Abstract
Amino acids are polymerized into peptides in the peptidyl transferase center of the ribosome. The nascent peptides then pass through the exit tunnel before they reach the extraribosomal environment. A number of nascent peptides interact with the exit tunnel and stall elongation at specific sites within their peptide chain. Several mutational changes in RNA and protein components of the ribosome have previously been shown to interfere with pausing. These changes are localized in the narrowest region of the tunnel, near a constriction formed by ribosomal proteins L4 and L22. To expand our knowledge about peptide-induced pausing, we performed a comparative study of pausing induced by two peptides, SecM and a short peptide, CrbCmlA, that requires chloramphenicol as a coinducer of pausing. We analyzed the effects of 15 mutational changes in L4 and L22, as well as the effects of methylating nucleotide A2058 of 23S rRNA, a nucleotide previously implicated in pausing and located close to the L4-L22 constriction. Our results show that methylation of A2058 and most mutational changes in L4 and L22 have differential effects on pausing in response to CrbCmlA and SecM. Only one change, a 6-amino-acid insertion after amino acid 72 in L4, affects pausing in both peptides. We conclude that the two peptides interact with different regions of the exit tunnel. Our results suggest that either the two peptides use different mechanisms of pausing or they interact differently but induce similar inhibitory conformational changes in functionally important regions of the ribosome.
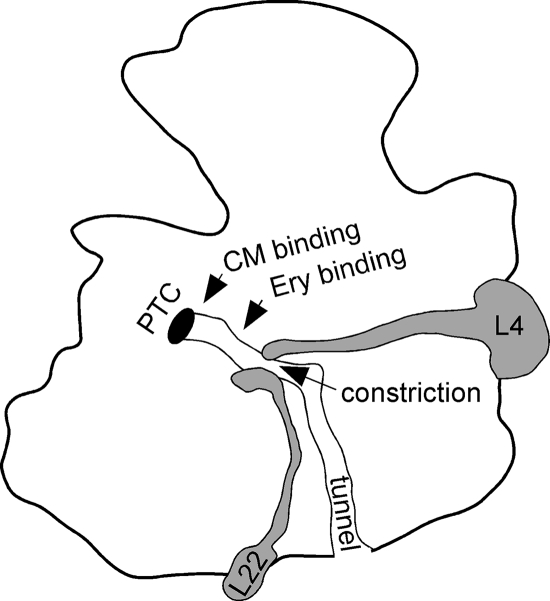
Peptide bond formation during translation takes place within the large subunit of the ribosome, at the peptidyl transferase center (PTC). In order to escape the ribosome, newly formed peptides must traverse a 100-Å-long channel referred to as the peptide exit tunnel. This tunnel is lined primarily by segments of 23S rRNA, although two ribosomal proteins, L4 and L22, also contribute part of the tunnel lining (1, 26). These two r-protein components form a constriction that results in the narrowest passage in the tunnel (Fig. 1). Mutational changes in both rRNA and r-protein components located near the constriction are important for bacterial resistance to macrolide-lincosamide-streptogramin antibiotics. These antibiotics bind to nucleotides A2058 and 2059 and other close-by portions of 23S rRNA (reviewed in reference 11).
FIG. 1.
Cartoon model of the 50S subunit of the ribosome of E. coli, showing the PTC, the peptide exit tunnel, the tips of r-proteins L4 and L22, and the approximate binding sites of antibiotics chloramphenicol (CM) and erythromycin (Ery).
The tunnel constriction has also been implicated in peptide-mediated pausing (7, 21). Peptide-mediated pausing occurs when sequence-specific interactions between the nascent peptide and the translating ribosome cause a stall in translation. SecM is a well-characterized example of a pause-inducing peptide. Mutations affecting the efficiency of SecM-induced pausing are found in the codons for a 17-amino-acid stretch of the peptide, termed the pause motif (21). This region is sufficient for inducing pausing when inserted into another protein sequence (10, 21). The SecM pausing response is also decreased by mutations altering the ribosome. Two of these changes, an A-to-G change at nucleotide 2058 in 23S rRNA and a 3-amino-acid deletion in r-protein L22 (L22-ΔMKR), are also known to confer resistance to erythromycin (Ery) (4, 23, 32, 36). In addition, several other L22 mutants shown to be deficient in SecM-mediated pausing do not confer Ery resistance (21). These previous studies, implicating r-protein L22 and nucleotide A2058 of 23S rRNA in SecM-mediated pausing, suggest that the region around the tunnel constriction is important for this pausing event. However, the well-studied K63E change in ribosomal protein L4, also located at the constriction, does not affect SecM-mediated pausing, although it does bestow resistance to erythromycin (reference 21 and this report).
Having recently isolated new erythromycin-resistant mutations in the genes for ribosomal proteins L4 and L22 of Escherichia coli (39), we wanted to take a more in-depth look at the relationship between erythromycin resistance and the efficiency of SecM-mediated pausing in these mutants. In addition, we wanted to expand this analysis to another class of pause-inducible peptides, those requiring an antibiotic for induction of genes bestowing resistance to the inducing antibiotic. CmlA is one such system. Pausing in this system is mediated by a leader peptide (CrbCmlA) encoded by a nine-codon open reading frame upstream of the cmlA gene (8, 28). Unlike the SecM system, pausing mediated by the CrbCmlA peptide requires, as a cofactor, sublethal levels of chloramphenicol, the drug to which the cmlA protein product confers resistance (2, 3). Pausing positions the ribosome on the mRNA to prevent formation of a hairpin that, in the absence of a paused ribosome, would occlude the cmlA translation initiation site. The result is chloramphenicol-induced synthesis of CmlA.
Our results indicate that all of the new Eryr mutational changes in L22 are deficient in SecM-mediated pausing, but none has a significant effect on chloramphenicol-induced CrbCmlA-mediated pausing. On the other hand, the mutational changes in L4 reduced CrbCmlA-mediated pausing, but only one mutant demonstrated a significant deficiency in SecM pausing. We also found that methylation of A2058 in 23S rRNA significantly reduces CrbCmlA-mediated pausing but has no effect on recognition of SecM. Our results suggest the SecM and CrbCmlA peptides induce pausing by interacting with different features of the exit tunnel.
MATERIALS AND METHODS
Primers.
The primers used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used
| Oligo no. | Name | Use | Sequence |
|---|---|---|---|
| O1460 | L22 forward | Amplify, sequence, or recombineer L22 gene | GAATTCGCACCGACTCGTAC |
| O1461 | L22 backward | Amplify, sequence, or recombineer L22 gene | GGTGTTCGCAAACCAGGTAGAG |
| O1462 | L4 forward | Amplify, sequence, or recombineer L4 gene | CTGGTTAAAGGTGCTGTCCC |
| O1455 | L4 backward | Amplify, sequence, or recombineer L4 gene | CCATCGCAGTAGACGCTTTTTC |
| O1893 | CrbCmlA forward | Amplify, clone crbcmlA-cmlA′ | CCCCGCGGCCGCGTTACGATTCAAATTCAATCATGAGAT |
| O1894 | CrbCmlA backward | Amplify, clone crbcmlA-cmlA′ | CCCCACTAGTCGTGGCGGCAAGGGAGTACCGCCAACTAAA |
| O1293 | secM forward | Amplify, clone secM pausing motif | ATTTCCAACGCGTTGACGCTCAGCGCGCTGCTGAC |
| O1294 | secM backward | Amplify, clone secM pausing motif | CGATGAGAGCTCCAGGGCCAGCACGGATGCCTTGC |
| O2040 | Complement 23S 2061-2078 | Measure methylation of 2058 | GTAAAGGTTCACGGGGTC |
| O1862 | ermC forward EcoRI | Amplify, clone ermC | TCTTCTGAATTCAGGAGGTCTTCTTCTATGAACGAGAAAAATATAAAACACAGTC |
| O1871 | ermC backward XbaI | Amplify, clone ermC | GTTCTCTAGATTACTTATTAAATAATTTATAGCTATTG |
| O1940 | lacZ forward | Amplify lacZα from pGEM5 | CTTTATGCTTCCGGCTCGTATGTTG |
| O1941 | lacZ backward | TCGTAACCGTGCATCTGCCAGTTTG |
Strains and plasmids.
Strains used in this study were derived from DY380, a DH10B derivative containing a λ prophage encoding phage proteins for homologous recombination (16). Strains were constructed as follows: DNA sequences encoding pausing peptides were amplified from the chromosome of E. coli K-12 for the secM/lacZ construct (primers O1293 and O1294) (Table 1) and from plasmid pDU1294 (described by Dorman and Foster [8]) for the crbcmlA/lacZ construct (primers O1893 and O1894 [Table 1]). The PCR fragments containing the pause sequences were cloned into pGEM5 to create in-frame fusions with lacZα. SacI and MluI sites were used for SecM, while SpeI and NotI were used for CrbCmlA. These pause sequence-lacZα fusions were then PCR amplified (O1940 and O1941 [Table 1]) and inserted into the chromosomal lacZ gene by homologous recombination. For the secM/lacZ fusion, we first constructed a Lac+ derivative of DY380 by converting its chromosomal lacZΔM15 gene to wild-type lacZ using a PCR product derived from the lacZα gene of pGEM5. The latter was synthesized using primers O1940 and O1941. We then replaced by recombineering the N-terminal end of lacZ with the secM/lacZ fusion on pGEM5, also synthesized using primers O1940 and O1941, and screened for white colonies. In the resulting strain, called DY380(SecM), the lacZ gene on the chromosome is replaced by an open reading frame with the following sequence: the N-terminal 5 codons of lacZ, 18 codons from pGEM5, 46 codons from secM (Thr121 through Pro166), 39 codons from pGEM5, and the rest of lacZ (beginning with codon 6) (see Fig. 2A, below). We inserted the crbcmlA/lacZ fusion into DY380 (containing lacZΔM15) and screened for blue colonies. The resulting strain, called DY380(CmlA), has on the chromosome an open reading frame containing the N-terminal 5 codons of lacZ, 30 codons from pGEM5, and the entire crb sequence (10 codons, including the termination codon), followed by the natural intergenic spacer between crb and cmlA, a second open reading frame containing the N-terminal 15 codons of cmlA, 27 codons from pGEM5, and lacZ starting with codon 6 (see Fig. 3A, below).
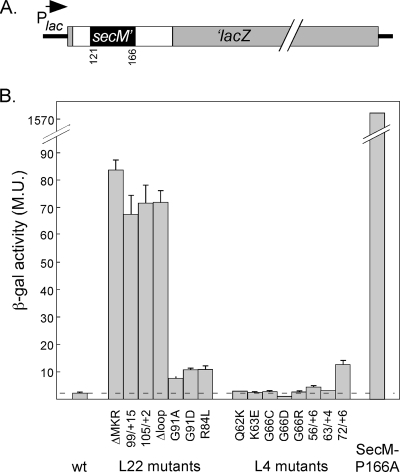
FIG. 2.
SecM-mediated pausing in L4 and L22 mutants. A. Map of the construct for quantifying efficiency of pausing. White boxed areas indicate sequences derived from the multicloning site of pGEM5. The SecM sequence is shown in black. Light gray boxes indicate sequences from the lacZ gene. For more details, see Materials and Methods. B. Quantitation of SecM/β-galactosidase fusion protein synthesis. The β-galactosidase activity was measured in strains containing the indicated L22 or L4 genes or by using a fusion protein construct containing a mutation in the SecM sequence (P166A) that inactivates the pausing peptide (21), as described in Materials and Methods. At least three independent cultures were analyzed for each mutant, and β-galactosidase assays were performed in duplicate for each culture. The standard error of the mean is indicated. M.U., Miller units.
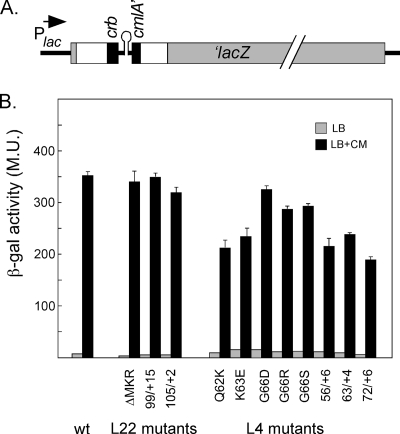
FIG. 3.
CrbCmlA-mediated pausing in L4 and L22 mutants. A. Map of the construct used for quantifying efficiency of pausing. White boxed areas indicate sequences derived from the multicloning site of pGEM5. This region is interrupted by the segment of DNA containing the Crb pausing peptide (black box), intergenic region, and the N terminus of cmlA (black box). Light gray boxes indicate sequences from the lacZ gene. For more details, see Materials and Methods. B. Quantitation of CmlA/β-galactosidase fusion protein synthesis. The β-galactosidase activity was measured in strains containing the indicated L22 or L4 genes, as described in Materials and Methods. The light gray bars indicate measurements from cells grown in the absence of antibiotic. The black bars indicate measurements from cells grown in the presence of chloramphenicol (0.8 μg/ml). At least three independent cultures were analyzed for each mutant, and β-galactosidase assays were performed in duplicate for each culture. The standard error of the mean is indicated for the induced values. M.U., Miller units.
L4 and L22 mutations were introduced into the chromosomes of DY380(SecM) and DY380(CmlA) by homologous recombination using PCR products generated with primers O1460 and O1461 for L22 and O1462 and O1455 for L4 (Table 1). For erythromycin resistance mutations (39), we selected recombinants on LB plates containing 400 μg/ml of erythromycin. Nonresistant mutants L22-G91A and L22-G91D were identified on the basis of their Lac+ phenotype. The L22-Δloop2 mutant was also identified as a Lac+ recombinant; in this mutant, amino acids 82 to 98 of the L22 tentacle have been replaced with two glycines (also, an unintentional base change in codon 101 changing Ser to Gly) (40).
The sequences of the lacZ fusions and mutant L4 and L22 sequences were confirmed by sequencing using ABI Big Dye v3.
Plasmid pErmC was constructed by amplifying the ermC gene from pE194 (15) (no. 1E7; obtained from the Bacillus Genetics Stock Center), using primers O1862 and O1871 (Table 1) and cloning into pBAD18 using EcoRI and KpnI. In the resulting construct, ErmC expression is induced in the presence of arabinose.
Plasmids used to express L22 under arabinose control were described previously (40).
β-Galactosidase assays.
β-Galactosidase assays were performed on cultures grown at 30°C in LB medium containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were inoculated at an optical density at 600 nm (OD600) between 0.04 and 0.05 and grown at 30°C to an OD600 of 0.4 to 0.6 (approximately 8 × 107 cells/ml). Where indicated, arabinose (0.2% final concentration) was used to induce ErmC synthesis. CrbCmlA-mediated translation pausing was analyzed by growing cells in the presence of chloramphenicol (0.8 μg/ml). Enzyme assays were performed basically as described previously (19) and read in a 96-well microtiter dish using a plate reader (12).
Plate screen for SecM-deficient mutants.
Wild-type DY380 or DY380(SecM)-containing plasmids with wild-type or mutant L22 downstream of the arabinose-inducible pBAD promoter were grown on LB solid medium containing 1 mM IPTG, 0.2% arabinose, and ampicillin at 200 μg/ml. Plates were incubated at 30°C and visually analyzed for colony color. Wild-type DY380 formed white colonies under these conditions, while colonies deficient for SecM pausing were various degrees of blue.
Ribosome preparations.
Cells were grown to an OD450 of 1.5 to 2.0 (about 3 × 108 cells per ml), harvested on ice, and centrifuged at 8,000 rpm for 10 min in a Beckman JLA 10.5 rotor. Cells were then resuspended in buffer A (20 mM HEPES-KOH pH 7.5, 6 mM MgCl2, 30 mM NH4Cl, 6 mM β-mercaptoethanol) and lysed using a French press at 16,000 lb/in2. Lysates were clarified by spinning at 22,000 rpm for 30 min in a Beckman MLA-80 rotor, and ribosomes were pelleted from the supernatant by spinning at 50,000 rpm for 4 h. The ribosome pellet was washed with buffer A and resuspended overnight in 400 μl buffer A. Ribosomes were salt washed by mixing 1 part crude ribosomes and 9 parts salt-wash buffer (20 mM HEPES-KOH pH 7.5, 6 mM MgCl2, 1 M NH4Cl, 6 mM β-mercaptoethanol), incubation on ice for 1 h, and centrifugation at 50,000 rpm for 4 h. Ribosomes were resuspended as described above.
Erythromycin binding.
Binding of erythromycin was determined as described by Zaman et al. (39). This method is modified from reference 30.
ErmC induction and analysis of A2058 methylation.
Cells containing pErmC were grown to mid-log phase in the presence or absence of 0.2% arabinose. Cultures were then harvested and ribosomes prepared as described above. rRNA was phenol-chloroform extracted from purified ribosomes followed by two ethanol precipitations. Methylation of A2058 was quantified using previously published methods (27, 33). Briefly, primer O2040 (Table 1) was 5′-32P end labeled and hybridized to 5 μg purified rRNA. The primer was extended in the presence of dTTP and ddCTP using reverse transcriptase. Reverse transcriptase reaction mixtures were then phenol-chloroform-isoamyl alcohol extracted, chloroform extracted, ethanol precipitated, and resuspended in 15 μl of H2O. Four μl was then run on a 20% denaturing polyacrylamide gel, and the gel was dried and visualized on a PhosphorImager (Molecular Dynamics). Band intensities were quantified using Image Quant software.
RESULTS
Effect of L22 mutants on SecM-mediated pausing.
For many years, only one mutation in the L4 gene and one mutation in the L22 gene had been shown to confer resistance to erythromycin in E. coli. We recently reported the isolation of new erythromycin-resistant ribosomal protein mutants (39). Eight mutations mapped in the gene for r-protein L4 and two mapped in L22. We were interested in determining if these new Eryr ribosomal protein mutational changes affect SecM pausing.
We constructed a derivative of strain DY380 (16) in which the SecM pause sequence is fused to the N terminus of the gene encoding β-galactosidase, resulting in the strain referred to as DY380(SecM) (Fig. 2A). Mutations in the ribosomal protein L22 gene conferring erythromycin resistance, originally isolated in E. coli strain AB301, were introduced into the chromosome of DY380(SecM) by homologous recombination and selected on erythromycin solid medium. β-Galactosidase assays were performed on both wild-type and mutant strains to measure the effect of L22 mutations on secM/lacZ expression. In this system, pausing directly affects the expression of the lacZ fusion protein (Fig. 2A). Thus, increased β-galactosidase activity indicates that ribosomes carrying a mutant protein are deficient in SecM pausing.
The original Eryr L22 mutant (which we call L22-ΔMKR) has a deletion that removes amino acids 82 to 84 from the so-called “tentacle,” the region of the protein that lines the peptide exit tunnel (31). One of the new mutants, L22-99/+15, has a 15-amino-acid insertion after amino acid 99, also in the tentacle. The other new L22 mutant, L22-105/+2, has a 2-amino-acid insertion after amino acid 105; this insertion is just downstream of the tentacle. Like L22-ΔMKR, both of our new L22 Eryr mutants accumulated significantly more β-galactosidase than wild-type DY380(SecM) (Fig. 2B), indicating that all three Eryr strains are defective in responding to the SecM pause signal.
As a comparison, we also introduced two other mutations into the L22 gene that had previously been shown to result in defective SecM pausing but not resistance to erythromycin (21). These mutants, L22-G91A and L22-G91D, also accumulated more β-galactosidase than the parent strain (Fig. 2B), although much less than the Eryr mutants. We also isolated two other mutants from DY380(SecM) that appeared spontaneously as bluer colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates and, upon sequencing, were found to have changes in L22: L22-R84L and a mutant containing two alterations, L22-A89V and R94H. As predicted from their plate phenotype, these two mutants also accumulated more enzyme than the parent (Fig. 2B and data not shown). These mutants were not resistant to Ery and, like the original Erys G91A and G91D mutants, were not as defective in SecM pausing as the Eryr mutants.
We have previously reported that L22 proteins containing deletions removing most or all of the tentacle are nevertheless still able to be incorporated into ribosomes that can be assembled into polysomes (40). As an initial screen of the effect these deletions have on the SecM response, we introduced into strain DY380(SecM) plasmids carrying two different tentacle deletion L22 genes. L22-Δloop1 lacks amino acids 85 to 95, and L22-Δloop2 lacks amino acids 82 to 98; both genes are under the control of the arabinose-inducible PBAD promoter (40). Cells expressing these mutant proteins from plasmids were also defective in SecM pausing, as judged by bluer colonies on X-Gal plates compared to cells expressing wild-type L22 (data not shown). To confirm these results, we substituted the wild-type chromosomal copy of L22 with the L22-Δloop2 tentacle deletion derivative. The resulting mutant had a significant defect in SecM-mediated pausing (Fig. 2B). It was also resistant to erythromycin, although the maximal erythromycin concentration tolerated by the mutant was only 100 μg/ml, compared to 200 μg/ml or more for our other L22 Eryr mutants. The isolation of this mutant strain, now harboring a single L22 gene encoding a protein with no tentacle, confirmed our previous report that the L22 tentacle is not necessary for ribosome function (40).
It should be noted that, although the L22 mutants just described had significantly higher levels of β-galactosidase than the wild-type parent, the enzyme levels were still much lower than the level we observed with a strain containing a defective SecM signal fused to lacZ. In this mutated pause sequence, a proline-to-alanine change at position 166 of SecM abolishes the pausing activity of SecM (21). This construct (SecM-P166A) accumulated more than 10 times as much β-galactosidase activity as the most defective L22 strain (Fig. 2B). We conclude that all of the L22 mutants tested here pause less efficiently in response to SecM than wild-type cells, although they still show significant pausing.
Effect of L4 mutants on SecM-mediated pausing.
We also introduced the canonical L4 erythromycin resistance mutation (L4-K63E) and seven new L4 mutations into strain DY380(SecM). All of these mutations map in the region of the gene encoding the tentacle of L4, the tip of which forms part of the lining of the peptide exit tunnel (34). Unlike the L22 Eryr mutations, mutations in the L4 gene had little or no effect on the cell's response to the SecM pause signal (Fig. 2B). The amino acid substitution mutants were essentially like the wild-type parent. The three insertion mutants showed slightly higher β-galactosidase activity, but only mutant L4-72/+6, which has a 6-amino-acid insertion after amino acid 72 of L4, showed increased β-galactosidase activity, comparable to the Erys L22 mutants. This is the first mutation in the L4 gene reported to affect the ribosome's response to SecM.
Effect of L4 and L22 Eryr mutants on pausing induced by the leader peptide of CmlA.
Translation of the leader upstream of CmlA causes ribosomal pausing in the presence of sublethal levels of chloramphenicol (2, 8, 17). We wanted to compare the effects of Eryr mutants on SecM-mediated pausing with their effects on chloramphenicol-induced pausing mediated by the CmlA leader peptide. Therefore, we constructed a strain in which the leader peptide (CrbCmlA), the intercistronic region between the Crb leader and cmlA, and first 15 codons of cmlA were inserted into the lacZ gene present on the chromosome, creating an in-frame fusion between cmlA and lacZ (Fig. 3A). Since pausing of ribosomes at a specific position in the Crb leader prevents the formation of the hairpin in the crb-cmlA intercistronic region, pausing exposes the cmlA translation initiation site and stimulates CmlA translation (28). Thus, the CmlA reporter differs from the SecM reporter in that decreased pausing results in decreased β-galactosidase activity. In the DY380(CmlA) construct, the Crb pausing sequence was moved from its natural state as a short 9-amino-acid open reading frame upstream of cmlA to a position 36 codons from the translation initiation site, yet the sequence still displayed efficient pausing (Fig. 3A).
Contrary to the results with SecM, we found that, in the presence of chloramphenicol, the mutational changes in L22 had little or no effect on cmlA-lacZ expression. On the other hand, most of the L4 mutants exhibited reduced levels of β-galactosidase activity and, hence, reduced responses to CrbCmlA-mediated pausing (Fig. 3B). The three amino acid substitutions at Gly-66 (G66D, G66R, and G66S) resulted in the weakest effects. More significant reductions were seen with L4 mutants Q62K, K63E, 56/+6 (6-amino-acid insertion after amino acid 56), and 63/+4 (4-amino-acid insertion after amino acid 63), which yielded β-galactosidase levels that were between 60 and 70% of the wild-type level. The greatest defect in CrbCmlA-mediated pausing was observed with the mutant L4-72/+6, which was induced by chloramphenicol to only half the wild-type level. Interestingly, this strain was the only one that exhibited defects with both pausing systems.
In the absence of chloramphenicol, all of the L4 mutants, except L4-72/+6, produced about twice as much β-galactosidase as the wild-type parent and the L22 mutants. As a result, the induction ratios for the L4 mutants ranged from 15- to 24-fold, while the wild-type parent and L22 mutants had induction ratios ranging from 43- to 85-fold. The relatively higher levels of β-galactosidase synthesis in the absence of chloramphenicol (compared to the wild-type parent) suggest that the mutational changes do not have a general inhibitory effect on β-galactosidase synthesis that could account for the reduced β-galactosidase levels after addition of the antibiotic. Consistent with this, we observed that mutational changes in L4 that resulted in reduced chloramphenicol induction levels had no effect on expression of a wild-type lacZ gene on the chromosome after induction with IPTG (data not shown). We also considered the possibility that chloramphenicol itself may have an inhibitory effect on synthesis of β-galactosidase in strains with mutated L4 genes, independent of its effect on stalling. However, addition of chloramphenicol at the sublethal concentrations used to induce pausing showed that the level of enzyme encoded by the IPTG-induced chromosomal lacZ gene was not affected by mutational alterations in L4 (data not shown). Finally, the deficiencies in Crb pausing are not due to increased resistance to chloramphenicol, since only the L4 G66D mutation resulted in increased (but slight) resistance to chloramphenicol (results not shown). We conclude that the decrease in β-galactosidase expression seen in the L4 strains compared to the wild-type and L22 strains reflects a decreased ability to respond to the CrbCmlA stalling peptide.
Methylation of A2058 and pausing.
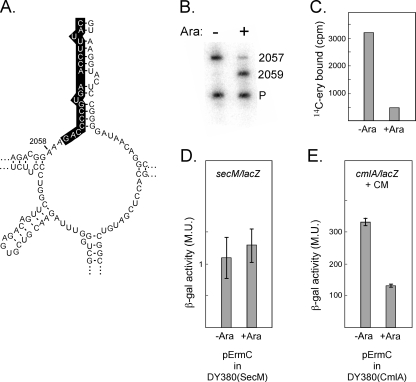
Previous studies have shown that an A-to-G mutation at nucleotide 2058 in 23S rRNA reduces the response to SecM-mediated pausing (21). The same mutation also eliminates the inhibitory effects of macrolide-lincosamide-streptogramin drugs (9, 32). Methylation of A2058 by the ErmC methylase also confers resistance to erythromycin (35). To test if ErmC methylation affects SecM-mediated pausing, we constructed a plasmid in which the gene encoding ermC was placed under the control of the inducible araBAD promoter; in this way the methylase is expressed only in the presence of arabinose. We confirmed methylation of A2058 in the presence of arabinose by reverse transcription of rRNA extracted from purified ribosomes (Fig. 4A); these experiments indicated that approximately 80% of the 23S rRNA molecules were methylated when ermC was induced (Fig. 4B). As expected, methylation strongly reduced binding of erythromycin to ribosomes (Fig. 4C). Strain DY380(SecM) containing this plasmid was then assayed for β-galactosidase activity in the presence and absence of arabinose. We found that, unlike the A2058G rRNA mutant, which exhibits decreased pausing (21), methylation of A2058 had no significant affect on SecM-mediated pausing (Fig. 4D). In contrast, arabinose induction of pErmC in the strain carrying the CrbCmlA pause system DY380(CmlA) reduced β-galactosidase activity by 60%, indicating that methylation of A2058 in 23S rRNA significantly inhibits chloramphenicol-induced CrbCmlA activity (Fig. 4E). Strains carrying the empty vector or expressing a control protein did not show this defect in the presence or absence of arabinose (data not shown).
FIG. 4.
Effect of methylation of A2058 on SecM- and CrbCmlA-mediated pausing and erythromycin binding. Wild-type cells containing a plasmid with an arabinose-inducible ErmC gene were grown in the absence or presence of arabinose. Aliquots were removed for quantitation of β-galactosidase activity, and the remainder of the cultures were used for ribosome preparation and primer extension and erythromycin binding analysis (see Materials and Methods for details). A. Secondary structure of 23S rRNA in the region of A2058. The oligo used for primer extension was complementary to the highlighted bases. B. Primer extension analysis of the methylation of A2058. P, primer; 2059, extension products terminated at nucleotide 2059 (indicating methylation blockage at A2058); 2057, extension products terminated at G2057 (no methylation). C. Binding of [14C]erythromycin to ribosomes from cells grown with and without arabinose. D. Quantitation of SecM/β-galactosidase fusion protein synthesis in cells grown with and without arabinose. Standard errors of the means are indicated. E. Quantitation of CmlA/β-galactosidase fusion protein synthesis in cells grown in the presence of arabinose. Standard errors of the means are indicated. M.U., Miller units.
DISCUSSION
Regulation of translation by cis-acting pause-inducing peptides involves interactions between the nascent peptide and ribosomal components lining the peptide exit tunnel (6, 7, 14, 17, 21). The precise mechanisms by which these interactions inhibit subsequent peptidyl transferase activity are still unclear. Previous studies of the SecM and TnaC systems implicated the region of the tunnel where a constriction is formed by r-proteins L4 and L22 (7, 21), but differential effects of ribosomal mutations suggested that the two systems may not work by exactly the same mechanism (18). In this work, we have shown that a large collection of L4 and L22 mutants as well as the methylation of the 23S rRNA nucleotide A2058 affect pausing in response to SecM and another pausing peptide, CrbCmlA. However, with the exception of one L4 mutant, we observed complete nonoverlap in the effects of the mutations on one system or the other. Our results not only add to our growing knowledge of the role played by ribosomal components near the constriction in the function of regulatory peptides but also highlight apparent differences in the mechanism by which pause-inducing peptides act.
Limited effects of L22 mutations.
It is important to note that the effects on SecM-mediated pausing attributed to mutations in the L22 gene are relatively small compared to the effect observed in cells carrying the P166A substitution in the SecM pausing peptide (numbering relative to the intact SecM protein), which eliminates pausing altogether (21). In the SecM system, the greatest effects of changes in the L22 protein resulted in accumulation of less than 10% of the β-galactosidase activity observed with the P166A mutation in the pausing peptide. The ribosomal mutants are not completely devoid of pausing function. If the ability to recognize specific signals in nascent peptides is a critical and integral part of ribosome function, perhaps a ribosome completely oblivious to a pause signal would be too dysfunctional to support growth. This idea is consistent with our observation that the L22 mutants we described here are able to support growth at doubling times no more than fourfold greater than the wild-type growth rate (data not shown) (39).
Changes in L22 and L4 affect SecM and CrbCmlA differently.
Previous studies identified ribosomal protein L22 as particularly important to the activity of both SecM and TnaC pausing peptides (7, 21). The experiments reported here show that mutations in the L22 gene that bestow erythromycin resistance (ΔMKR, 99/+15, 105/+2, and Δloop2) have significantly stronger effects on pausing than do mutations with no detectable change in the level of erythromycin sensitivity (G91A, G91D, and R84L). Presumably, mutations that confer resistance to erythromycin have a more severe conformational effect on the region(s) of the tunnel involved in SecM recognition and Ery inhibition. However, our experiments cannot determine whether the same tunnel determinants are involved in both.
Contrary to pausing mediated by the SecM peptide (21, 38), pausing mediated by the CrbCmlA peptide is affected by alterations in the L4 protein but not by mutations altering L22. Another difference between the two pausing peptides is that only CrbCmlA-mediated pausing requires a coinducer, chloramphenicol. It would seem that chloramphenicol-induced alterations in ribosome structure must be combined with peptide-induced changes in order to establish the pausing condition.
Mechanism of pausing.
All but one (L4-72/+6) of the L4 mutants showed an increased basal level expression of the CmlA/β-galactosidase fusion protein when compared to wild-type or L22 mutants, indicating an increase in the amount of time ribosomes occupied the CrbCmlA pause site in these mutants. Possibly, these elevated basal levels result from an increased sensitivity of the mutant ribosomes to the CrbCmlA pause sequence in the absence of the inducer chloramphenicol. This would agree with previous reports that a synthetic peptide of the CrbCmlA 8-mer is able to induce changes in ribosome structure in vitro in the absence of chloramphenicol (14). If certain L4 mutants are indeed more sensitive to the CrbCmlA peptide alone, it is not clear why these same mutants show reduced levels of induction in the presence of chloramphenicol. It is not likely that these mutations inhibit binding of the drug at its normal binding site, since only one of the L4 mutants exhibiting increased basal levels of CmlA/β-Gal (G66D) also showed an increased resistance to chloramphenicol. It was previously shown that chloramphenicol-resistant mutations do not impair CrbCmlA-mediated pausing and, conversely, mutations that impair pausing do not affect chloramphenicol's inhibitory effects (24). Our results in conjunction with these previously published observations suggest that chloramphenicol may bind differently to the ribosome during general inhibition of protein synthesis and during CrbCmlA-mediated pausing.
Our genetic experiments agree with previous reports indicating that the constriction in the vicinity of A2058 and the tips of L4 and L22 (Fig. 5) is important for translation pausing induced by the nascent peptide. However, if the constriction indeed does gate the tunnel as previously suggested (21), the L22 tentacle is not likely an essential part of this gate. This conclusion is based on the observation that deletion of the entire L22 tentacle only mildly affects pausing. It is important to note that the L22-Δloop2 gene in the strain used in the experiments shown in Fig. 2 and 3 is in the chromosomal S10 operon and hence is the only source of L22. This is different from our previous construct in which the strain contained a wild-type L22 gene in addition to the plasmid-based L22-Δloop mutant gene (40). Based on the experiments reported here, we can therefore unambiguously conclude that the L22 tentacle is not essential for protein synthesis or pausing. Moreover, the haploid L22-Δloop2 mutant grows only a little slower than the wild-type parent (data not shown), indicating that ribosomes carrying the tentacle-less L22 function well in protein synthesis.
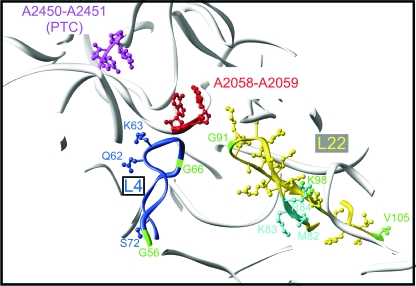
FIG. 5.
Structure of the E. coli ribosome exit tunnel. The figure shows a slab view through the ribosome tunnel from the PTC past the region of the tunnel lined on one side by the L22 tentacle. Nucleotides A2450 and A2451 (magenta) are used as markers for the PTC. Also shown are A2058 and A2059 (red), marking the erythromycin binding site and the entry to the beginning of the narrow part of the tunnel. Nascent peptides are synthesized at the PTC and then migrate past nucleotides A2058 and A2059 and through the constriction formed by the tips of r-proteins L4 and L22 before moving further into the tunnel lined on one side by the L22 tentacle. The tentacles of L4 (blue) and L22 (yellow) are shown with selected side chains marking positions of amino acid substitutions, insertions, and deletions analyzed in this study. Of particular interest is the Δloop2 deletion in L22, in which all residues from M82 through K98 were replaced with two glycine residues. In this mutant, much of the L22 contribution to the tunnel lining is presumably eliminated. For more details about mutants, see the text and also references 39 and 40.
The positions of the various L22 mutations indicate that for at least some of the mutations, the effects are indirect. Perhaps the most potent argument for this idea is that the complete deletion of the L22 tentacle, which contributes to the tunnel lining over almost two-thirds of the tunnel length (Fig. 5), has no stronger an effect on SecM pausing than do the other L22 mutations. If a direct interaction(s) between the pausing peptide and L22 were essential, we would expect that the deletion of the L22 tentacle would have eliminated pausing entirely. Furthermore, a mutation in the globular portion of L22 outside the tunnel (105/+2) affects pausing more severely than point mutations within the tentacle located in the tunnel.
The idea that ribosomal protein mutations can have indirect effects on tunnel function also arises from the short length of the CrbCmlA pause peptide, which is only 8 amino acids long. If this peptide were fully extended, it could reach as far as about 25 Å from the PTC (assuming 3 to 3.4 Å per residue). If the nascent CrbCmlA peptide forms a more helical structure, it might reach only as far as 12 Å (assuming 1.5 Å per residue). That range of reach suggests that the nascent peptide could interact with nucleotide A2058, but it is not likely to form an extensive interaction with the L4/L22 constriction, which is 20 to 35 Å from the PTC (18) (Fig. 5). Thus, it seems inescapable that the effects of the L4 mutants on CrbCmlA-mediated pausing are indirect. Indirect effects of mutations on ribosome function are also compatible with effects of the L4-K63E mutation on 50S-mediated peptidyl transferase activity and 30S-mediated decoding (22, 36); both of these activities involve regions of the ribosome that are distant from amino acid 63 of L4. Several other L4 and L22 mutations, located across extensive portions of the ribosome and none of which are located close to the PTC or other ribosome functional centers, also affect the rate of peptide chain elongation (39). Furthermore, a 2-amino-acid deletion in Streptococcus pneumoniae L4 results in resistance to not only erythromycin but also chloramphenicol (37), which binds closer to the PTC than erythromycin (25) (Fig. 1).
The SecM peptide, which is 17 amino acids long, has the potential to form interactions in a much more extended ribosome landscape than does the 8-amino-acid CrbCmlA peptide, even though the SecM peptide apparently condenses during pausing (38). Interestingly, the six C-terminal amino acids of the SecM peptide are sufficient to induce a pause (5, 29), albeit not as efficiently as that caused by full-length SecM. Thus, interactions in the tunnel close to the PTC may establish a minimal pause. It has been hypothesized that essential interactions between the C-terminal portion of SecM and the ribosome tunnel between the PTC and A2058 are enhanced by interactions between the N-terminal part of the SecM pausing peptide and L22 in the more distant portions of the tunnel (21). Perhaps chloramphenicol in some way substitutes for the effect of the N-terminal portion of the SecM pausing peptide in facilitating CrbCmlA-mediated pausing.
Previous reports have proposed that interaction between SecM and nucleotide A2058 of 23S rRNA is important for SecM-mediated pausing (20, 21). While the SecM peptide may interact with A2058, as suggested by the effect of the A2058G mutation, any putative interaction does not mimic the interaction of erythromycin with A2058, since we showed that methylation of A2058 by the ErmC methylase inhibits the binding of erythromycin but has no affect on SecM activity. The difference between the effects of A2058G mutation and methylation of the same nucleotide on SecM-mediated pausing suggests that either SecM and erythromycin interact differently with this nucleotide or that the effects of the A2058G mutation on SecM pausing are indirect.
It has been reported that methylation of A2058 has no effect on the activity of another chloramphenicol-induced regulatory peptide, Cat-86 (13), a pausing peptide related to CrbCmlA. However, our results show that methylation of A2058 dramatically reduces CrbCmlA-mediated pausing. While Cat-86 only has a length of 5 amino acids as opposed to 8 for CrbCmlA, it is quite intriguing that two peptides which are regulated by the same drug would have different responses to the same rRNA modification. This could indicate that CrbCmlA and Cat-86 interact differently with the ribosome and therefore possibly have different mechanisms of inhibition.
In summary, the comparison of SecM and Crb pausing reveals new diversity in the mechanisms of nascent peptide-induced inhibition of PTC function. Further studies should contribute to our understanding not only of pausing but also of ribosome assembly and function in general.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289905-920. [DOI] [PubMed] [Google Scholar]
- 2.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 1734493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns, J. L., C. E. Rubens, P. M. Mendelman, and A. L. Smith. 1986. Cloning and expression in Escherichia coli of a gene encoding nonenzymatic chloramphenicol resistance from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 29445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chittum, H. S., and W. S. Champney. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 1766192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier, J., C. Bohn, and P. Bouloc. 2004. SsrA tagging of Escherichia coli SecM at its translation arrest sequence. J. Biol. Chem. 27954193-54201. [DOI] [PubMed] [Google Scholar]
- 6.Cruz-Vera, L. R., M. Gong, and C. Yanofsky. 2006. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA 1033598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Vera, L. R., S. Rajagopal, C. Squires, and C. Yanofsky. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell 19333-343. [DOI] [PubMed] [Google Scholar]
- 8.Dorman, C. J., and T. J. Foster. 1985. Posttranscriptional regulation of the inducible nonenzymatic chloramphenicol resistance determinant of IncP plasmid R26. J. Bacteriol. 161147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douthwaite, S., and B. Vester. 2000. Macrolide resistance conferred by alterations in the ribosome target site, p. 431-439. In EDITORS (ed.), The ribosome: structure, function, antibiotics, and Cellular interactions. ASM Press, Washington, DC.
- 10.Evans, M. S., K. G. Ugrinov, M. A. Frese, and P. L. Clark. 2005. Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nat. Methods 2757-762. [DOI] [PubMed] [Google Scholar]
- 11.Franceschi, F., Z. Kanyo, E. C. Sherer, and J. Sutcliffe. 2004. Macrolide resistance from the ribosome perspective. Curr. Drug Targets Infect. Disord. 4177-191. [DOI] [PubMed] [Google Scholar]
- 12.Griffith, K. L., and R. E. Wolf, Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290397-402. [DOI] [PubMed] [Google Scholar]
- 13.Gu, Z., R. Harrod, E. J. Rogers, and P. S. Lovett. 1994. Properties of a pentapeptide inhibitor of peptidyltransferase that is essential for cat gene regulation by translation attenuation. J. Bacteriol. 1766238-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrod, R., and P. S. Lovett. 1995. Peptide inhibitors of peptidyltransferase alter the conformation of domains IV and V of large subunit rRNA: a model for nascent peptide control of translation. Proc. Natl. Acad. Sci. USA 928650-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol. 150804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 17.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mankin, A. S. 2006. Nascent peptide in the “birth canal” of the ribosome. Trends Biochem. Sci. 3111-13. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 20.Mitra, K., C. Schaffitzel, F. Fabiola, M. S. Chapman, N. Ban, and J. Frank. 2006. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol. Cell 22533-543. [DOI] [PubMed] [Google Scholar]
- 21.Nakatogawa, H., and K. Ito. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108629-636. [DOI] [PubMed] [Google Scholar]
- 22.O'Connor, M., S. T. Gregory, and A. E. Dahlberg. 2004. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 325750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardo, D., and R. Rosset. 1977. Properties of ribosomes from erythromycin resistant mutants of Escherichia coli. Mol. Gen. Genet. 156267-271. [DOI] [PubMed] [Google Scholar]
- 24.Rogers, E. J., N. P. Ambulos, Jr., Z. Gu, and P. S. Lovett. 1993. Parallel induction strategies for cat-86: separating chloramphenicol induction from protein synthesis inhibition. Mol. Microbiol. 81063-1069. [DOI] [PubMed] [Google Scholar]
- 25.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413814-821. [DOI] [PubMed] [Google Scholar]
- 26.Schuwirth, B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310827-834. [DOI] [PubMed] [Google Scholar]
- 27.Sigmund, C. D., M. Ettayebi, A. Borden, and E. A. Morgan. 1988. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 164673-690. [DOI] [PubMed] [Google Scholar]
- 28.Stokes, H. W., and R. M. Hall. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 2610-19. [DOI] [PubMed] [Google Scholar]
- 29.Sunohara, T., K. Jojima, H. Tagami, T. Inada, and H. Aiba. 2004. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 27915368-15375. [DOI] [PubMed] [Google Scholar]
- 30.Teraoka, H. 1970. A reversible change in the ability of Escherichia coli ribosomes to bind to erythromycin. J. Mol. Biol. 48511-515. [DOI] [PubMed] [Google Scholar]
- 31.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121257-270. [DOI] [PubMed] [Google Scholar]
- 32.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 451-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vester, B., L. H. Hansen, and S. Douthwaite. 1995. The conformation of 23S rRNA nucleotide A2058 determines its recognition by the ErmE methyltransferase. RNA 1501-509. [PMC free article] [PubMed] [Google Scholar]
- 34.Voss, N. R., M. Gerstein, T. A. Steitz, and P. B. Moore. 2006. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 360893-906. [DOI] [PubMed] [Google Scholar]
- 35.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittmann, H. G., G. Stoffler, D. Apirion, L. Rosen, K. Tanaka, M. Tamaki, R. Takata, S. Dekio, and E. Otaka. 1973. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 127175-189. [DOI] [PubMed] [Google Scholar]
- 37.Wolter, N., A. M. Smith, D. J. Farrell, W. Schaffner, M. Moore, C. G. Whitney, J. H. Jorgensen, and K. P. Klugman. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 493554-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolhead, C. A., A. E. Johnson, and H. D. Bernstein. 2006. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell 22587-598. [DOI] [PubMed] [Google Scholar]
- 39.Zaman, S., M. Fitzpatrick, L. Lindahl, and J. Zengel. 2007. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 661039-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zengel, J. M., A. Jerauld, A. Walker, M. C. Wahl, and L. Lindahl. 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosomal assembly or L4-mediated autogenous control. RNA 91188-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]