (10). The resulting tryptic peptides were analyzed using an Applied Biosystems 4700 MALDI-TOF mass spectrometer as described above.
FA.
A 6-carboxyfluorescein-labeled DNA fragment containing the ohrA operator site was generated by annealing 5′-6-carboxyfluorescein-TACAATTAAATTGTATACAATTAAATTGTA-3′ (Integrated DNA Technologies) and its unlabeled complementary strand. Fluorescence anisotropy (FA) measurements (λex = 495 nm, slit width = 15 nm; λem = 520 nm, slit width = 20 nm, integration time = 1 s) were performed with 50 nM DNA and 300 nM OhrR in 3 ml of 20 mM Tris (pH 8.0) containing 150 mM NaCl and 5% (vol/vol) glycerol. FA values were recorded automatically every 10 s by use of a Perkin-Elmer LS55 luminescence spectrometer. The g factor for the experiments was 1.07 ± 0.01.
RESULTS
Conversion of the B. subtilis 1-Cys OhrR into a 2-Cys OhrR.
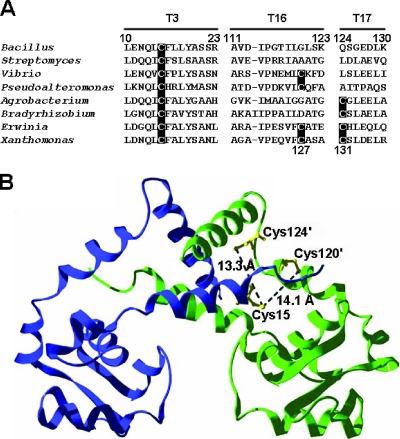
Cysteines corresponding to B. subtilis OhrR C15 are highly conserved among OhrR orthologs in bacteria (Fig. 1A) and are thought to define the site of initial protein oxidation (the peroxidatic cysteine). Many OhrR orthologs contain one or more additional cysteine residues near the C terminus and upon oxidation are thought to generate intersubunit disulfide bonds, by analogy with what is seen for the Xanthomonas campestris OhrR protein (11, 14). It is not known whether or not there are other significant differences in the structures or functions of the 1-Cys and 2-Cys OhrR proteins apart from the presence of an additional Cys residue in the C-terminal domain.
FIG. 1.
(A) Alignment of OhrR homologs. The sequences corresponding to tryptic peptides T3, T16, and T17 of B. subtilis OhrR are shown with Cys residues highlighted. The first two sequences shown are representative of the 1-Cys family of OhrR proteins, and the second pair are sequences that have a second Cys at the position corresponding to the redox active C127 of Xanthomonas campestris (last line). The next two have a more distal Cys residue corresponding to C131 in Xanthomonas. The last two sequences contain two Cys residues in this carboxyl-terminal region. Bacillus subtilis, CAA05594; Streptomyces coelicolor A3(2), CAB87337; Vibrio cholerae MZO-2, ZP_01977344; Pseudoalteromonas atlantica T6c, YP_659686; Agrobacterium tumefaciens strain C58, AAK86653; Bradyrhizobium sp. strain BTAi1, YP_001236314; Erwinia carotovora subsp. atroseptica SCRI1043, CAG76066; Xanthomonas campestris pv. Phaseoli, AAK62673. (B) B. subtilis OhrR (6) was modeled with the G120C and Q124C mutations. The predicted positions and distances between C15 and C120′ and C124′ are shown. Monomers are colored blue and green.
Comparisons of both primary sequences (Fig. 1A) and protein structures (6, 11) suggest that G120 and Q124 in B. subtilis OhrR correspond to the two carboxyl-terminal cysteine residues (C127 and C131) in the X. campestris ortholog. Upon oxidation, C127 forms an intersubunit disulfide with C22 (corresponding to C15 in the B. subtilis protein). The role of C131 in X. campestris is not known, although it is apparently not involved in OhrR oxidation (14). However, in Agrobacterium tumefaciens OhrR, there is only one carboxyl-terminal Cys residue at the position corresponding to X. campestris C131. Therefore, if this protein works by a 2-Cys mechanism, it presumably generates an intersubunit disulfide involving this residue.
To determine if the presence of carboxyl-terminal cysteine residues would alter the products of B. subtilis OhrR oxidation, we generated G120C and Q124C substitution mutants and the corresponding double mutant (GQ). By modeling of the G120C and Q124C mutants on the B. subtilis OhrR structure, the distances between sulfur atoms in C15 and the two introduced cysteine residues in G120C and Q124C were estimated to be approximately 14Å and 13Å, respectively (Fig. 1B). This is comparable to the 15.5-Å separation between the γS atoms of C122 and C127′ (C127 of the other OhrR subunit) in reduced X. campestris OhrR (11).
Peroxide sensing by the 2-Cys OhrR G120C variant in vitro.
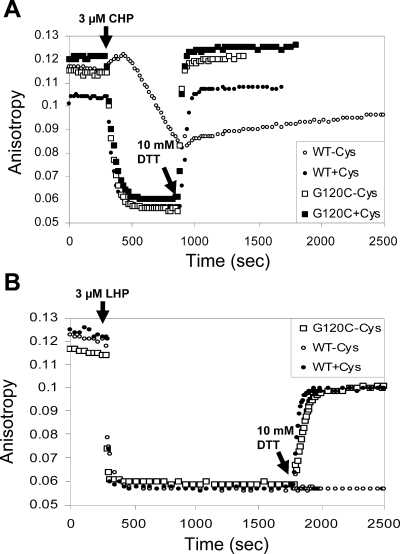
To investigate the mechanisms of peroxide sensing by the G120C mutant, the Q124C mutant, and the GQ double mutant proteins, we used a FA-based DNA-binding assay to monitor the kinetics and reversibility of OhrR oxidation in vitro as described previously (10). As previously reported, purified OhrR is oxidized to an inactive form only slowly (over many minutes) in the absence of low-molecular-weight thiols. This corresponds to rapid oxidation to the protein sulfenate (which still binds DNA) followed by the slow cyclization to the inactive protein sulfenamide (10). In contrast, in the presence of 1 mM cysteine, inactivation is rapid and fully reversible upon the subsequent addition of 10 mM DTT. The OhrR G120C mutant is inactivated rapidly upon the addition of 3 μM CHP, and full activity is restored upon the addition of 10 mM DTT (Fig. 2A). Significantly, and in contrast with what is seen for wild-type OhrR, the inactivation of the OhrR G120C mutant is fully reversible even in the absence of added cysteine. A similar pattern of reversible oxidation was noted for the GQ double mutant, but the Q124C mutant protein displayed a more complex pattern: activity could be largely restored by 10 mM DTT but the corresponding reduction kinetics were biphasic, with a rapid reduction of one half of the protein and a slower reduction of the other half (data not shown).
FIG. 2.
In vitro oxidation of mutant OhrR proteins is accompanied by disulfide bond formation. (A) FA assays monitoring inactivation of wild-type (WT) and G120C treated with 3 μM CHP in the presence or absence of 1 mM Cys. Oxidant was added at 5 min (arrow), and 10 mM DTT was added at 15 min. (B) The same analysis was performed using 3 μM LHP. Oxidant was added at 5 min, and 10 mM DTT was added at 30 min.
If OhrR is treated with 3 μM LHP instead of 3 μM CHP, the protein is much more sensitive to overoxidation (17). In the absence of added thiols, wild-type OhrR is quantitatively overoxidized to the sulfinic and sulfonic acid derivatives and the activity of this overoxidized protein cannot be restored even by incubation with 10 mM DTT. As shown previously (17), even 1 mM cysteine only partially protects wild-type OhrR against overoxidation and ∼70% of the DNA-binding activity can be recovered upon reduction by DTT (corresponding to the fraction of the protein protected by S cysteinylation) (Fig. 2B). Notably, the 2-Cys OhrR variant, the G120C mutant, is protected against overoxidation to the same extent even in the absence of any added cysteine (Fig. 2B).
Intermolecular disulfide bond formation by 2-Cys OhrR variants in vitro.
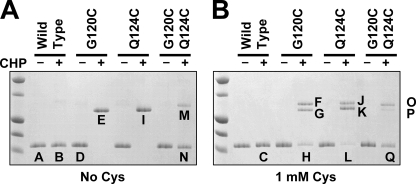
To monitor the extent of intersubunit disulfide bond formation under these conditions, wild-type OhrR and the three mutant proteins (G120C, Q124C, and the GQ double mutant) were treated with CHP in the presence or absence of 1 mM cysteine and the products were analyzed by nonreducing SDS-PAGE (Fig. 3). In the absence of cysteine, the G120C and Q124C proteins were quantitatively converted to disulfide-linked dimers by CHP treatment (Fig. 3A, bands E and I). In contrast, oxidation of the GQ mutant protein (containing two carboxyl-terminal cysteine residues) resulted in two bands approximately corresponding in size to monomeric and dimeric OhrR (Fig. 3A, bands M and N). When these same reactions were performed in the presence of 1 mM cysteine, the conversion to the dimeric form was still observed for both the G120C and Q124C mutants: two distinct bands appeared in the size range corresponding approximately to dimeric OhrR (Fig. 3B). These results suggest that the formation of disulfide-linked dimers is highly efficient upon oxidation in vitro and that the majority of the oxidized proteins form disulfide-linked dimers even in the presence of 1 mM cysteine.
FIG. 3.
(A) Disulfide bond formation in OhrR as monitored by nonreducing SDS-PAGE. Samples of protein either were left untreated or were treated with 3 μM CHP for 10 min prior to analysis. (B) Reactions were performed as for panel A, except that the reaction mixtures contained 1 mM cysteine. Protein bands indicated by letters were selected for structural analysis using MALDI-TOF MS, as shown in Fig. 4 and 5. The first lane in each panel contains molecular mass standards corresponding to 15, 20, 25, 37, and 50 kDa (bottom to top).
Analysis of OhrR oxidation products.
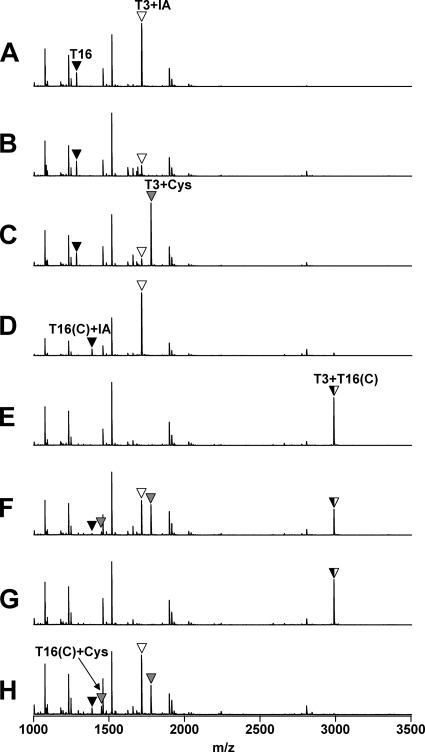
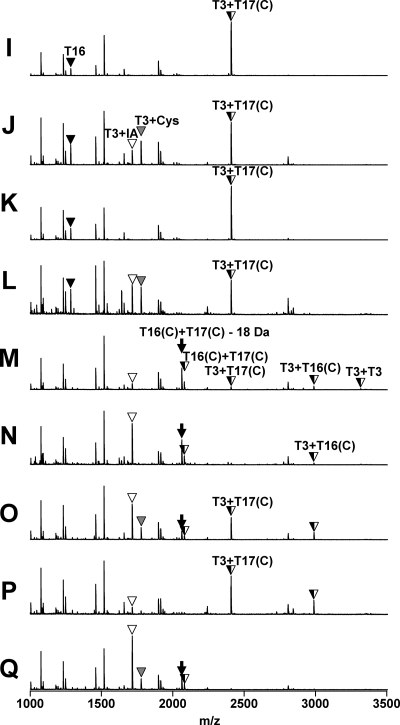
To elucidate the nature of the covalent changes resulting from oxidation, the bands from the CHP-treated samples and selected control (untreated) bands were excised from the polyacrylamide gels, digested with trypsin, and analyzed by MALDI-TOF MS (Fig. 4 and 5). As expected, in the wild-type control samples the peptide containing C15 (T3; Fig. 1A) was fully alkylated by IA, and this peptide was greatly reduced in intensity after oxidation by CHP (Fig. 4A and B). In the absence of cysteine, the oxidation of C15 leads largely to the formation of the protein sulfenamide (10), whereas in the presence of 1 mM cysteine, this peptide is S cysteinylated (Fig. 4C). In the absence of cysteine, the G120C mutant protein was oxidized at both active sites, resulting in a complete loss of the IA-modified tryptic peptides and the appearance of the expected disulfide-linked product peptide (Fig. 4E). Similarly, oxidation of the Q124C protein led to the appearance of the expected disulfide-linked peptide product [designated T3+T17(C); Fig. 5].
FIG. 4.
Analysis of structural changes accompanying oxidation for wild-type and G120C OhrR proteins. Samples A through H (from Fig. 3) were analyzed by MALDI-TOF MS after tryptic digestion. The T3 peptide (Fig. 1A) with a reduced Cys residue was modified with IA and is indicated by a white triangle. S-cysteinylated T3 peptide is indicated with a gray triangle. Note that the mass of the T16 peptide is larger in the G120C mutant protein as predicted [indicated as T16(C)+IA].
FIG. 5.
Analysis of structural changes accompanying protein oxidation for OhrR Q124C and the G120C Q124C double mutant. Samples I through Q (from Fig. 3) were analyzed by MALDI-TOF MS after tryptic digestion. The T3 peptide (Fig. 1A) with a reduced Cys residue was modified with IA and is indicated by a white triangle. S-cysteinylated T3 peptide is indicated with a gray triangle. Note that the mass of the T16 peptide is larger in the G120C mutant protein as predicted [indicated as T16(C)+IA]. Note that the disulfide-linked peptides indicated as T16(C)+T17(C) are detected as two peaks of m/z values of 2,060 and 2,078 Da, as described in the text.
We next analyzed the oxidation products from reaction mixtures containing 1 mM cysteine, a concentration thought to be higher (by perhaps three- to fivefold) than the concentration of free thiols in the cytosol of B. subtilis (5, 12). Under these oxidation conditions, all three mutant proteins gave rise to three distinct oxidation products, two corresponding in size to dimeric OhrR and one to monomeric protein (Fig. 3B). In the G120C reaction, the most slowly migrating product (Fig. 3B, band F) corresponded to a single protein cross-link (C15-C120), as judged by the appearance of the T3+T16(C) peptide (Fig. 4F). The remaining C15-containing peptides appeared as either alkylated (T3+IA) or S-cysteinylated (T3+Cys) peaks with approximately equal intensities (Fig. 4F). This suggests that this dimeric form of the protein contains two monomers linked by one disulfide bond. The mass spectrum of the slightly faster-migrating band (Fig. 3B, band G) was very similar, although the T3+IA and T3+Cys peptides were missing (Fig. 4G), consistent with the hypothesis that this protein contains two intersubunit disulfides rather than one. The monomeric protein band (Fig. 3B, band H) contained no C15-C120 disulfide, as evidenced by the absence of the peak corresponding to T3+T16 peptide. Instead, this band contained T3 and T16(C) peptides in either alkylated (+IA) or S-cysteinylated (+Cys) form. A similar distribution of products was noted for the Q124C protein: the dimeric products contained a major peak corresponding to the C15-C124 disulfide [T3+T17(C)], and the slower-migrating band additionally contained evidence of both reduced and S-cysteinylated peptide containing C15 (Fig. 5). Since this concentration of CHP is sufficient for full oxidation of wild-type OhrR, we suggest that the presence of monomeric, reduced protein in these reactions likely results from reduction of disulfide bonds by the excess of free cysteine. Indeed, peptides corresponding to S cysteinylation at both C15 and C120 were clearly detected (Fig. 4H). The ability of 1 mM cysteine to reduce S-cysteinylated OhrR has been documented previously (10), and it seems likely that the reduction of intersubunit protein disulfides is equally facile.
Oxidation of the GQ double mutant led to a more complex mixture of products (Fig. 5). The upper band contained peptides corresponding to both the C15-C120 [T3+T16(C)] and C15-C124 [T3+T17(C)] peptides, suggesting that this band contains two monomers cross-linked by either one of these possible intersubunit disulfide bonds. In addition, this dimeric protein contains peptides corresponding to both reduced C15 (T3+IA) and a cross-link between the two carboxyl-terminal cysteine residues (T16+T17). The latter product appears as a major peak at 2,060 Da and a smaller peak at 2,078 Da. These two peptides appear to arise from intramolecular disulfide bond formation, consistent with the proximity of these two introduced cysteine residues in the protein structure (Fig. 1B). Indeed, the major peak (2,060 Da) corresponds in mass to a single (T16+T17) peptide containing an internal disulfide linkage. Since the sequence in this region is C120LSKC124, it is understandable that an intramolecular disulfide bond could hinder trypsin cleavage at the intervening Lys residue. The lower-abundance 2,078-Da peak corresponds to the cleaved but disulfide-linked peptides resulting from trypsin-mediated hydrolysis of the K123-C124 linkage. Since the monomeric products of GQ oxidation (as judged by mobility during nonreducing SDS-PAGE) (Fig. 3, bands N and Q) also contain both the 2,060- and 2,078-Da products in similar ratios (Fig. 5N and Q), it is clear that this is an intramolecular disulfide bond. We suggest that the C120-C124 disulfide bond forms by thiol-disulfide exchange reactions subsequent to the formation of an intersubunit disulfide involving C15. A thiol exchange reaction involving the adjacent, unreacted carboxyl-terminal cysteine would generate the observed intrasubunit disulfide with regeneration of reduced C15.
The B. subtilis 2-Cys variants form intersubunit disulfide bonds in vivo.
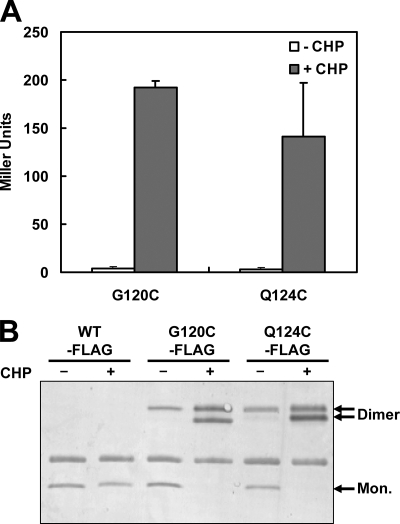
To monitor function in vivo, we expressed the G120C and Q124C mutant proteins in an ohrR mutant strain from the xylose-inducible xylA promoter. Both the G120C and Q124C mutants complemented the ohrR null mutation and allowed full repression of a PohrA-cat-lacZ reporter fusion. Upon the addition of CHP, both the G120C and Q124C mutant strains were fully derepressed for PohrA-cat-lacZ expression (Fig. 6A).
FIG. 6.
In vivo oxidation of the G120C and Q124C OhrR proteins. (A) β-Galactosidase activity assays of OhrR activity in strains expressing either the G120C or the Q124C variant. OhrR activity was monitored using a PohrA-cat-lacZ reporter fusion. Empty and filled bars represent the samples that were left untreated (-CHP) and treated with 100 μM CHP for 15 min (+CHP), respectively. (B) Immunoblot analysis of disulfide bond formation upon CHP oxidation. Cells were left untreated (−) or were treated with 100 μM CHP for 1 min (+) before immunoprecipitation in a buffer containing IA. Mon., monomeric.
We used immunoblot analysis to determine if the mutant proteins formed intermolecular disulfides in vivo upon oxidation. For this experiment, we used strains that express epitope-tagged G120-FLAG or Q124-FLAG, which are both fully functional as judged by β-galactosidase assays (data not shown). Upon CHP treatment, both mutant proteins were completely oxidized, as judged by the loss of monomeric protein and conversion to dimeric forms shown by nonreducing SDS-PAGE (Fig. 6B). Treatment with DTT prior to electrophoresis converted the proteins back to monomeric form (data not shown), consistent with the formation of one or more intersubunit disulfide bonds. Based on our in vitro analyses, we suspect that the upper, dimeric protein band has a single-disulfide cross-link between two OhrR monomers, whereas the lower, dimeric band is doubly cross-linked. We note that appearance of the singly cross-linked protein dimer band was not dependent on the addition of oxidant, although the intensity of this band was increased with CHP treatment. This suggests that some disulfide bond formation may occur even in untreated cells, although not to a level sufficient to lead to derepression (Fig. 6A).
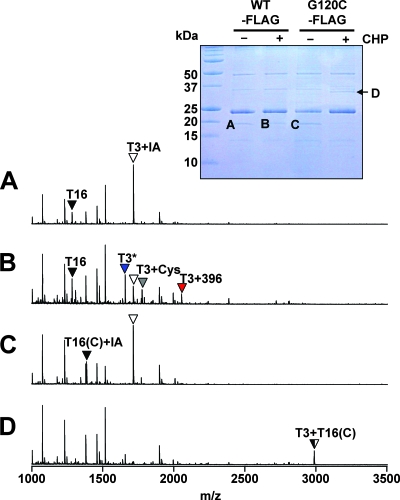
To identify the nature of the covalent changes in the oxidized G120C-FLAG protein, we immunopurified protein corresponding to monomeric OhrR and the CHP-oxidized proteins (Fig. 7, inset), treated the proteins with trypsin, and analyzed the resulting peptides using MALDI-TOF MS. As expected, in the wild-type control, the untreated OhrR was fully reduced, as judged by the alkylation of the C15-containing peptide by IA present in the cell disruption buffer (Fig. 7A). Upon oxidation, wild-type OhrR is converted into a mixture of S-thiolated products (T3+Cys and T3+396) and the protein sulfenamide (T3*) (Fig. 7B), as previously shown (10).
FIG. 7.
MALDI-TOF analysis of oxidation products for OhrR G120C in vivo. The inset shows the Coomassie-stained gel of immunoprecipitated OhrR-FLAG samples used for analysis. Cells were left untreated (−) or were treated with 100 μM CHP for 1 min (+) before immunoprecipitation of OhrR-FLAG proteins in a buffer containing IA. Proteins were resolved by nonreducing 16% Tris-Tricine SDS-PAGE. The Coomassie-stained band of ∼24 Da corresponds to the light chain of the anti-FLAG antibody. The indicated bands (A to D) were used for MALDI-TOF MS analysis after digestion with trypsin. The resulting MALDI-TOF spectra are shown as spectra A through D, with peaks labeled as described for Fig. 4. Upon oxidation, wild-type OhrR forms mixed disulfides with a 398-Da thiol (shown as T3+396; red arrow) and with cysteine (T3+Cys; gray arrow). In addition, some of the protein is converted to an IA-resistant form (presumably the sulfenamide), as indicated by the blue arrow (T3*).
In the G120C protein, the two cysteine-containing peptides (T3 and T16) were alkylated by IA. This indicates that both cysteine residues were reduced inside the cells, as expected (Fig. 7C). Analysis of the protein eluted from the CHP-dependent dimer band (band D) revealed a loss of these two IA-modified peptides and the appearance of a new peptide with a mass corresponding precisely to those of the disulfide-linked T3+T16 peptides (Fig. 7D). We did not detect any evidence of protein S thiolation under these conditions. These results indicate that both active sites have been oxidized and that each protein dimer (in band D) contains two intersubunit disulfide bonds. Together, these immunoblotting and MS studies establish that the B. subtilis G120C OhrR mutant can be inactivated by a mechanism analogous to that documented for X. campestris OhrR and that a significant fraction of the resulting oxidized protein contains two intersubunit disulfide bonds per dimer (Fig. 6B).
The 2-Cys G120C OhrR variant is resistant to protein overoxidation.
As shown previously (10) and confirmed here (Fig. 7B), OhrR forms mixed disulfides upon oxidation by CHP, with lesser amounts of sulfenamide-containing and overoxidized products (sulfinic and sulfonic acid derivatives). In contrast, the G120C variant forms intersubunit disulfide bonds both in vivo (Fig. 6B) and in vitro (Fig. 3). Thus, intersubunit disulfide bond formation is apparently much faster than the reactions to form mixed disulfides at the concentrations of low-molecular-weight thiols in the cell.
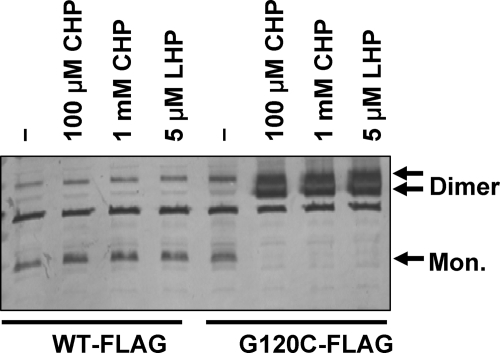
Next, we asked whether the presence of a second cysteine residue in the protein could protect OhrR against overoxidation. Protein overoxidation presumably occurs when the initially formed C15 sulfenate reacts with another molecule of oxidant prior to disulfide bond formation. We previously demonstrated that high concentrations of CHP (e.g., 1 mM) and even low amounts of LHP (5 μM) lead to significant amounts of OhrR overoxidation (17). In contrast, treatment of cells expressing the OhrR G120C variant with these same harsh oxidation conditions led to quantitative conversion to dimeric forms of OhrR (Fig. 8), consistent with the reversible oxidation noted for Fig. 2B. These results suggest that OhrR proteins that employ a 2-Cys mechanism are significantly protected against overoxidation.
FIG. 8.
The G120C OhrR protein can form an intersubunit disulfide bond under conditions that irreversibly overoxidize the wild-type protein. Immunoblot analysis of G120C-FLAG treated with strong oxidants Cells (0.45 ml) either were left untreated (−) or were treated with 100 μM CHP, 1 mM CHP, or 5 μM LHP (as indicated) for 2 min before sonication in the presence of 10% trichloroacetic acid. The precipitated proteins were separated by nonreducing SDS-PAGE and analyzed using anti-FLAG antibody to visualize the monomeric (Mon.) and disulfide-cross-linked (dimer) forms of OhrR. Note that in contrast to that shown in Fig. 6, this immunoblot was done using crude-cell extracts rather than immunoprecipitated samples.
DISCUSSION
Members of the OhrR family contain in their amino-terminal domain a cysteine residue that is absolutely conserved and is critical for organic hydroperoxide sensing. One or more additional cysteine residues are present in many OhrR orthologs (Fig. 1A), and these proteins are thought to employ a 2-Cys inactivation pathway in which oxidation results in intersubunit disulfide bond formation, as shown previously for X. campestris OhrR (11, 14). The results presented here demonstrate that the introduction of additional cysteine residues into the carboxy-terminal domain of the 1-Cys OhrR protein from B. subtilis converts this protein into a peroxide sensor that efficiently uses the 2-Cys pathway. Oxidation effectively inactivates the 2-Cys variants in vivo (Fig. 6A) and results in intersubunit disulfide bond formation, with the majority of the oxidized protein containing two disulfide linkages per dimer (Fig. 6B and 7D).
Analysis of oxidation products under a variety of reaction conditions provides insights into the possible advantages of the 2-Cys over the 1-Cys mechanism. The results presented here suggest that the introduced C120 residue efficiently captures the initially oxidized protein sulfenate as a disulfide even in the presence of 1 mM cysteine. Since the intracellular concentration of low-molecular-weight thiols in B. subtilis is estimated as between 0.2 and 0.5 mM (5, 12), the 2-Cys OhrR proteins may have an advantage relative to the wild-type (1-Cys) OhrR in preventing protein overoxidation. This expectation is supported by the finding that the G120C protein efficiently forms dimers under harsh oxidation conditions that lead to the overoxidation of wild-type protein (Fig. 8 and reference 17). Whether this advantage would also be relevant for cells that contain much higher levels of free thiols is not yet known, and the concentrations of glutathione in some bacterial species are estimated to be greater than 1 mM (1). Our results also suggest a possible role for the presence of a third cysteine residue in some OhrB orthologs, as is present in X. campestris OhrR. This cysteine residue may play a role in regenerating the active site thiolate, with the generation of an intrasubunit disulfide. Indeed, a C120-C124 intrasubunit disulfide was clearly visualized in the double mutant protein upon oxidation. Formation of this disulfide can replace an intersubunit with an intrasubunit disulfide, thereby regenerating a monomeric species (Fig. 3, bands N and Q) that now contains a reactive (alkylatable) C15 residue (Fig. 5N and Q). Little is yet known regarding the enzymatic pathways that might mediate the reduction of oxidized OhrR back to its fully reduced form. It is possible, for example, that an intrasubunit disulfide may be a preferred substrate for protein rereduction.
Given the apparent advantages of a 2-Cys over a 1-Cys reaction mechanism, particularly in an organism with a relatively low level of intracellular thiols, it is puzzling that B. subtilis and related species retain OhrR orthologs that function exclusively by a 1-Cys mechanism. Recent results demonstrate that the B. subtilis OhrR is overoxidized by strong oxidants such as LHP and therefore functions as a sacrificial repressor (17). Moreover, the ability of OhrR to be protected against overoxidation is also very sensitive to the levels of reduced thiols in the cytosol: oxidation of cellular thiols with diamide prior to treatment with CHP precludes S thiolation and leads to protein overoxidation (10). Thus, one possible advantage of the 1-Cys reaction pathway is that a more robust genetic response may result from the irreversible protein inactivation that accompanies exposure to harsh oxidizing conditions.
Acknowledgments
This work was supported by a grant from the National Science Foundation (MCB-0640616).
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54439-461. [DOI] [PubMed] [Google Scholar]
- 2.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 27811570-11578. [DOI] [PubMed] [Google Scholar]
- 3.Derre, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38335-347. [DOI] [PubMed] [Google Scholar]
- 4.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA 996690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gusarov, I., and E. Nudler. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. USA 10213855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong, M., M. Fuangthong, J. D. Helmann, and R. G. Brennan. 2005. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20131-141. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S. O., K. Merchant, R. Nudelman, W. F. Beyer, Jr., T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109383-396. [DOI] [PubMed] [Google Scholar]
- 8.Kullik, I., M. B. Toledano, L. A. Tartaglia, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 1771275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, C., S. M. Lee, P. Mukhopadhyay, S. J. Kim, S. C. Lee, W. S. Ahn, M. H. Yu, G. Storz, and S. E. Ryu. 2004. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 111179-1185. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. W., S. Soonsanga, and J. D. Helmann. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA 1048743-8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newberry, K. J., M. Fuangthong, W. Panmanee, S. Mongkolsuk, and R. G. Brennan. 2007. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28652-664. [DOI] [PubMed] [Google Scholar]
- 12.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1781990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paget, M. S., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 3791-121. [DOI] [PubMed] [Google Scholar]
- 14.Panmanee, W., P. Vattanaviboon, L. B. Poole, and S. Mongkolsuk. 2006. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 1881389-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seraphin, B., and S. Kandels-Lewis. 1996. An efficient PCR mutagenesis strategy without gel purification step that is amenable to automation. Nucleic Acids Res. 243276-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soonsanga, S., M. Fuangthong, and J. D. Helmann. 2007. Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J. Bacteriol. 1897069-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soonsanga, S., J.-W. Lee, and J. D. Helmann. 2008. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol. Microbiol. 68978-986. [DOI] [PubMed] [Google Scholar]
- 18.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2188-194. [DOI] [PubMed] [Google Scholar]
- 19.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 1834405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]