Abstract
The two recently characterized Streptococcus pneumoniae strains—R6Chi and R6Cho−—that have lost the unique auxotrophic requirement of this bacterial species for choline differ in their mechanisms of choline independence. In strain R6Chi the mechanism is caused by a point mutation in tacF, a gene that is part of the pneumococcal lic2 operon, which is essential for growth and survival of the bacteria. Cultures of lic2 mutants of the encapsulated strain D39Chi growing in choline-containing medium formed long chains, did not autolyze, had no choline in their cell wall, and were completely avirulent in the mouse intraperitoneal model. In contrast, while the Cho− strain carried a complete pneumococcal lic2 operon and had no mutations in the tacF gene, deletion of the entire lic2 operon had no effect on the growth or phenotype of strain Cho−. These observations suggest that the biochemical functions normally dependent on determinants of the pneumococcal lic2 operon may also be carried out in strain Cho− by a second set of genetic elements imported from Streptococcus oralis, the choline-independent streptococcal strain that served as the DNA donor in the heterologous transformation event that produced strain R6Cho−. The identification in R6Cho− of a large (20-kb) S. oralis DNA insert carrying both tacF and licD genes confirms this prediction and suggests that these heterologous elements may represent a “backup” system capable of catalyzing P-choline incorporation and export of teichoic acid chains under conditions in which the native lic2 operon is not functional.
The cell wall and membrane teichoic acid of the bacterial pathogen Streptococcus pneumoniae is unusual in that it contains in its structure phosphorylcholine residues, which are involved with a wide variety of physiological and ecological functions (5, 6). S. pneumoniae—as a species—is also unique in its dependence on an exogenous source of choline for growth (15).
Choline is taken up from the culture medium (or from the in vivo environment) and converted to CDP-choline by the sequential activity of proteins encoded by determinants organized into the lic1 operon (31). The protein products of licB, licA, and licC catalyze cellular uptake (5) and conversion of intracellular choline to phosphorylcholine in an ATP-dependent reaction (28) followed by the conversion of phosphorylcholine in a CTP-dependent reaction to CDP-choline (2, 14, 17), which is assumed to be the substrate of an additional enzyme(s) that catalyzes the incorporation of P-choline residues (one or two, depending on the particular S. pneumoniae strain) into the teichoic acid chains. The genetic determinants involved appear to be part of a second operon (lic2) adjacent to lic1 on the pneumococcal chromosome (11, 12, 31).
While the unique auxotrophic requirement for choline can be fulfilled by other structurally different amino alcohols (24, 27), the normal physiological properties of the bacterium require the trimethylamino group of choline. S. pneumoniae strains growing in medium in which choline has been replaced by ethanolamine show numerous abnormalities: they form long chains, they do not autolyze, and they are resistant to bacteriophage and cannot undergo genetic transformation (24). Interestingly, these are exactly the same abnormalities shown by three recently isolated choline-independent strains of S. pneumoniae when cultured in choline-free medium (4, 19, 30).
Two of these choline-independent strains were laboratory mutants isolated by an enrichment procedure in which the parental strain was serially passaged in a culture medium, the choline component of which was replaced by gradually decreasing concentrations of ethanolamine. This procedure eventually yielded mutants R6Chi (4) and JY2190 (30), which could grow in medium completely lacking the amino alcohol component. The mechanism of choline independence in JY2190 remains unknown at the present time. On the other hand, genetic and biochemical studies of R6Chi allowed the identification of a single G→T point mutation in spr1150—one of the genes of the lic2 operon—as the molecular basis of choline independence in this mutant. The gene spr1150 (renamed tacF) was proposed to encode a polysaccharide transmembrane transferase (“flippase”) that catalyzes transport of teichoic acid chains to the outer surface of the pneumococcal plasma membrane (4).
The mechanism of choline independence in the third extensively studied S. pneumoniae strain, R6Cho− (19), is not known. R6Cho− shared many of the physiological properties of R6Chi but had a more complex origin: it was isolated as the product of a heterologous genetic cross in which the recipient was S. pneumoniae strain R6 and the source of donor DNA was Streptococcus oralis, a streptococcal species that contains choline in its teichoic acid but has no auxotrophic requirement for it (19).
The purpose of the studies described here was to compare the status and essentiality of genes in the lic1 and lic2 operons in R6Chi and R6Cho−, in order to better understand the mechanism of the physiological abnormalities of these choline-free strains of pneumococci, particularly the dramatic reduction in their virulence-related properties (4, 9, 12).
MATERIALS AND METHODS
Strains and bacterial growth.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains of S. pneumoniae were grown in a casein-based semisynthetic medium (C+Y) and Cden medium (25) with 5 μg/ml choline or without choline at 37°C without aeration. Escherichia coli strains containing the pJDC9 plasmid (3) or its derivatives were grown in Luria-Bertani (LB) medium in the presence of 1 mg/ml erythromycin (Sigma). Strains of S. pneumoniae containing pJDC9 were grown in C+Y medium or Cden medium in the presence of 1 μg/ml erythromycin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Streptococcus pneumoniae strains | ||
| R6 | Penicillin-susceptible laboratory strain derived from R36A | RUa collection |
| R6Cho− | Choline-independent derivative of R6 | 19 |
| R6Chi | Spontaneous mutant of S. pneumoniae strain R6 carrying a G→T point mutation at nucleotide position 700 in spr1150 | 4 |
| R6Cho−spr1150ID | Insertion/duplication mutant of R6Cho−, with inactivated spr1150 | This study |
| R6Cho−ΔlicD1D2 | licD1-licD2 deletion mutant of R6Cho− | This study |
| R6Cho−Δlic2 | The lic2 operon deletion mutant of R6Cho− | This study |
| D39 | Type 2 capsular virulent S. pneumoniae strain | RU collection |
| D39Cho− | Choline-independent derivative of D39 | 12 |
| D39Cho−ΔlicD1D2 | licD1-licD2 deletion mutant of D39Cho− | This study |
| D39Cho−Δlic2 | The lic2 operon deletion mutant of D39Cho− | This study |
| D39Chi | Choline-independent derivative of D39 carrying G700T point mutation in the tacF gene | 4 |
| D39ChiΔlicD1D2 | licD1-licD2 deletion mutant of D39Chi | This study |
| Plasmids | ||
| pJDC9 | E. coli plasmid; Ermr | 3 |
| pR410 | E. coli plasmid; Kanr | 21 |
| pGEM3Z | E. coli plasmid; Ampr | 29 |
| pSPR1150ID | Plasmid pJDC9 carrying an internal DNA fragment of tacF amplified with spr1150IDF-RI and spr1150IDR-BamHI primers | This study |
| pΔD1D2DU3 | Plasmid pGEM3Z carrying an insert composed of upstream flank of the licD1 gene and downstream flank of the licD2 gene | This study |
| pΔD1D2DU31 | Plasmid pGEM3Z carrying an insert composed of upstream flank of the licD1 gene/kanamycin resistance cassette and downstream flank of the licD2 gene | This study |
| pΔLIC2 | Plasmid pGEM3Z carrying an insert composed of upstream flank of the spr1150 (tacF) gene/kanamycin resistance cassette and downstream flank of the licD2 gene | This study |
RU, Rockefeller University.
DNA manipulations.
All routine DNA manipulations were performed by using standard methods (18). PCR primers were synthesized at the custom primer facility of Invitrogen. Chromosomal DNA from strains was isolated as described earlier (13), and PCR products and DNA recovered after restriction endonuclease digestions were purified using the QIAquick PCR purification kit (Qiagen). Long-range PCR was performed using iProof High-Fidelity DNA polymerase (Bio-Rad Laboratories) according to the manufacturer's instructions. DNA sequencing was done at Genewiz (South Plainfield, NJ) and at the Nano + Bio Center, University of Kaiserslautern. Nucleotide sequences were analyzed by using DNAStar and CloneManager software.
Transcription of the lic2 operon.
The R6Cho− strain was grown in C+Y until the culture reached an optical density at 590 nm (OD590) of 0.4. At this time steady-state RNA was isolated using the Trizol reagent. A sample of 10 μg denatured RNA was loaded on a 1.5% formaldehyde denaturation agarose gel. The RNA from the gel was transferred to a Super Nitran membrane using the Turbo Blot system. Cross-linked membrane was next hybridized to the [α-32P]dCTP-labeled combined probe of licD1-licD2 genes. Hybridization was followed by washing of the membrane, and the transcript of the lic2 operon was detected by autoradiography.
Inactivation of tacF (spr1150) by insertion duplication in the background of R6Cho−.
An insertion duplication mutant of spr1150 was constructed in the genetic background of R6Cho− (Fig. 1). An internal 640-bp DNA element of tacF was PCR amplified from R6 genomic DNA using primers spr1150IDF-RI (Table 2) and spr1150IDR-BamHI (Table 2). The product was digested with EcoRI and BamHI and ligated to pJDC9, digested with the same restriction enzymes. The ligation mixture was transferred into competent E. coli cells and plated on LB agar containing 1 mg/ml erythromycin. The resulting plasmid, pSPR1150ID, contained the desired insert (tacF) as determined by restriction analysis and DNA sequencing. After transformation of S. pneumoniae R6Cho−, transformants were selected on blood agar plates (BAP) supplemented with 1 μg/ml erythromycin. One such erythromycin-resistant transformant was named R6Cho−spr1150ID. Genomic DNA from R6Cho−spr1150ID was used to confirm the insertion duplication event by PCR amplification and sequencing of the PCR product from both ends.
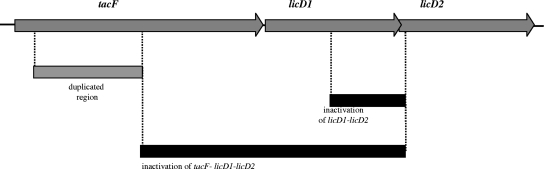
FIG. 1.
Schematic representation of the S. pneumoniae R6 lic2 operon and inactivation constructs. The genetic organization of the tacF, licD1, and licD2 genes forming the lic2 operon is presented. Black bars show the extent of deletions in strains R6Cho−ΔlicD1D2 and R6Cho−Δlic2. The indicated parts of the genes were replaced by a kanamycin resistance gene. The internal region of tacF used to inactivate the gene by insertion-duplication in strain R6Cho−spr1150ID is shown by a gray bar. The same constructs were applied to inactivate genes in a different genetic background.
TABLE 2.
Primers used in this study
| Primer | Sequence | Reference |
|---|---|---|
| spr1150IDF-RI | CCGAATTCACTATGGTTACTTCAACTCAGTC | This study |
| spr1150IDR-BamHI | CCGGATCCTCGCTGAGCTATGGTATAGTAAC | This study |
| delD2ForPstI | CCCTGCAGGACTATATTGATGAGACTTGTAAG | This study |
| delD2RevHindIII | CGATAAGCTTTAATGCTATGACTATACCACTC | This study |
| PRO16 | CCTGAATTCTTAAAATGAAACAACTAACCGT | 21 |
| PRO17 | GAAGGGATCCTCAAAGCGATCTATAGGGAAAAT | 21 |
| DAM301 | CGCGCAAGCTGGGGATCCG | 21 |
| DAM347 | CCGAATTCTAGGTACTAAAACAATTCATCCAGTAA | 21 |
| Uo5_pheA_for | GCCTATTCATCAGCAGTTGATGGTGGTTCC | This study |
| Uo5_hom_PCR_down | CTTGTCACCCTCTTTGCCATCTTGAAGGATTTGC | This study |
Inactivation of licD1-licD2 in R6Cho−, D39Cho−, and D39Chi strains.
The licD1-licD2 genes were inactivated in the R6Cho− genetic background with the help of plasmid pΔD1D2DU31 containing the kanamycin resistance gene aphIII as well as the flanking regions for deletion. The 3′ end of licD1 and the 5′ end of licD2 including the translation initiation region were deleted, leading to a complete loss of expression of functional proteins. The genetic organization of licD1 and licD2 and the extent of the deletion are shown in Fig. 1.
A 657-bp downstream fragment containing ′licD2 was amplified by PCR from R6 genomic DNA using the primers delD2ForPstI and delD2RevHindIII (Table 2). The purified PCR product and plasmid pGEM3Z (29) were digested with PstI and HindIII, ligated, and transformed into competent E. coli cells. Transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin, 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and 8 μg/ml IPTG (isopropyl-β-d-thiogalactopyranoside). White colonies were processed for plasmid isolation and restriction analysis. A recombinant plasmid with the desired insertion was used to clone an upstream fragment harboring licD1′ for homologous recombination. The upstream fragment was PCR amplified from R6 genomic DNA using primers PRO17 and PRO16 (31). The PCR product and recombinant (′licD2) vector were digested with EcoRI-BamHI, purified, ligated, and transformed into E. coli. Transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin. Plasmid pΔD1D2DU3 was confirmed to release a 1.2-kb insert when digested with EcoRI-HindIII corresponding to the combined upstream (licD1′) and downstream (′licD2) fragments.
Plasmid pΔD1D2DU3 (Table 1) then served to clone the kanamycin resistance gene aphIII into the XbaI site located between the upstream and downstream recombination fragments. The resistance gene was PCR amplified from plasmid pR410 (Table 1) using primers DAM301 and DAM347 (Table 2). Plasmid pΔD1D2DU3 was digested with XbaI and end filled with E. coli DNA polymerase I (Klenow fragment). Subsequently, the blunt-end kanamycin cassette was ligated to pΔD1D2DU3 and E. coli cells were transformed with the ligation mixture. Transformants conferring kanamycin resistance were selected on LB agar supplemented with 50 μg/ml kanamycin. Plasmid pΔD1D2DU31 was confirmed to carry the kanamycin resistance gene aphIII between licD1′ and ′licD2 (Fig. 1). This 2.1-kb insert was PCR amplified with primer pair PRO16 and delD2RevHindIII (Table 2), and the gel-purified amplicon served as donor DNA to transform competent R6Cho− cells. Kanamycin-resistant transformants were selected on BAP supplemented with 400 μg/ml kanamycin and confirmed to have the constructed insertion/deletion in licD1-licD2 by both PCR analysis and Southern hybridization experiments. The resulting strain was designated R6Cho−ΔlicD1D2. In order to inactivate licD1 and licD2 in an encapsulated S. pneumoniae strain, genomic DNA isolated from R6Cho−ΔlicD1D2 was used to transform competent cells of strain D39Cho− (expressing capsular polysaccharide 2) to generate transformant D39Cho−ΔlicD1D2, which was confirmed to carry the insertion/deletion in the licD1-licD2 genes.
Similarly, D39ChiΔlicD1D2 was constructed by using genomic DNA isolated from R6Cho−ΔlicD1D2 to transform competent D39Chi cells. Transformants were selected on blood agar-kanamycin plates and were confirmed for deletion of licD1-licD2.
Inactivation of the entire lic2 operon.
The lic2 operon was deleted in the R6Cho− genetic background using plasmid pΔD1D2DU31 as a vector. In order to achieve this, the upstream recombination fragment (licD1′) had to be replaced by a tacF fragment (Fig. 1). This fragment was PCR amplified on R6 genomic DNA with the use of spr1150IDF-RI and spr1150IDR-BamHI primer pairs (Table 2) and Pfu DNA polymerase. The PCR product was digested with EcoRI-BamHI and ligated to the large fragment of EcoRI-BamHI-digested pΔD1D2DU31. Competent E. coli cells were then transformed with the ligation mixture, and transformants were selected on LB agar supplied with the desired amounts of ampicillin and kanamycin. The resulting plasmid was designated pΔLIC2. The insert carried on pΔLIC2 (tacF′-aphIII-′licD2) was PCR amplified with Pfu DNA polymerase and spr1150IDF-RI and delD2RevHindIII primers. Transformation was performed by using the gel-purified insert as donor and R6Cho− as recipient. Transformants (R6Cho−Δlic2) were selected on BAP supplemented with 400 μg/ml kanamycin and were confirmed to carry the insertion/deletion by PCR analysis, DNA sequencing, and Southern hybridization experiments. R6Cho−Δlic2 genomic DNA was then transferred to D39Cho−, and transformants were selected on BAP containing kanamycin. The transformant D39Cho−Δlic2 was confirmed to carry the insertion/deletion by PCR analysis and Southern hybridization experiments.
Detection of choline in purified cell walls.
Choline incorporation into the cell wall was determined in R6Cho−, R6Cho−ΔlicD1D2, R6Cho−Δlic2, D39Chi, and D39ChiΔlicD1D2. All strains were grown in Cden medium supplemented with 80 μCi and 5 μg/ml of [3H]choline to an OD590 of 0.6. Cell walls were purified (19), and 100 μg purified cell walls was digested with muramidase and processed for quantification of radioactive counts.
Intraperitoneal (i.p.) mouse virulence model.
All strains with the D39Cho− and D39Chi backgrounds were grown in C+Y medium to an OD590 of 0.6. Cells were resuspended in pyrogen-free 0.9% sodium chloride solution, 10-fold serial dilutions were prepared, and groups of 8-week-old female CD1 mice (10 mice per bacterial concentration) were injected in the peritoneal cavity with 0.5 ml of the inoculum containing 103 to 107 CFU. Mouse survival was monitored.
Intranasal colonization of mice.
Strains D39Cho−, D39Cho−ΔlicD1D2, D39Cho−Δlic2, D39Chi, and D39ChiΔlicD1D2 were grown to an OD590 of 0.6 and centrifuged to pellet bacterial cells. Bacteria were resuspended in pyrogen-free saline (0.9% NaCl) to obtain a bacterial concentration of 108 CFU/ml. Groups of 8-week-old CD1 female mice (10 per strain) were anesthetized by i.p. injection of 75 μl of a xylazine and ketamine mixture (12). Suspensions of bacteria (10 μl) were inoculated through the nostrils with a 10-μl blunt-ended Hamilton syringe. Mice were sacrificed 48 h after inoculation by i.p. injection of 100 μl pentobarbital sodium (Nembutal). Bacteria colonizing the nasopharynx were collected by expelling 50 μl saline solution through the trachea. Viable counts were determined on BAP supplemented with 5 μg/ml gentamicin.
RESULTS AND DISCUSSION
The status of the lic1 operon in R6Chi and R6Cho−.
Confirmation of the existence of a lic1 operon in R6Cho− (12) and R6Chi (4) was provided by isolation and characterization of steady-state mRNA which showed that the five genes spr1148, spr1149, licA, licB, and licC formed a polycistronic message the molecular size of which corresponded to the combined size of these genes (12).
Genes of the lic1 operon were shown to be essential for growth and survival of all isolates of S. pneumoniae except in strains R6Cho− and R6Chi. Strain R6Cho− and R6Chi with inactivated licA, licB, or licC continued to grow in choline-containing growth medium in the form of long autolysis-resistant chains of cells. Formation of long autolysis-resistant chains was also the phenotype of R6Chi and R6Cho−, which carried functional genes in the lic1 operon but were growing in choline-free medium. In vitro, addition of choline to the medium of such cultures resulted in a rapid breakup of chains and return of the autolysis-prone phenotype. This choline-dependent reversion was blocked in mutants of the lic1 operon in both R6Cho− and R6Chi (4, 12).
In vivo, inactivation of the same lic1 operon genes in derivatives of both R6Cho− and R6Chi expressing a type 2 capsular polysaccharide caused a drastic reduction in virulence presumably for the same reason, i.e., by preventing utilization of choline and preserving a choline-free cell surface (4, 12).
The status of the lic2 operon in R6Chi and R6Cho−.
CDP-choline, the final product of proteins encoded by genes of the lic1 operon, was proposed to serve as the substrate of one or two P-choline transferases encoded by the genes licD1 and licD2 (31)—hypothetical choline transferases which catalyze the incorporation of P-choline into teichoic acid precursors or teichoic acid polymers. The genetic organization of licD1, licD2, and a third gene, spr1150, on the chromosomes of strains R6 and TIGR4 suggested that these genes may form a single lic2 operon. In order to confirm this, we performed steady-state mRNA analysis on extracts prepared from strain R6Cho− grown in a choline-containing medium. The RNA preparation was hybridized to an [α-32P]dCTP product prepared from the PCR product of licD1-licD2. Northern blot analysis demonstrated the presence of a single 3.2-kb polycistronic RNA which corresponds to the combined sizes of the genes spr1150 (renamed tacF), licD1, and licD2 (data not shown).
The structure and function of tacF in the choline-independent strain R6Cho−.
The mechanism of choline independence in mutant R6Chi is a single G→T point mutation at base position 700 in tacF, the first gene in the lic2 operon (4). The protein product of tacF was shown to have structural features characteristic of polysaccharide transmembrane transferases (flippase). It was proposed that the flippase of parental-type pneumococci can transport lipid-linked teichoic acid units to the outer surface of the pneumococcal plasma membrane only if the teichoic acid chains carry P-choline residues. This structural requirement appears to be lost in the tacF G700T point mutant, which can also transport choline-free teichoic acid chains and thus provides the mutant R6Chi with a choline-independent phenotype (4).
We sought to test whether R6Cho− also carried the G700T transversion within the tacF gene. Three types of experiments were performed.
In the first experiment tacF along with its promoter was PCR amplified with Pfu DNA polymerase on R6Cho− genomic DNA and processed to decipher the nucleotide sequence. Results of DNA sequencing of the tacF gene from R6 and R6Cho− revealed that the R6Cho− sequence was 100% identical to that of the parental strain R6, i.e., strain R6Cho− did not carry the G700T transversion.
In a second experiment, the tacF gene of R6Cho− along with its promoter was cloned in pMSP3535 (1) and transformed into strain R6. If overexpression of tacF of R6Cho− were responsible for the choline independence of this strain, then R6 transformants carrying copies of plasmid-borne tacF from R6Cho− should become choline independent and grow in a choline-free environment. Erythromycin-resistant transformants were tested for growth in a choline-free environment. None of the transformants obtained could grow in choline-free Cden medium.
Finally, we inactivated tacF in strain R6Cho− by an insertion duplication strategy (Fig. 1) using the set of insertion duplication primers described in Materials and Methods. Transformant R6Cho−spr1150ID remained fully viable and continued to exhibit physiology similar to that of parental strain R6Cho− when grown in choline-containing medium. Attempts to inactivate spr1150 (renamed tacF) with the same strategy in the background of R6Chi were unsuccessful, indicating the essentiality of this gene. The essentiality of tacF (spr1150) in the parental strain R6 has already been documented (20).
Inactivation of the genes of the lic2 operon: impact on the growth and phenotype of R6Cho− and R6Chi.
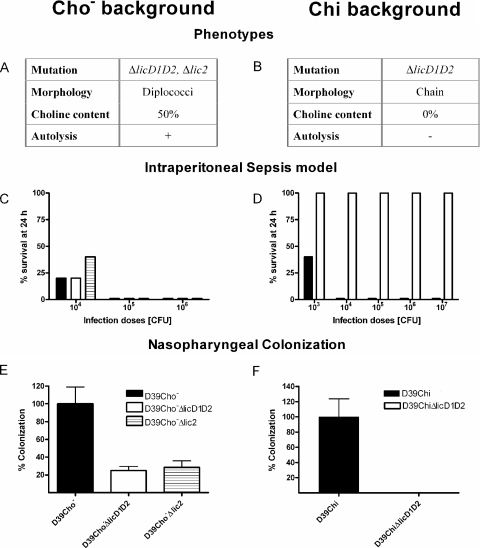
Deletion mutants of licD1 and licD2 were constructed in the background of R6Cho− (see Materials and Methods; also Fig. 1). Cultures of the deletion mutant R6Cho−ΔlicD1D2 grew in choline-containing medium in the form of diplococci or short chains, autolyzed in the stationary phase, and retained sensitivity to deoxycholate-induced lysis (Fig. 2A).
FIG. 2.
Effects of mutations in the lic2 operon on the virulence and phenotypes of the two choline-independent strains D39Chi and D39Cho−. Strains D39Chi, D39ChiΔlicD1D2, D39Cho−, D39Cho−ΔlicD1D2, and D39Cho−Δlic2 were tested and compared in the mouse nasopharyngeal colonization assay (E and F) and in the mouse i.p. virulence assay (C and D). Phenotypes of strains R6Chi (B) and R6Cho− (A) carrying mutations in the lic2 operon. Strains were grown in choline-containing medium.
Next we proceeded to delete the entire lic2 operon—including not only licD1 and licD2 but tacF (spr1150) as well (Fig. 1)—to yield strain R6Cho−Δlic2. The strain R6Cho−Δlic2 has remained fully viable and retained the normal physiology: growth in the form of autolysis-prone pneumococci with diplococcal morphology, indistinguishable from the phenotype of the parental strain R6Cho− when grown in choline-containing medium (Fig. 2A).
In sharp contrast to the licD1D2 deletion mutant of R6Cho−, inactivation of the same genes in R6Chi (R6ChiΔlicD1D2) produced a striking and abnormal phenotype: the mutant cells grew in long chains and became completely resistant to autolysis even when grown in choline-containing medium (Fig. 2B). Thus, in R6Chi, the inactivation of licD1 and licD2—putative P-choline transferases—produced the same phenotype as did inactivation of genes in the lic1 operon (12). These observations strongly suggest that this deletion mutant must produce a choline-free teichoic acid even when grown in medium in which choline is available. Chemical determination of the choline content of such mutants has confirmed this prediction (see below).
In conclusion, inactivation of licD1D2 caused completely different phenotypes in the two choline-independent strains, strongly suggesting that the mechanism of choline independency should be different in the two strains.
Choline content of the cell walls in lic2 mutants of R6Cho− and R6Chi.
We determined the choline content of cell walls purified from strains R6Cho− and R6Cho−ΔlicD1D2 (and R6Cho−Δlic2) and from R6Chi and R6ChiΔlicD1D2—each culture growing in choline-containing medium which was also supplemented with [3H]choline. An aliquot of 100 μg purified cell walls was digested with muramidase and processed to determine choline content as described in Materials and Methods. Results shown in Fig. 2A indicate that the cell walls of the R6Cho−ΔlicD1D2 mutants had reduced choline content, corresponding to about 50% of the choline content of the parental strain R6 grown under the same conditions. Apparently, this reduced choline content in the cell walls was sufficient for the cells to maintain a normal (diplococcal and autolysis-prone) phenotype (Fig. 2A).
In contrast, deletion of licD1-licD2 in the Chi mutant resulted in a completely choline-free cell wall (Fig. 2B), which paralleled the abnormal (chain-forming and autolysis-defective) phenotype of the bacteria.
Impact of inactivation of genes in the lic2 operon on the virulence of D39Cho− and D39Chi.
The availability of strains D39Cho−ΔlicD1D2 and D39Cho−Δlic2—containing just 50% of the normal amount of surface-bound choline—gave us a tool to determine the impact of cell wall choline content on virulence. Cultures of D39Cho−, D39Cho−ΔlicD1D2, D39Cho−Δlic2, D39Chi, and D39ChiΔlicD1D2 were grown in choline-containing medium to an OD590 of 0.6. Bacteria were pelleted, washed, and resuspended in pyrogen-free saline; various bacterial concentrations were inoculated into the peritoneal cavity of 8-week-old CD1 female mice; and the survival rate of the animals was determined. As expected, D39Cho− and D39Chi showed similar high degrees of virulence (Fig. 2C and D).
Interestingly, deletion of licD1-licD2 as well as of the entire lic2 operon in D39Cho−, which caused a reduction of the choline content of cell walls to about half of that of the parent strain D39Cho−, had only a minor effect on the virulence potential of this strain (Fig. 2C).
In contrast, D39ChiΔlicD1D2, in which the cell wall choline content was reduced to zero, exhibited a dramatic drop in virulence: mice inoculated with 107 CFU of D39ChiΔlicD1D2 showed 100% survival even after 24 h whereas mice inoculated with 104 CFU of D39Chi died within 24 h (Fig. 2D).
Impact of inactivation of genes of the lic2 operon on the colonizing capacity of D39Cho− and D39Chi.
Strains D39Cho−, D39Cho−ΔlicD1D2, D39Cho−Δlic2, D39Chi, and D39ChiΔlicD1D2 were grown in choline-containing medium to an OD590 of 0.6. Suspensions of bacteria (10 μl) at a concentration of 108 CFU/ml were inoculated through the nostrils, and mice were sacrificed after 48 h as described in the work of Kharat and Tomasz (12). Colonizing bacteria were enumerated on BAP supplemented with 5 μg/ml gentamicin.
While the approximately 50% reduction in the amount of surface-bound choline in strains D39Cho−ΔlicD1D2 and D39Cho−Δlic2 had little effect on virulence in the i.p. assay (Fig. 2C), the colonizing capacity of these mutants was reduced significantly to 25% and 28% of the control levels, respectively (Fig. 2E).
The completely choline-free D39ChiΔlicD1D2 strain failed to colonize (Fig. 2F). This complete loss of colonizing capacity was comparable to the suppression of colonizing capacity already described for lic1 mutants of both D39Cho− (12) and D39Chi (4), neither one of which had any detectable choline residues in their cell walls.
R6Cho− has a large deletion in the vicinity of the spr1226 gene.
Earlier studies showed that, during the process of the genetic cross that led to the acquisition of choline independence, the transformant R6Cho− lost a SmaI recognition site (19) and the lost SmaI recognition sequence was mapped within spr1226 (12). Interestingly, spr1225, the gene located upstream of spr1226 in the R6 genome, is annotated as a licD1 paralog and the same gene in the TIGR4 genome is annotated as licD3 (11, 23). The spr1225 gene is not an exact copy of licD1 or licD2 but bears significant homology to licD1 of R6 and TIGR4. The function of the spr1225 gene is not known. To distinguish this gene clearly from licD1 in the lic2 locus, we adopted the TIGR4 annotation and designated it licD3.
We sought to test the status of spr1225 in R6Cho−. Genomic DNA obtained from R6 and R6Cho− was digested with various restriction endonucleases and subjected to Southern blot analysis using an [α-32P]dCTP-labeled probe. No hybridization signals were detected with R6Cho− DNA (data not shown), indicating that licD3 has been deleted in R6Cho−.
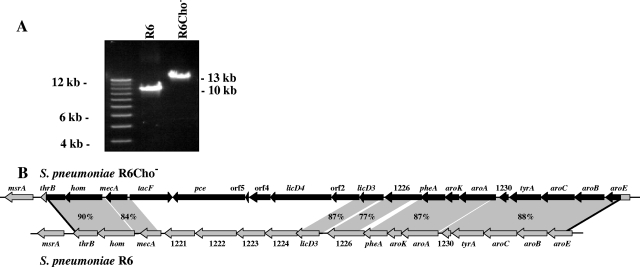
Since both spr1226 and licD3 (spr1225) were deleted in the R6Cho− strain, we sought to investigate the extent of the deletion in R6Cho− by designing probes for pheA and aroK upstream of spr1226 and for spr1224, hom, and msrA downstream of licD3 (Fig. 3B). In these Southern blot experiments no signals were obtained from R6Cho− DNA with spr1224 and hom probes and only a weak signal with pheA (data not shown). Clear hybridization was detected with msrA and aroK probes. These results demonstrate that a region of at least 10 kb has been deleted in the licD3 region of strain R6Cho−. In addition, weak hybridization signals may indicate replacement of R6 genes by similar but not identical S. oralis homologues.
FIG. 3.
(A) PCR analysis of the licD3 locus in S. pneumoniae R6 and the choline-independent derivative R6Cho−. Long-range PCR products using primers located in hom and pheA (see panel B) with genomic DNA of R6 and R6Cho− are shown on a 1% agarose gel. (B) Schematic representation of the licD3 locus in S. pneumoniae R6 and the choline-independent derivative R6Cho−. The genomic organization of the licD3 region of S. pneumoniae R6 (bottom) is shown along with the same region of S. pneumoniae R6Cho− after integration of S. oralis ATCC 35037 DNA (top). Endogenous R6 genes are shown by gray arrows and are indicated by gene names or “spr” numbers (11, 12, 31). Acquired S. oralis genes are shown by black arrows. The sites of recombination are indicated by black lines. Replacement of S. pneumoniae R6 DNA by S. oralis ATCC 35037 DNA occurred between thrB (position 1.214.973 in the R6 genome) and aroE (position 1.231.893). The degree of identity (percent) between S. pneumoniae and S. oralis genes is indicated. Some of the gene products, whose genes showed no significant similarity to R6, could be identified by their protein similarity. Designations orf2, orf4, and orf5 were given according to their positions in the presumed licD3 operon of S. oralis. Identities of R6 genes spr1221 to spr1224 to S. oralis genes could not be assessed because the whole-genome sequence of S. oralis ATCC 35037 is not available. S. oralis homologues of R6 genes spr1226 and spr1230 were given the same number to highlight similarity. They are not likely to occupy the same position in the S. oralis genome. The S. oralis nucleotide sequence is available from GenBank (accession number EU675999).
Identification of the inserted S. oralis DNA in strain R6Cho−.
In the course of the Southern blotting experiments, a preliminary genome sequence of strain S. oralis Uo5 (16) became available. The genomic region of this strain corresponding to the observed deletion in R6Cho− suggested that the genes that were not detected in the Southern blot experiments could have been replaced by a large S. oralis region, which included lic genes. The sequences surrounding this S. oralis locus appear to be similar enough to allow its integration into the pneumococcal genome by homologous recombination. In order to document this large replacement in strain R6Cho−, long-range PCR experiments were performed using primers located inside a number of genes in this genomic region. Primers were designed to match genes of both strains, R6 and S. oralis Uo5, which should improve their chances of also matching DNA from S. oralis ATCC 35037, the strain used to produce strain R6Cho−. Several PCR products were obtained by this approach, all indicating that a large replacement of R6 DNA by S. oralis DNA occurred. As an example, the 13-kb product amplified from R6Cho− DNA with primers located in hom and pheA is shown in Fig. 3. The corresponding fragment of R6 is only 10 kb in size (Fig. 3A).
The nucleotide sequence of this 13-kb PCR product revealed that genes implicated in choline metabolism were integrated in R6Cho−, replacing endogenous R6 genes (Fig. 3B). A new copy of licD3 is found right next to the spr1226 homologue followed by a gene encoding a putative glycosyltransferase (orf2). A second licD gene, tentatively designated licD4; genes (orf4 and orf5) for two hypothetical proteins; and the phosphocholine esterase gene pce (26) are additionally present in this locus. The LicD4 protein has a large extension of about 450 amino acids at its N terminus, which is not found in other LicD proteins. A tacF homologue encoding teichoic acid flippase is found downstream of pce and on the opposite strand (Fig. 3B).
The DNA sequence of the hom-pheA long-range PCR fragment of R6Cho− also revealed that the DNA is completely of S. oralis origin. Therefore, integration of S. oralis DNA occurred beyond the borders of this fragment. DNA sequencing by primer walking eventually identified the site of crossing over within thrB and aroE (Fig. 3B). Thus, a large 20,249-bp fragment of S. oralis DNA replaced the 16,920 bp of the R6 genome during the transformation event leading to the loss of choline auxotrophy—a property unique to the species S. pneumoniae.
The mechanisms of choline independence in R6Chi and R6Cho−.
The experimental results described in this communication indicate that the mechanisms of choline independence in R6Chi and R6Cho− are different in several respects.
R6Chi carries a mutated form of the first gene of the lic2 operon, tacF, and this mutation appears to be the molecular basis of the choline-independent phenotype. A functional lic2 operon remains essential for the survival and the choline-independent phenotype of this strain.
R6Cho− also appears to have retained the pneumococcal lic2 operon as indicated by the capacity of the strain to produce a polycistronic message that corresponds precisely to the size of the pneumococcal lic2 operon. While this pneumococcal operon is no longer essential for the survival of bacteria, it may remain functional under some conditions of growth.
The survival and physiological properties of R6Cho− derivatives from which the entire lic2 operon was deleted clearly indicate that genes of the lic2 operon play no roles in the choline-independent phenotype of this strain. Therefore, one could predict that the acquisition of genetic elements from the heterologous S. oralis strain included a functional equivalent of tacF and a second gene that could replace the phosphorylcholine transferase function of the pneumococcal licD1. The nucleotide sequence of the S. oralis insertion in R6Cho− is fully consistent with this prediction, as it revealed homologues of tacF and licD (Fig. 3B).
The most likely candidate to carry out the phosphorylcholine transferase reaction in strain Cho− is the protein encoded by the imported S. oralis gene licD4. Its C-terminal domain is most similar to the LicD1 and LicD2 proteins of S. pneumoniae, showing 35% and 32% identical residues, respectively. Another LicD domain is found in the gene product of orf4, but similarity is rather limited and incomplete. The LicD3 protein is also less likely to be involved in phosphorylcholine transfer, since the very similar LicD3 of R6 (93% identity) plays no role in this process. This is confirmed by our finding that strain D39ChiΔlicD1D2 is free of surface-bound choline despite the fully functional licD3 gene. Gene inactivation studies will be needed to unambiguously identify the phosphorylcholine transferase(s) encoded in this region.
The second component expected to be present in the heterologous S. oralis DNA is tacF, encoding the teichoic acid flippase. While the endogenous R6 gene is part of the lic2 operon located about 60 kb from the licD3 locus, tacF of S. oralis is located immediately downstream of the presumptive licD3 operon (Fig. 3B) and appears to be a single transcriptional unit. In contrast to TacF of R6, S. oralis flippase is able to export teichoic acids with or without attached choline phosphoryl groups as indicated by the ability of S. oralis to grow in choline-containing and choline-free medium as well (10). Both enzymes have 47% residues in common.
In addition to tacF and licD genes, a gene (orf2) for a glycosyltransferase, a small gene (orf5) encoding a protein of unknown function, and pce specifying phosphocholine esterase (26) are also contained in the acquired S. oralis DNA. Interestingly, the glycosyltransferase of orf2 is similar (42% identical amino acids) to that of spr1223, which was lost upon S. oralis DNA insertion (Fig. 3B). Since pce of R6 is located outside the licD3 region, R6Cho− is equipped with two phosphocholine esterases with 45% identical residues.
Both choline-independent mutants, R6Cho− and R6Chi, share the common physiological abnormalities of chain formation and defective autolysis when grown in choline-free medium. These phenotypes are understandable in terms of the known structural requirements of two pneumococcal enzymes, LytB and LytA, both of which require choline residues in the cell wall for their activity (7, 8, 22). However, we found that 50% surface-bound choline—as seen in the lic2 deletion mutants of R6Cho−—was still sufficient to guarantee a functioning autolytic system, allow complete daughter cell separation, and ensure the wild-type diplococcal morphology of the bacteria.
Another surprising—and novel—observation described in this report was the drastic and selective reduction in the colonizing capacity of strains D39Cho−ΔlicD1D2 and D39Cho−Δlic2 carrying only a single choline residue per teichoic acid chain, which was not accompanied by a comparable reduction in the level of virulence of these strains as measured in the i.p. mouse model of disease. These findings suggest that different stages of pneumococcal pathogenesis may have differential sensitivities to the number of choline residues available at the cell surface.
Acknowledgments
Partial support for these studies was provided by the Irene Diamond Foundation (to Alexander Tomasz). Florian Gehre was supported by a grant from the American Austrian Foundation. Work at the University of Kaiserslautern was sponsored in part by the EU program Intafar (LSHM-CT-2004-51238) and by the Stiftung Rheinland-Pfalz für Innovation (D. Denapaite).
We thank the Nano + Bio Center (NBC), University of Kaiserslautern, for DNA sequencing and bioinformatic support.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44183-190. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, H. A., and C. Kent. 2001. The CTP:phosphocholine cytidylyltransferase encoded by the licC gene of Streptococcus pneumoniae: cloning, expression, purification, and characterization. Biochim. Biophys. Acta 153485-95. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64155-164. [DOI] [PubMed] [Google Scholar]
- 4.Damjanovic, M., A. S. Kharat, A. Eberhardt, A. Tomasz, and W. Vollmer. 2007. The essential tacF gene is responsible for the choline-dependent growth phenotype of Streptococcus pneumoniae. J. Bacteriol. 1897105-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer, W. 2000. Phosphocholine of pneumococcal teichoic acids: role in bacterial physiology and pneumococcal infection. Res. Microbiol. 151421-427. [DOI] [PubMed] [Google Scholar]
- 6.Fischer, W., T. Behr, R. Hartmann, J. Peter-Katalinic, and H. Egge. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide). Eur. J. Biochem. 215851-857. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 311275-1281. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, P., M. Paz Gonzalez, E. Garcia, J. L. Garcia, and R. Lopez. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33128-138. [DOI] [PubMed] [Google Scholar]
- 9.Gehre, F., S. Leib, D. Grandgirad, J. Kummer, A. Buhlmann, F. Simon, R. Gaumann, A. Kharat, M. Tauber, and A. Tomasz. 2008. Essential role of choline for pneumococcal virulence in an experimental model of meningitis. J. Intern. Med. 264143-154. [DOI] [PubMed] [Google Scholar]
- 10.Horne, D. S., and A. Tomasz. 1993. Possible role of a choline-containing teichoic acid in the maintenance of normal cell shape and physiology in Streptococcus oralis. J. Bacteriol. 1751717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharat, A. S., and A. Tomasz. 2006. Drastic reduction in the virulence of Streptococcus pneumoniae expressing type 2 capsular polysaccharide but lacking choline residues in the cell wall. Mol. Microbiol. 6093-107. [DOI] [PubMed] [Google Scholar]
- 13.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 712758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak, B. Y., Y. M. Zhang, M. Yun, R. J. Heath, C. O. Rock, S. Jackowski, and H. W. Park. 2002. Structure and mechanism of CTP:phosphocholine cytidylyltransferase (LicC) from Streptococcus pneumoniae. J. Biol. Chem. 2774343-4350. [DOI] [PubMed] [Google Scholar]
- 15.Rane, L., and Y. Subbarow. 1940. Nutritional requirements of the pneumococcus: I. Growth factors for types I, II, V, VII, VIII. J. Bacteriol. 40695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichmann, P., A. Konig, J. Linares, F. Alcaide, F. C. Tenover, L. McDougal, S. Swidsinski, and R. Hakenbeck. 1997. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J. Infect. Dis. 1761001-1012. [DOI] [PubMed] [Google Scholar]
- 17.Rock, C. O., R. J. Heath, H. W. Park, and S. Jackowski. 2001. The licC gene of Streptococcus pneumoniae encodes a CTP:phosphocholine cytidylyltransferase. J. Bacteriol. 1834927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. R. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Severin, A., D. Horne, and A. Tomasz. 1997. Autolysis and cell wall degradation in a choline-independent strain of Streptococcus pneumoniae. Microb. Drug Resist. 3391-400. [DOI] [PubMed] [Google Scholar]
- 20.Song, J. H., K. S. Ko, J. Y. Lee, J. Y. Baek, W. S. Oh, H. S. Yoon, J. Y. Jeong, and J. Chun. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19365-374. [PubMed] [Google Scholar]
- 21.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 675190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swiatlo, E., L. McDaniel, and D. Briles. 2004. Choline binding proteins, p. 49-60. In E. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 23.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 24.Tomasz, A. 1968. Biological consequences of the replacement of choline by ethanolamine in the cell wall of Pneumococcus: chanin formation, loss of transformability, and loss of autolysis. Proc. Natl. Acad. Sci. USA 5986-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomasz, A. 1964. Studies on the competence (for genetic transformation) of Diplococcus pneumoniae in a synthetic medium, abstr. G87. Abstr. 64th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 26.Vollmer, W., and A. Tomasz. 2001. Identification of the teichoic acid phosphorylcholine esterase in Streptococcus pneumoniae. Mol. Microbiol. 391610-1622. [DOI] [PubMed] [Google Scholar]
- 27.Ware, D., J. Watt, and E. Swiatlo. 2005. Utilization of putrescine by Streptococcus pneumoniae during growth in choline-limited medium. J. Microbiol. 43398-405. [PubMed] [Google Scholar]
- 28.Whiting, G. C., and S. H. Gillespie. 1996. Incorporation of choline into Streptococcus pneumoniae cell wall antigens: evidence for choline kinase activity. FEMS Microbiol. Lett. 138141-145. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 30.Yother, J., K. Leopold, J. White, and W. Fischer. 1998. Generation and properties of a Streptococcus pneumoniae mutant which does not require choline or analogs for growth. J. Bacteriol. 1802093-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 311477-1488. [DOI] [PubMed] [Google Scholar]