Abstract
Only little information on a particular class of myoviruses, the SPO1-like bacteriophages infecting low-G+C-content, gram-positive host bacteria (Firmicutes), is available. We present the genome analysis and molecular characterization of the large, virulent, broad-host-range Listeria phage A511. A511 contains a unit (informational) genome of 134,494 bp, encompassing 190 putative open reading frames (ORFs) and 16 tRNA genes, organized in a modular fashion common among the Caudovirales. Electron microscopy, enzymatic fragmentation analyses, and sequencing revealed that the A511 DNA molecule contains linear terminal repeats of a total of 3,125 bp, encompassing nine small putative ORFs. This particular genome structure explains why A511 is unable to perform general transduction. A511 features significant sequence homologies to Listeria phage P100 and other morphologically related phages infecting Firmicutes such as Staphylococcus phage K and Lactobacillus phage LP65. Equivalent but more-extensive terminal repeats also exist in phages P100 (∼6 kb) and K (∼20 kb). High-resolution electron microscopy revealed, for the first time, the presence of long tail fibers organized in a sixfold symmetry in these viruses. Mass spectrometry-based peptide fingerprinting permitted assignment of individual proteins to A511 structural components. On the basis of the data available for A511 and relatives, we propose that SPO1-like myoviruses are characterized by (i) their infection of gram-positive, low-G+C-content bacteria; (ii) a wide host range within the host bacterial genus and a strictly virulent lifestyle; (iii) similar morphology, sequence relatedness, and collinearity of the phage genome organization; and (iv) large double-stranded DNA genomes featuring nonpermuted terminal repeats of various sizes.
Listeria monocytogenes is a gram-positive, opportunistic pathogen that can cause a wide spectrum of diseases, such as meningitis, septicemia, abortion, and gastroenteritis, associated with mortality rates as high as 25 to 30% (17, 62). Listeria bacteria are ubiquitously found in nature and can enter the food chain at many points (26, 66). They are able to proliferate over a wide range of external conditions, including low temperatures and high salt concentrations, thus posing severe problems to the food industry (2, 22, 15, 48).
Almost all of the Listeria bacteriophages described to date are temperate (38); only very little is known about virulent phages infecting this host. Listeria phages are valuable sources of biological information and useful tools for the study, differentiation, manipulation, and control of these bacteria (6, 33, 41). Phage A511 and some of its components have been particularly useful in phage typing schemes (35, 61), as reporter phage (39), with respect to their endolysins (16), and also for the control of Listeria contamination of foods (7; S. Guenther, D. Huwyler, S. Richard, and M. J. Loessner, submitted for publication). In contrast to temperate Listeria phages A118 and PSA (36, 71), A511 is a virulent (i.e., strictly lytic) phage, unable to integrate itself into the bacterial chromosome. The virus features a nonflexible, contractile tail and an isometric capsid and belongs to the A1 morphological group of the Myoviridae (72) within the order Caudovirales. Of particular interest is its extremely broad host range within the Listeria genus; it can infect approximately 95% of the common L. monocytogenes strains of serovars 1/2 and 4 (35, 61) and 100% of L. ivanovii strains tested (unpublished results). Thus, A511 is an ideal candidate for reporter phage construction for rapid detection of viable host bacteria in food samples (39). In agreement with the species- and serovar-independent host range was the finding that A511 probably uses conserved peptidoglycan structures as primary recognition molecules, possibly in conjunction with other, non-serovar-specific cell surface carbohydrates (67).
Based on morphology and host bacteria, A511 belongs to the group of SPO1-like phages proposed recently (9), the prototype of which is Bacillus subtilis phage SPO1. This phage and its close relative SP82 have been extensively studied, with respect to life cycle (65, 68), mode of DNA replication and transcription (11, 18, 49), and genome organization (12, 19, 23, 31, 53, 57). Common aspects of the SPO1-like phages (9) are that they infect low-G+C-content, gram-positive hosts (Firmicutes), have a strictly lytic (virulent) lifestyle, have similar morphologies, and have relatively large genomes with some sequence similarity.
Although A511 and the related Listeria phage P100 are useful tools for detection and control of Listeria, very little is known on the molecular level. Only a small portion of the A511 genome, namely, the lysis genes (42) and a late gene region specifying the major capsid and tail components (40), has previously been sequenced. Therefore, the aim of this study was to determine the complete genome sequence of A511 and analyze the genome structure and encoded gene products. We also wanted to compare A511 to P100 and other SPO1-like phages, such as Staphylococcus phage K (47) and Lactobacillus phage LP65 (9). We found that A511 has a genome of 137.6 kb in physical size, which features large, invariable terminal repeats of 3.1 bp, and that this genome structure is shared by phages P100 and K (featuring terminal repeats of 6 kb and 20 kb) and probably also the other members of this group, such as LP65, Twort, SPO1, φEF24C (60), and others. We conclude that A511 represents a good model for the morphology and genome structure of the SPO1-like phages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study were Listeria ivanovii WSLC 3009, Escherichia coli XL1-Blue MRF′ (Stratagene, La Jolla, CA), E. coli DH5αMRC (Invitrogen, Basel, Switzerland), and Staphylococcus aureus strain PSK (ATCC 19685). Bacteria were grown at 30°C or 37°C under constant agitation in brain heart infusion (Difco, Detroit, MI) (for Listeria and Staphylococcus) or Luria-Bertani broth (Oxoid, Hampshire, United Kingdom) (for E. coli).
Phage DNA isolation.
Phages were propagated in liquid cultures and purified by polyethylene glycol precipitation and CsCl gradient centrifugation as described previously (40, 52). For DNA extraction, phages were dialyzed twice for 2 h against a 1,000-fold excess of SM buffer (52) at 4°C. DNA extraction was performed as described elsewhere (40), using proteinase K and EDTA, followed by extraction with phenol, phenol-chloroform, and chloroform, followed by ethanol precipitation. DNA was dissolved in Tris-EDTA buffer. All samples were tested for their suitability for subsequent restriction by digestion with EcoRI (Fermentas, St. Leon-Rot, Germany).
Restriction analysis.
Restriction enzymes have been used according to the manufacturer's instructions (New England Biolabs or Fermentas). Five hundred nanograms of phage DNA was used for each reaction. Digests were analyzed electrophoretically as described below. For the extraction of A511 DNA fragments from gels, approximately 150 μg of DNA was digested with SwaI (Fermentas), electrophoresed (UltraPure low-melting-point agarose; Invitrogen), and stained with ethidium bromide. Bands corresponding to the desired fragments were recovered, and DNA was extracted from the agarose by using a gel extraction kit (Qiagen, Hilden, Germany).
Bal 31 nuclease treatment.
A total of 40 μg of A511 DNA was digested with Bal 31 (0.5 units per μg) (New England Biolabs) at 30°C. Samples were removed immediately before and 5, 10, 20, 40, and 60 min after the addition of the enzyme. All samples were purified by phenol-chloroform extraction and ethanol precipitation as described elsewhere (52).
Electrophoresis.
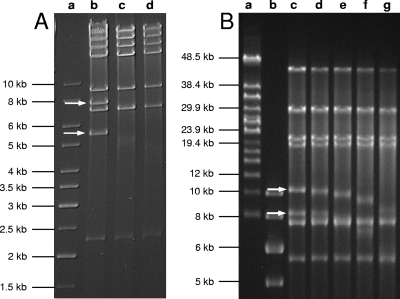
Conventional agarose gel electrophoresis was performed with a GNA200 horizontal electrophoresis apparatus (GE Healthcare) at 5 V/cm in 1× Tris-acetate-EDTA buffer. Pulsed-field gel electrophoresis (PFGE) was performed with 0.5× Tris-borate-EDTA as the running buffer (CHEF DR III; Bio-Rad). The following protocols were used depending on the desired gel resolution. (i) For the separation of DNA fragments 1 to 40 kb in size, a 0.9% agarose gel (Megabase agarose; Bio-Rad) was used with an initial switch time of 0.5 s and a final switch time of 1.7 s at 8 V/cm, at an angle of 120 degrees and 14°C buffer temperature. Run time was 4.5 h. (ii) For the electrophoresis run shown in Fig. 6B, the following settings were used: 1.2% agarose gel, an initial switch time of 1 s, a final switch time of 6 s at 5 V/cm, an angle of 120 degrees, 14°C buffer temperature, and 16-h run time. (iii) For the separation of full-length phage genomic DNA (see Fig. 2A), 1% agarose was used, with an initial switch time of 1 s, a final switch time of 25 s at 6 V/cm, an angle of 120 degrees, 14°C buffer temperature, and 21-h run time. For DNA size standards, we used a 1-kb DNA ladder (Fermentas), Lambda Mix 19 (Fermentas), and MidRange I PFG marker (New England Biolabs).
FIG. 6.
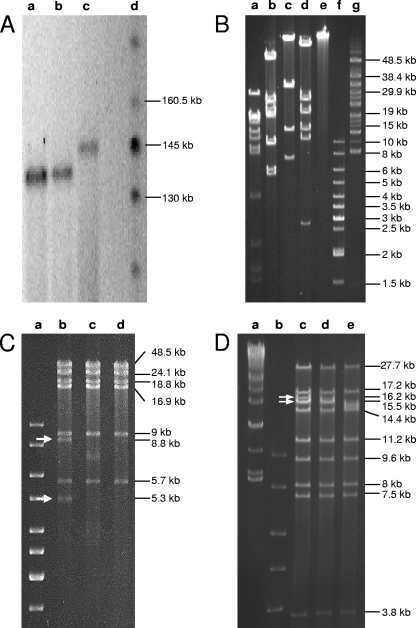
Structure analysis of phage P100 and K genomic DNAs. (A) PFGE of SwaI-digested phage P100 genomic DNA pretreated with Bal 31 for the indicated time intervals. Lanes: a, DNA length standard; b, untreated control DNA digested with SwaI; c, P100 with Bal 31 treatment for 5 min; d, P100 with Bal 31 treatment for 10 min. (B) PFGE of PflMI-digested phage K genomic DNA pretreated with nuclease Bal 31 for the indicated time intervals. Lanes: a and b, DNA length standards; c, untreated control DNA digested with PflMI; d, K with Bal 31 treatment for 5 min; e, K with Bal 31 treatment for 10 min; f, K with Bal 31 treatment for 20 min; g, K with Bal 31 treatment for 40 min. The terminal fragments disappearing over time are indicated.
FIG. 2.
Enzymatic analysis of A511 genomic DNA. (A) PFGE separation of full-length phage DNAs. Lanes: a, A511; b, P100; c, K; d, DNA length standard. The image has been inverted for better visualization. (B) PFGE of phage A511 DNA digested with XcmI (a), SwaI (b), ClaI (c), EcoRI (d), undigested control DNA (e), and DNA length standards (f and g). (C) PFGE of SwaI-digested A511 DNA after treatment with nuclease Bal 31 for the indicated time intervals. Lanes: a, DNA length standard; b, untreated control DNA digested with SwaI; c, A511 with Bal31 treatment for 5 min; d, A511 with Bal 31 treatment for 10 min. Restriction fragments disappearing over time are indicated by arrows. (D) PFGE of XcmI-digested phage A511 genomic DNA after Bal 31 treatment for indicated time intervals. Lanes: a and b, DNA length standards; c, untreated control DNA (XcmI digested); d, A511 with Bal 31 treatment for 5 min; e, A511 with Bal 31 treatment for 10 min. Arrows point to the terminal fragments disappearing over time.
Genome sequencing.
For the preparation of phage DNA libraries, purified A511 DNA was fragmented by using a nebulizer (GATC Biotech, Constance, Germany). Fragments of the desired length were recovered from agarose gels and ligated into EcoRV-digested pBluescript II SK(−) (Stratagene). Ligation products were transformed into E. coli DH5αMRC via electroporation. Blue-white screening on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing plates was used to identify plasmid-bearing clones. The inserts of isolated plasmids were sequenced, using M13 forward and M13 reverse primers. Obtained nucleotide sequences were edited and aligned using Vector NTI Advance version 10.3 (Invitrogen) and DNASIS MAX version 2.6 (Miraibio, San Francisco, CA) software. Gaps between remaining contigs were closed by a primer walking strategy using purified genomic DNA as the template. Primers were derived from the contig sequences as they became available (13). Sequences with insufficient redundancy (<3-fold coverage) or sequences containing ambiguities were verified by primer walking with appropriately designed primers or by sequencing PCR amplification products encompassing the region of interest.
PCRs were performed using Taq polymerase (Fermentas) by using the following temperature profiles: 2 min initial denaturation at 95°C followed by 30 cycles at 95°C for 15 s, 60°C for 35 s, and 72°C for 3 min, followed by a final elongation step at 72°C for 8 min. PCR products were purified (GenElute PCR cleanup kit; Sigma) and sequenced. The specific genome end fragments purified from SwaI digests were used as templates for direct runoff sequencing of termini. All primers used are listed in Table 1.
TABLE 1.
Oligonucleotide primers used for amplification and sequencing of the genome end fragments
| Primer | Sequence (5′-3′)a | Length(s) of target fragment(s) (bp)b |
|---|---|---|
| P1 | CAGGGGGTACCTTCGAGGCTAGC | 5,321 and 8,803 |
| P8_comp | GGTAATTATCCCCACAACCCCGC | 5,321 and 8,803 |
| P2inv | CGGGTGTTTACAAGGTTATTGGTGTG | 5,321 |
| P3 | ACGCACGTAGATACTTGTCATCATCG | 5,321 and 8,803 |
| P8inv | GCGGGGTTGTGGGGATAATTACC | 5,321 |
| P3inv | CGATGATGACAAGTATCTACGTGCGT | 5,321 |
| P4b | GCATATCCATGGTCTGCCTC | 5,321 |
| P11_comp | CCACCTACAATACCTGCCGATTCC | 5,321 |
| P10inv | GAGGTGACTCGGCGGAGTCTGC | 5,321 |
| P0 | CAGGGGGTACCTTCGAGGCTAGC | 8,803 |
| P28c_P25_int | CTGAAAACAAAAATTACGAGTTAGG | 8,803 |
| P28c_P25_int_rev | ATCCTCATCCTTGTGTTCTCC | 8,803 |
| P29c_P23_int | GAGTTAAGAAAAAAAAACTAGGAGCG | 8,803 |
| P29c_P23_int_rev | ATCGTTTTCTACTGTAGCATCCC | 8,803 |
| P22_P2_int | GGGAGAAAGTTAAGCCTATGC | 8,803 |
| P22_P2_int_rev | ACAGCTTGGTTGTCTATACTACC | 8,803 |
| P9_comp | CCTGTGTAGTTACCTCTTCCCGC | 8,803 |
| P2 | CACACCAATAACCTTGTAAACACCCG | 8,803 |
| P22_lead | GGGAAATGCAGAGGGGTGGGACAC | 8,803 |
| P31c | ATCTACTATTTTTTGGACTACCTGC | 8,803 |
| P23_lead | GACGGTACAGGAAACGAATATCC | 8,803 |
| P30c | CGCTTGTTCGTCTTCGTAAGTACC | 8,803 |
| P24_lead | GCGTGGATATTCAAATGGACATGC | 8,803 |
| P29c | TGCTTATTATCTAAAGTCAACAC | 8,803 |
| P25_lead | GGAAATGGAAGACTTGGCAGAGG | 8,803 |
| P28c | GCGGTCTAGTTCATAGGCTAGTTCG | 8,803 |
| P26 | CCTAGAAAGATTAGCGGAAGGGG | 8,803 |
| P27 | GTAAGAGGAAATGCAACTACAGAGC | 8,803 |
| P_runoff5321_1 | CCTTTCTTATCCTGCACTTGG | 5,321 |
| P_runoff5321_2 | AGAAACTTACTATGGAACAACG | 5,321 |
| P_runoff8803_1 | CGAAAACATTATGGCAAGTGTAGG | 8,803 |
| P_runoff8803_2 | TGTTGTTTTCCCCTGTTAGCTCG | 8,803 |
Primer sequences were deduced from core regions of SwaI-containing restriction fragments.
The column indicates whether primers were used for sequencing of the 5,321-bp SwaI fragment, the 8,803-bp SwaI fragment, or both fragments.
Electron microscopy.
Phages were negatively stained with 2% uranyl acetate, 2% Na-phosphotungstic acid, or 2% ammonium molybdate (56). The samples were observed in a Philips CM100 (100-kV acceleration voltage; FEI Company, Hillsboro, OR) equipped with a TVIPS Fastscan charge-coupled-device camera (Tietz Systems, Gauting, Germany) or in a Tecnai G2 Spirit instrument (at 120 kV) equipped with an Eagle charge-coupled-device camera (FEI Company).
For electron microscopy of phage DNA, samples were prepared by use of the microdiffusion technique (32) or by being spread over a water surface with formamide and carbonate buffer as described earlier (55). For length measurement, the DNA was photographed on 35-mm film and traced using a LM4 length-measuring device (Brühl, Nürnberg, Germany).
Mass spectrometry and peptide mass fingerprinting.
Phage proteins were separated by horizontal sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gradient gels (ExcelGel SDS gradient, 8 to 18%; GE Healthcare, Germany) as previously described (37, 72). For a molecular size marker, we used Dalton Mark VII-L (Sigma). Gels were stained with a colloidal Coomassie dye (PhastBlue R; GE Healthcare). Protein bands were excised from the SDS-PAGE gels and digested with trypsin, and eluted supernatants containing the peptide fragments were pooled and vacuum dried as described earlier (71). Peptides were dissolved in 5% formic acid, and matrix-assisted laser desorption ionization-mass spectrometry using a Bruker Reflex III time of flight mass spectrometer with a nitrogen UV laser and dual-channel plate detector (Bruker Daltonic, Bremen, Germany) was performed as previously described (71). An acceleration voltage of 20 kV was used, and the detector was adjusted to 1.7 kV.
Bioinformatics.
Nucleotide and amino acid sequence analyses and interpretation were performed using Vector NTI Advance (Invitrogen) and DNASIS MAX (Miraibio). Pairwise sequence alignments were done using the BLASTn, BLASTp, and tBLAST programs available at the NBCI website (1) or the DNASIS built-in BLAST engine. Multiple sequence alignments were conducted by ClustalW (DNASIS or the ClustalW tool online [http://www.ebi.ac.uk/clustalw/]). tRNAs were predicted using tRNAScan SE (43). Dot plots were generated with the EMBOSS dotpath program from the EMBOSS website (http://www.ch.embnet.org/EMBOSS) by using a word size of 8. Promoters were predicted using the Neural Network Promoter Prediction tool of the Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/promoter.html) or the HUSAR bioinformatics suite, available at the German Cancer Research Center website (http://genome.dkfz-heidelberg.de/). Prokaryotic factor-independent RNA polymerase terminator prediction was performed according to the method of Brendel and Trifonov (3) by using the HUSAR bioinformatics suite or by hairpin loop analysis (DNASIS).
Nucleotide sequence accession numbers.
The A511 sequence was deposited in GenBank under accession number DQ003638. Sequences of P100 (DQ004855), K (AY176327), and LP65 (AY682195) were retrieved from the same source, whereas the preliminary sequence of SPO1 was obtained from the Pittsburgh Bacteriophage Institute (http://pbi.bio.pitt.edu).
RESULTS
Morphologies of phages A511, P100, and K are highly similar.
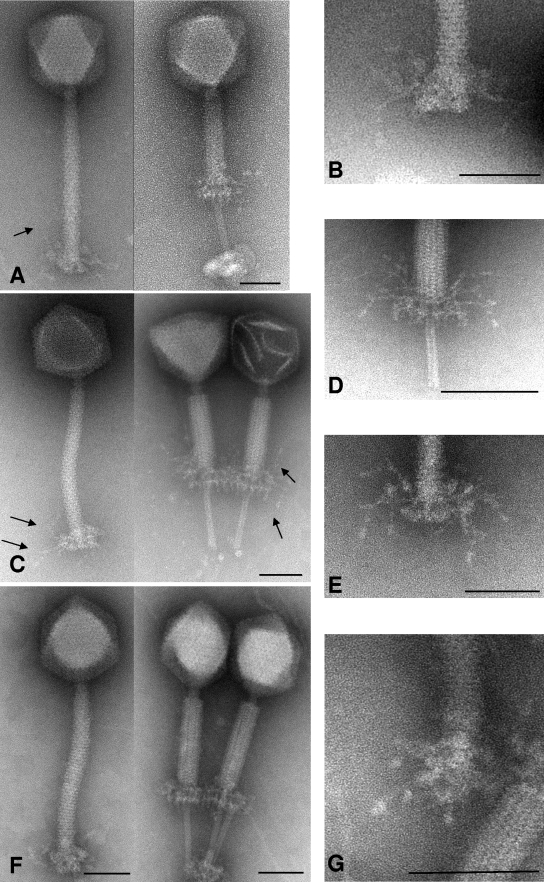
Transmission electron micrographs of negatively stained phages A511, P100, and K were obtained from purified phages (Fig. 1). Based on morphologies, all three phages belong to the SPO1-like group of the Myoviridae in the order Caudovirales; they feature a long, contractile, nonflexible tail and an isometric head. Virtually no morphological differences among the three phages A511, P100, and K were observed. The dimensions for phage K appeared to be slightly larger than those reported earlier (head diameter [flat-flat] = 93.83 nm [standard deviation {SD} = 4.02 nm; n = 20]; tail length = 218.92 nm [SD = 7.96 nm; n = 20]; tail diameter = 20.07 nm [SD = 2.24 nm; n = 20]). This may be due to variations in the negative staining techniques used (51). A511 is slightly smaller and features a head diameter of 87.36 nm (SD = 3.65 nm; n = 20), tail length of 199.44 nm (SD = 4.2 nm; n = 15), and tail diameter of 20.37 nm (SD = 2.17 nm; n = 20), confirming previous data (72). Phage P100 is virtually identical in its dimensions to A511 (head diameter = 89.55 nm [SD = 2.37 nm; n = 15]; tail length = 198.24 nm [SD = 5.48 nm; n = 15]; tail diameter = 19.0 nm [SD = 0.78 nm; n = 15]).
FIG. 1.
Electron micrographs of A511, P100, and K. (A) Phage A511 in a native and contracted state, negatively stained with uranyl acetate. Arrows indicate the thin, long tail fibers (whiskers) originating at the base plate. The inner tube extends approximately 70 nm beyond the contracted outer tail shaft and base plate. (B) Close-up view of A511 base plate stained with ammonium molybdate. (C) Phage P100, in noncontracted (left panel [ammonium molybdate]) and contracted status (right panel [stained with phosphotungstic acid]). Arrows indicate tail fibers. In the right panel, the lighter phage capsid (left phage) is packed with DNA, whereas the darker capsid (right phage) is empty as a result of DNA ejection. (D and E) Close-up views of P100 tail in contracted and noncontracted states (stained with phosphotungstic acid) including the long, flexible tail fibers. (F) Phage K with uncontracted (left) and contracted tail (right), stained with ammonium molybdate. (G) Close-up view of the K adsorption apparatus (stained with uranyl acetate) from diagonal below; the symmetrical arrangement of tail fibers is clearly visible. Bars represent 50 nm.
For the first time, we are able to document the presence of long tail fibers with a sixfold symmetry attached to the base plate regions of A511, P100, and K (Fig. 1). These whiskers are distinct from the short tail spikes present on the lower side of the base plate structure, which are best visible in images of contracted phage tails (e.g., Fig. 1D), and also exhibit a sixfold symmetry. Interestingly, the base plates of all three phages seem to undergo quite dramatic conformational changes during tail contraction, allowing better visualization of its structure and components (Fig. 1A, C, D, and F). As a result of contraction triggering, the base plate moves upwards and the tail tube is exposed and extends beneath the base plate and tail sheath. Similar observations have been made for another SPO1-like phage, LP65 of Lactobacillus (9), which also appears to resembles A511 in morphology and approximate dimensions. Via the tips of exposed tail tubes in the contracted state, virions frequently adhere to some unstructured material, probably representing cell wall debris from lysed host cells (Fig. 1F).
The A511 genome features nonpermuted, redundant ends.
A total of 574 individual sequences were obtained by using the shotgun approach, and remaining gaps in the contigs were closed by primer walking directly on the phage DNA. Both strands were sequenced at least twice, and an approximately fourfold overall coverage was sufficient to precisely assemble the phage genome. The 134,494-bp informational genome (see below) has a G+C content of 36.1 mol%, which agrees well with values for other Listeria phages (36, 71) and the host bacterium (5, 17, 46).
We were surprised to find that experimentally obtained restriction enzyme fragment patterns were not in agreement with those predicted from the sequence (Fig. 2B). In most cases, one predicted fragment was missing or the fragment was significantly shorter than calculated. At the same time, one or two unexpected (additional) fragments appeared. For example, ClaI and EcoRI digestion yielded additional fragments of 7,000 bp and 12,000 bp, respectively. Digestion with SwaI revealed that a 10,999-bp fragment was missing, but two others of approximately 5,300 bp and 8,800 bp appeared (the approximately 9-kb band in Fig. 2B is a double band). In all cases, the addition of the individual fragment sizes indicated a genome length of more than the obtained 134.5-kb unit sequence.
Heat treatment of the DNA fragments before electrophoresis did not alter the fragment patterns (data not shown), excluding the possibility of single-stranded, overlapping, and cohesive genome ends. Physical analysis by partial denaturation spreading of A511 DNA followed by electron microscopy suggested that the A511 DNA is not permuted and all molecules are identical in size and feature identical ends (results not shown). The size of A511 DNA measured on transmission electron micrographs was 137.8 ± 2.9 kb (n = 49), which is significant and indicated a redundancy of approximately 3.3 kb (data not shown). Further evidence for a larger DNA molecule was provided by pulsed-field gel electrophoresis of full-length A511 DNA (Fig. 2A), which clearly showed that the packaged A511 DNA is longer than the 134-kb unit sequence. This finding also applied to the DNA of phages P100 and K.
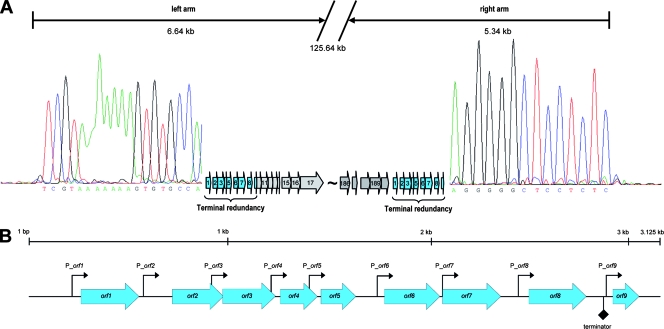
DNA treatment with Bal 31, an exonuclease that degrades double-stranded linear DNA from both ends simultaneously, and subsequent cleavage of the DNA by restriction enzymes revealed that only two specific restriction fragments were simultaneously shortened over time (Fig. 2), and these fragments therefore must represent the physical genome ends. Figure 2C shows an SwaI digest of Bal 31-treated DNA. The 8,803- and 5,321-bp fragments disappeared after 20 min. Figure 2D shows an XcmI digest of Bal 31-treated DNA; it was again obvious that two fragments disappear over time. These fragments were not predicted by in silico analysis of the unit genome. We concluded that A511 could not have a circularly permuted genome, because this would result in an even, simultaneous degradation of all restriction fragments (36). The two SwaI restriction fragments likely representing the genome ends (Fig. 2C) were used as templates for PCR, which was performed using a set of primers designed to amplify overlapping stretches of 500 to 1,500 nucleotides (nt) in the regions of interest (Table 1). The sequencing of the products enabled the determination of the core sequences of the left and right genome arms. Finally, runoff sequencing of the left and right ends (5,321- and 8,803-bp SwaI fragments, respectively) revealed the sequence of both ends and the extent of the terminal redundancy of 3,125 bp. When the reassembled genome was used for in silico restriction patterns prediction, results were then in perfect agreement with our experimental data (Fig. 2B).
Annotation of the A511 genome.
The exact length of the full-size DNA molecule packaged in the phage head is 137,619 bp, including the 3,125-bp terminal redundancy. The 134,494-bp unique (or unit) sequence is the designated informational genome. A total of 190 putative open reading frames (ORFs) and a cluster of 16 tRNA genes could be identified (see Table S1 in the supplemental material). Most ORFs feature a conserved, near-consensus ribosome binding site motif AGGAGGTG (17, 54), located 6 to 10 bp upstream of the translation initiation codon ATG, TTG, or GTG. The sizes of predicted proteins ranged from 3.7 kDa (gp187) to 145.8 kDa (gp104). However, putative functions could be assigned to only 39 of the predicted gene products (20.5%), based on BLAST analyses and amino acid sequence similarities (see Table S1 in the supplemental material). ORF numbering starts with bp 1 at the left genome end (see Table S1 in the supplemental material), and earlier designations (40, 42) were modified accordingly. With the terminal redundancy, nine duplicates (orf1 to orf9) are present in addition to the 190 unique putative ORFs. Most ORFs (149) use ATG start codons, whereas the remaining 41 ORFs feature GTG or TTG codons.
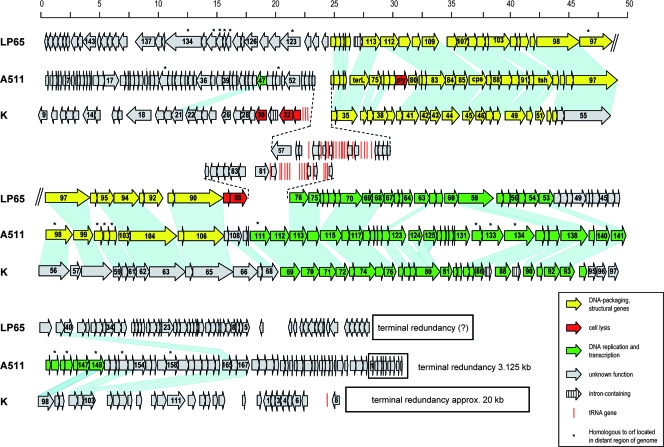
Overall, the A511 genome can be structured into three blocks featuring opposite transcription directions (orf1 to orf27, orf28 to orf69, and orf70 to orf190) (Fig. 3). Local sequence alignments revealed a further subdivision of the genome into functional modules. Figure 3 shows the genomic map of A511, aligned with those of phages K (47) and LP65 (9). The order of the genes in the early and late gene cluster appears relatively conserved, with few insertions or deletions. Some rearrangements among the layouts of these three phages are obvious, especially among the lysis genes and the tRNA gene clusters. Sequence alignments predicted that the A511 module encoding structural proteins, phage assembly proteins, and DNA packaging factors (late genes) is represented by orf70 to orf106. This region largely corresponds to the structural gene module of both LP65 and K; with sequence identities from 20 to 60% (see Table S1 in the supplemental material). Noteworthy is the unusual (compared to those of most other phages) location of the A511 endolysin gene, close to the large subunit of the terminase and the portal protein.
FIG. 3.
Alignment of genome maps of A511, Staphylococcus phage K, and Lactobacillus phage LP65. Predicted ORFs are represented by arrows drawn approximately to scale (in kb) and numbered consecutively. ORFs whose products show significant amino acid homologies are linked by shading. The terminal redundancies are indicated by boxes (putative for LP65). ORFs marked with an asterisk are homologous to ORFs located in distant regions of the genomes and could not be linked graphically (see Table S1 in the supplemental material for further details).
Researchers predicted two potential promoter structures, which face in opposite directions and feature near-consensus −10 and −35 motifs in comparison to those of housekeeping genes from Listeria (17), upstream of the late gene cluster specifying structural proteins in the intergenic space between orf69 and orf70 (genome coordinates 33,448 to 33,562).
Another cluster is formed by the genes encoding proteins with genome replication and decoding functions (orf111 to orf148); it is preceded by a possible promoter 35 to 74 bp upstream of the orf111 start. Encoded are replisome components, such as DNA polymerase (encoded by orf137 and orf138), helicase (encoded by orf111), primase (encoded by orf113 and orf117), and a putative sigma factor (encoded orf143). The integration host factor (gp136) is a member of the DNA binding protein family that binds and bends DNA, and is required in many cellular processes, including transcription, recombination, and higher-order nucleoprotein complex assembly (44). Again, significant similarities to products of K, P100, LP65, and others were observed in this cluster (Fig. 3; see Table S1 in the supplemental material). However, the order of the early genes varies among these phages, and the boundaries of this module are well defined.
Some bacterial viruses break down host nucleic acids to generate the building blocks required for large-scale synthesis of their own DNA. Three putative corresponding gene products involved in nucleotide metabolism are present in A511 (gp123 to gp125). The genes encode the alpha and beta subunits of ribonucleotide reductase, an enzyme responsible for the last step in the conversion of ribonucleosides into deoxyribonucleosides (27, 58).
A total of 16 tRNA genes is found clustered within the leftwards-oriented genes (orf28 to orf69), (Fig. 3; see Table S1 in the supplemental material). A511 uses the same tRNA species (same anticodons) as the Listeria host, but the nucleotide sequences of the tRNA molecules are mostly different, with the exception of tRNAGln. Moreover, frequencies of the individual A511 tRNA anticodons are somewhat different from those published for its bacterial host (17), and this reflects a slightly different codon usage. Some amino acids (e.g., Lys, Asn, Thr, and Gln) seem to be overrepresented among A511 structural proteins. Thus, the corresponding tRNAs may serve to avoid bottlenecks in large-scale synthesis of structural components. Phages K and LP65, as well as other phages of gram-positive and gram-negative bacteria, such as T4 (45) and T5 (64), also contain tRNA genes. However, their precise role is not clear.
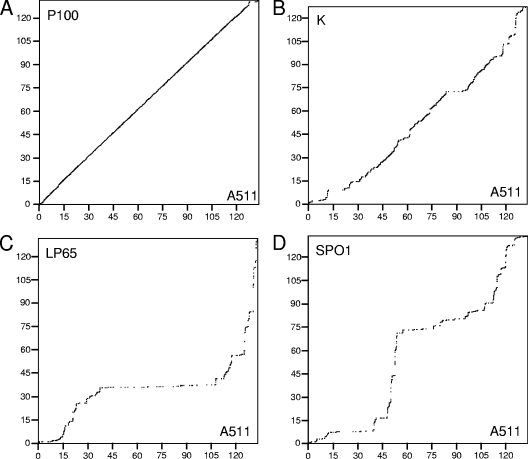
Whole-genome dot plot alignments comparing A511 to P100, K, LP65, and SPO1 are shown in Fig. 4. The extensive collinearity of A511 to P100 (Fig. 4A) underlines the close relationship of these phages. At the protein level, 167 predicted A511 proteins feature strong homologies (≥70% amino acid identity), whereas 23 gene products have no counterpart in P100. All tRNA genes identified in A511 are also present in P100. Surprisingly, A511 is also largely collinear to K, whereas the missing collinearity between A511 and LP65 or SPO1 suggests a major divergence in genome evolution.
FIG. 4.
Dot plot alignment of the A511 genome and related phages. Nonredundant, full-length dot plot alignments of A511 against three other phages were calculated using EMBOSS dotpath with a word size of 8 nt. The A511 informational genome sequence is represented by the horizontal x axis, and the scale ranges from 0 to 135 kb. The genome sequences of P100, K, LP65, and SPO1 were permuted in order to achieve maximum fit with the A511 genome. Therefore, the indicated genome coordinates (kb) for these phages do not correspond to the published genome sequence. (A) A511 aligned with P100 (131,384 bp). (B) A511 compared to K (127,395 bp). (C) A511 compared to LP65 (131,522 bp). (D) A511 aligned with SPO1 (133,391 bp).
In general, A511 shows homologies only to other members of the SPO1-like phages, whereas virtually no relatedness to any temperate phages was found. Only eight putative proteins have significant sequence similarity with proteins from temperate phages, mainly from Listeria phages, and six of the A511 early genes feature weak to moderate homologies to phages of enterobacteria (mainly T4 and T5). They encode proteins involved in DNA modification and repair (gp106, gp111, gp113, gp114, and gp115) and a putative serine-threonine phosphatase (gp28).
Information content of the terminal redundancy.
The 3.1-kb redundant terminal region of A511 contains nine very small putative ORFs (42 to 95 codons) (Fig. 5) with no sequence similarities of their putative products to any known protein (except the P100 homologues). In silico analyses using Pfam, Prosite, and Interpro Scan did also not reveal any specific characteristics or allocations to protein families or possible functions.
FIG. 5.
A511 genome ends and terminal redundancies. (A) Terminal runoff sequencing chromatograms of both ends of the phage genome (see the text) are shown to the left and right of the redundancies. The drop-offs indicate the physical ends of the sequenced strand of the A511 DNA molecule. The left-end sequence chromatogram has been inverted for reasons of clarity and shows the sequence of the complementary strand of the DNA. (B) Detailed map of the terminal redundancy region, including orf1 to orf9 and showing the location and orientation of predicted promoter (see Table 2) and transcription terminator sequences.
These nine ORFs are ordered sequentially, with comparatively large intergenic spaces of up to 170 bp. Surprisingly, each gene appears to features its own promoter sequence and ribosome binding site motif (Table 2). While the known Listeria σB motif (28) does not correspond to the −10 and −35 sequences of these promoters, they feature conserved −10 and −35 motifs known to direct expression of Listeria housekeeping genes (17) and motifs recognized by Bacillus σA (29). Thus, it is possible that their expression may be Listeria σA dependent. A strong, Rho-independent stem-loop transcription terminator with a free energy of −52.8 kJ/mol is located in the space between orf8 and orf9 (Fig. 5), which suggests that expression of this module may be independent of expression of the remaining A511 genes.
TABLE 2.
Putative promoters and Shine-Dalgarno sequences in the redundant terminal region (orf1 to orf9) of A511a
| Promoter or sequence typeb | −35 sequence | Spacer (bp)c | −10 sequence | Ribosome binding site sequence |
|---|---|---|---|---|
| Consensus | TTGACA | 17 ± 1 | TATAAT | AGGAGGTG |
| P_orf1 | TTGACt | 18 | TATAAT | gGGAGGTG |
| P_orf2 | TTGACt | 28 | TATAAa | AGGAGGTt |
| P_orf3 | TTagCA | 21 | TATtAT | AGGgGGaa |
| P_orf4 | cTGAgt | 19 | TcaAAa | AGGAGGaa |
| P_orf5 | TTagtA | 22 | TATttAT | AGGAGtaG |
| P_orf6 | gGAggA | 25 | TtgaAAT | AtGAGGgG |
| P_orf7 | TTACt | 15 | TtTAAT | AGGAGGTa |
| P_orf8 | TTGACt | 28 | TATAAT | AGGtGGTa |
| P_orf9 | TTcAac | 19 | TATAta | AGAGGgG |
Lowercase letters in the sequence indicate variations from the consensus sequence.
Promoters are named according to the ORFs they precede.
The lengths of the spacers between the −10 and −35 core motifs are given.
Similar genome structures in other SPO1-like phages.
Restriction patterns of P100 revealed the same differences between predicted and observed patterns (data not shown), and Bal 31-digestion of P100 DNA also indicated a terminally redundant, linear genome with invariable, noncohesive ends (Fig. 6A). The physical genome size determined by PFGE was 137 kb, very similar to that of A511 (Fig. 2A). Since the unit genome is 3.1 kb smaller than that of A511 (131,384 bp versus 134,494 bp) (7), the redundancy in P100 amounts to approximately 6 kb, more extensive than that in A511. This is also reflected in the abrupt disruption in collinearity between A511 and P100 and suggests a locally different genome organization.
We then wanted to see whether other non-Listeria SPO1-like phages also possess such genome structures. Staphylococcus phage K (with a 127,395-bp genome) was assumed to have a linear, nonpermuted genome without cohesive ends (51). However, we found that the terminal redundancy of K was quite extensive. PFGE of full-length phage DNA (Fig. 2A) and Bal 31-based genome analysis (Fig. 6B) indicated a redundant region of about 20 kb, corresponding to approximately the nucleotide positions 124000 to 5000 of the current genome map (47). This region encompasses orf117, orf118, and orf1 to orf7, which are unrelated to any other genes with the exception of orf6, which encodes a putative serine/threonine phosphatase. The likely genome ends are located in the region preceding the tRNA genes and structural component module (47). This arrangement is similar to what we found for A511, and it also aligns with the K genome in the region from nucleotide positions 119121 to 122530 with an overall similarity of 43.2%.
A511 structural proteins.
Proteins of the purified phage particles were separated by thin-layer SDS-PAGE and analyzed by mass spectrometry. Peptide fingerprints permitted allocation of 16 proteins bands to A511 gene products (Fig. 7). Four bands apparently contained more than one protein. Peptides corresponding to the major capsid (Cps or gp86) and tail sheath (Tsh or gp93) proteins (40) were also found in other bands of lower mass (∼45 kDa for Tsh, and ∼45 kDa and ∼32 kDa for Cps); possibly due to proteolytic cleavage or degradation. Similar observations were reported for Staphylococcus phage 812 (14), closely related to K. The A511 portal protein (gp83), with a predicted mass of 61.6 kDa, was also found in two bands with significantly lower molecular masses, suggesting posttranslational processing. An unusual finding was that bands 13 and 15 both contain products of orf145, which is not located in the late gene cluster but among genes specifying proteins probably involved in DNA replication and control.
FIG. 7.
Structural proteins of A511. An SDS-PAGE profile representing the structural protein profile of A511 is shown on the left; masses of the molecular size markers are indicated (in kDa). Assignment of the bands to gene products was based on matrix-assisted laser desorption ionization-mass spectrometry peptide fingerprinting; functional assignments of proteins and their probable location in the A511 virus are indicated on the right (the A511 image shown on the right was taken after negative staining with uranyl acetate and cropped from a scan of the original micrograph).
DISCUSSION
We have determined the sequence and genome structure of the broad-host-range, virulent Listeria myovirus A511. The nonredundant unit (informational) genome spans 134,494 bp. Genes are arranged in two major and clearly distinct clusters, encoding structural components and DNA replication functions. Such modular genome organization is very common among tailed phages and can be regarded as a universal paradigm in bacteriophage biology.
Other myoviruses of low-G+C-content host bacteria, such as Staphylococcus aureus phage K (47), Lactobacillus plantarum bacteriophage LP65 (9), and Enterococcus faecalis phage φEF24C (60), infect noncompatible hosts and should therefore be genetically isolated from each other and from A511. Therefore, the similarities regarding overall genome composition and the sequence homologies found among these phages are intriguing. Comparison of the genomic maps of A511, K, and LP65 revealed a flexible organization comprising life cycle-specific gene clusters of these virulent phage genomes, even more flexible than those of the temperate phages (36, 71). For example, cell lysis genes and tRNA gene clusters of A511, K, and LP65 are located at completely different linear coordinates within their genomes, highlighting the mosaicism among these phage genomes. However, the overall conserved organizations in large parts of the early and late gene clusters of A511, LP65, and K suggest a modular distribution and acquisition of these elements during the evolution of these viruses.
The A511 phage DNA molecule features two identical 3,125-bp terminal sequences, amounting to a total unit genome of 137,619 bp packaged into the phage capsid. The genes located within the redundancy are the first to enter and be expressed in the infected cell. This agrees well with the observation that they appear to be optimized for efficient early expression in the host environment, since the transcriptional units and individual genes feature near-consensus bacterial promoter sequences and ribosome binding sites, respectively. For SPO1, it has been proposed that the corresponding genes represent a “host-takeover module” (57), which paves the way for efficient phage replication. However, A511 gp1 to gp9 show no homology to SPO1 products or any other known proteins, and their precise role during the initial stages of the infection process remains to be determined.
Phages A511, P100, and K are virtually indistinguishable by morphology (Fig. 1), and, with respect to dimensions, mode of tail contraction, and base plate structure, also match with LP65 (9). This underlines the relationship of SPO1-like phages with host ranges including different bacterial genera and agrees well with the finding that morphology correlates well with similarities among the structural-component-related genes and proteins. In contrast, only little similarity was seen among the genes directing DNA replication and many other unknown functions, which suggests a more host-specific adaptation of these elements when we consider divergent evolution of these viruses alongside their host bacteria. Interestingly, six of these genes feature weak to moderate homologies to coliphages T4 and T5. Similar findings have also been reported for LP65 (9). These genes represent the only link of A511 to phages of gram-negative bacteria, underlining the relatively isolated position of the SPO1-like phages within the Myoviridae.
Clearly, high-frequency recombination among phages infecting common hosts occupying similar ecological niches will result in a high degree of mosaicism in local phage populations. Horizontal exchange of genetic modules of different sizes also takes place across the entire phage population, although at a much lower frequency (4, 20, 21). Homologous recombination certainly plays an important role in module exchange among viruses infecting related host bacteria. This is common in lysogens, from which the incoming phage can draw from the gene pool of prophages or cryptic phage remnants residing in the bacterial genomes (4, 8).
The increasing numbers of the available phage sequences, genome maps, and comparative alignments indicate that promiscuous mix-and-match of genetic information is a common feature also among virulent phages such as those in the SPO1 group, which are able to exchange genetic material only during the first stages of the infection process. Considering that these phages are generally able to infect only one genus of host bacteria but mostly feature a rather broad host range within this taxonomic group, one has to conclude that their relationship is based on divergence from a common ancestor infecting less-diversified host bacteria and/or from sporadically occurring intergeneric transduction events, followed by subsequent (illegitimate) recombination. Interestingly, A511 appears more closely related to Staphylococcus phages than to phages infecting Bacillus; the phylogeny of the host bacteria would suggest a different situation. However, the number of available sequences is much too limited for a reasonable phylogenetic analysis.
None of the known SPO1-like phages infects bacteria from more than one genus, although their hosts are all grouped into the phylum Firmicutes and are therefore more or less genetically linked. This is especially true for the hosts of the phages described in this paper, which all belong to the class Bacilli and the orders Bacillales (Listeria, Staphylococcus, and Bacillus) and Lactobacillales (Lactobacillus). Interestingly, it is not known whether SPO1-like phages for other classes within this phylum (e.g., Clostridia) exist.
It has been demonstrated that bacteriophages with circularly permuted genomes can transduce genetic markers, whereas phages featuring invariable genome ends do not (22, 36, 71). The inability of A511 to transduce can now be explained by the structure of its genome, and this explanation likely holds true for other SPO1-like phages. Initiation of DNA packaging by terminase is probably dependent on recognition of a specific (yet unidentified) DNA sequence motif (pac site) (24), which prevents accidental packaging of nonphage bacterial or plasmid DNA.
The presence of terminally redundant, nonpermuted genomes in myoviruses infecting gram-positive bacteria was reported for Bacillus phage SPO1 (deduced from restriction maps and DNA-DNA hybridization) (50) and supposed for phage SP82 (10, 34, 49, 50). The similarity of Lactobacillus phage LP65 to SPO1-like myoviruses was mentioned previously (9), but the authors of that study did not investigate the genome structure. We have shown that, besides that of A511, the DNA molecules of both P100 and K have redundant end regions. Interestingly, the extent of terminal redundancy in these otherwise similar phages varies greatly, from 3.1 kb in A511 to approximately 20 kb in K. In fact, our data indicate that this is a general feature of the SPO1-like phages. The sequence of Enterococcus faecalis phage φEF24C has been published very recently (60) and features significant homologies to A511 (47% overall nucleic acid identity and 64 proteins with >33.7% identity [expectation values of <0.001]). Although φEF24C was classified as being a SPO1-like phage, the authors of that publication assumed that it has a circularly permuted genome. Considering the findings presented here, however, it is not unlikely that this phage also features redundant genome ends. In contrast, the giant Bacillus thuringiensis phage 0305ϕ8-36, which also features 6.5-kb terminal redundancy (59), probably represents a lineage of its own. It is clearly different from the SPO1-like phages with respect to morphology, genome size, and its complete lack of sequence homologies.
In conclusion, it can be anticipated that many of the yet-uncharacterized myoviruses mentioned in the literature, those infecting not only Listeria, Staphylococcus, and Bacillus but also other genera such as Enterococcus and Brochothrix (25, 30), will fall into this group of broad-host-range, virulent SPO1-like phages featuring large, terminally redundant genomes. A511 represents a typical example and suitable model to study this evolutionarily very successful class of bacterial viruses.
Supplementary Material
Acknowledgments
We are grateful to Siegfried Scherer (Technical University Munich, Germany) and the BMBF-funded PathoGenoMik Competence Network (Würzburg, Germany) for financial support to J.D. and M.J.L.
We thank Parviz Sabour (Guelph Food Research Centre, Canada) for the gift of phage K, Hans-Wolfgang Ackermann (Quebec, Canada) for providing an A511 micrograph (Fig. 7), and Sherwood Casjens (University of Utah) for helpful comments and suggestions.
Footnotes
Published ahead of print on 20 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahk, J., and E. H. Marth. 1990. Listeriosis and Listeria monocytogenes, p. 247-257. In D. O. Cliver (ed.), Foodborne diseases. Academic Press, San Diego, CA.
- 3.Brendel, V., and E. N. Trifonov. 1984. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 124411-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchrieser, C., C. Rusniok, F. Kunst, P. Cossart, and P. Glaser. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35207-213. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, A. 2003. The future of bacteriophage biology. Nat. Rev. Genet. 4471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlton, R. M., W. H. Noordman, B. Biswas, E. D. de Meester, and M. J. Loessner. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43301-312. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed bacteriophages. Curr. Opin. Microbiol. 8451-458. [DOI] [PubMed] [Google Scholar]
- 9.Chibani-Chennoufi, S., M. L. Dillmann, L. Marvin-Guy, S. Rami-Shojaei, and H. Brussow. 2004. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 1867069-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cregg, J. M., and C. R. Stewart. 1978. Terminal redundancy of “high frequency of recombination” markers of Bacillus subtilis phage SPO1. Virology 86530-541. [DOI] [PubMed] [Google Scholar]
- 11.Cregg, J. M., and C. R. Stewart. 1977. Timing of initiation of DNA replication in SPO1 infection of Bacillus subtilis. Virology 80289-296. [DOI] [PubMed] [Google Scholar]
- 12.Curran, J. F., and C. R. Stewart. 1985. Cloning and mapping of the SPO1 genome. Virology 14278-97. [DOI] [PubMed] [Google Scholar]
- 13.Dorscht, J. 2007. Comparative genomics of Listeria bacteriophages. Ph.D. thesis. Technical University of Munich, Freising, Germany.
- 14.Eyer, L., R. Pantucek, Z. Zdrahal, H. Konecna, P. Kasparek, V. Ruzickova, L. Hernychova, J. Preisler, and J. Doskar. 2007. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 764-72. [DOI] [PubMed] [Google Scholar]
- 15.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaeng, S., S. Scherer, H. Neve, and M. J. Loessner. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 662951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294849-852. [DOI] [PubMed] [Google Scholar]
- 18.Glassberg, J., M. Franck, and C. R. Stewart. 1977. Multiple origins of replication for Bacillus subtilis phage SPO1. Virology 78433-441. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich-Blair, H., V. Scarlato, J. M. Gott, M. Q. Xu, and D. A. Shub. 1990. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell 63417-424. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6506-511. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix, R. W., G. F. Hatfull, and M. C. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154253-257. [DOI] [PubMed] [Google Scholar]
- 22.Hodgson, D. A. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35312-323. [DOI] [PubMed] [Google Scholar]
- 23.Hoet, P., M. Coene, and C. Cocito. 1983. Comparison of the physical maps and redundant ends of the chromosomes of phages 2C, SP01, SP82 and phi e. Eur. J. Biochem. 13263-67. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118365-388. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis, A. W., L. J. Collins, and H. W. Ackermann. 1993. A study of five bacteriophages of the Myoviridae family which replicate on different gram-positive bacteria. Arch. Virol. 13375-84. [DOI] [PubMed] [Google Scholar]
- 26.Jemmi, T., and R. Stephan. 2006. Listeria monocytogenes: food-borne pathogen and hygiene indicator. Rev. Sci. Tech. 25571-580. [PubMed] [Google Scholar]
- 27.Jordan, A., and P. Reichard. 1999. Ribonucleotide reductases. Annu. Rev. Biochem. 6771-98. [DOI] [PubMed] [Google Scholar]
- 28.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 1855722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenney, T. J., and C. P. Moran, Jr. 1991. Genetic evidence for interaction of sigma A with two promoters in Bacillus subtilis. J. Bacteriol. 1733282-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 1025174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landthaler, M., N. C. Lau, and D. A. Shub. 2004. Group I intron homing in Bacillus phages SPO1 and SP82: a gene conversion event initiated by a nicking homing endonuclease. J. Bacteriol. 1864307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang, D., and M. Mitani. 1970. Simplified quantitative electron microscopy of biopolymers. Biopolymers 9373-379. [DOI] [PubMed] [Google Scholar]
- 33.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 1844177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrie, J. M., and H. R. Whiteley. 1977. A physical map of bacteriophage SP82 DNA. Gene 2233-250. [Google Scholar]
- 35.Loessner, M. J. 1991. Improved procedure for bacteriophage typing of Listeria strains and evaluation of new phages. Appl. Environ. Microbiol. 57882-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35324-340. [DOI] [PubMed] [Google Scholar]
- 37.Loessner, M. J., I. B. Krause, T. Henle, and S. Scherer. 1994. Structural proteins and DNA characteristics of 14 Listeria typing bacteriophages. J. Gen. Virol. 75701-710. [DOI] [PubMed] [Google Scholar]
- 38.Loessner, M. J., and C. E. Rees. 2005. Listeria phages: basics and applications, p. 362-379. In M. K. Waldor, D. I. Friedman, and L. A. Sankar (ed.), Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 39.Loessner, M. J., C. E. Rees, G. S. Stewart, and S. Scherer. 1996. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl. Environ. Microbiol. 621133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loessner, M. J., and S. Scherer. 1995. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J. Bacteriol. 1776601-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 623057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 161231-1241. [DOI] [PubMed] [Google Scholar]
- 43.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch, T. W., E. K. Read, A. N. Mattis, J. F. Gardner, and P. A. Rice. 2003. Integration host factor: putting a twist on protein-DNA recognition. J. Mol. Biol. 330493-502. [DOI] [PubMed] [Google Scholar]
- 45.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 6786-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 322386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 1862862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliver, H. F., M. Wiedmann, and K. J. Boor. 2007. Environmental reservoir and transmission into the mammalian host, p. 111-138. In H. Goldfine and H. Shen (ed.), Listeria monocytogenes (pathogenesis and host response), 1st ed. Springer Science, New York, NY.
- 49.Panganiban, A. T., and H. R. Whiteley. 1981. Analysis of bacteriophage SP82 major “early” in vitro transcripts. J. Virol. 37372-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pero, J., N. M. Hannett, and C. Talkington. 1979. Restriction cleavage map of SP01 DNA: general location of early, middle, and late genes. J. Virol. 31156-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rees, P. J., and B. A. Fry. 1981. The morphology of staphylococcal bacteriophage K and DNA metabolism in infected Staphylococcus aureus. J. Gen. Virol. 53293-307. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, 3rd ed., vol. 1-3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 53.Sampath, A., and C. R. Stewart. 2004. Roles of genes 44, 50, and 51 in regulating gene expression and host takeover during infection of Bacillus subtilis by bacteriophage SPO1. J. Bacteriol. 1861785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schurr, T., E. Nadir, and H. Margalit. 1993. Identification and characterization of E. coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 214019-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spiess, E., and R. Lurz. 1988. Electron microscopic analysis of nucleic acids and nucleic acid-protein complexes. Methods Microbiol. 20293-323. [Google Scholar]
- 56.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200351-365. [DOI] [PubMed] [Google Scholar]
- 57.Stewart, C. R., I. Gaslightwala, K. Hinata, K. A. Krolikowski, D. S. Needleman, A. S. Peng, M. A. Peterman, A. Tobias, and P. Wei. 1998. Genes and regulatory sites of the “host-takeover module” in the terminal redundancy of Bacillus subtilis bacteriophage SPO1. Virology 246329-340. [DOI] [PubMed] [Google Scholar]
- 58.Stubbe, J. 1998. Ribonucleotide reductases in the twenty-first century. Proc. Natl. Acad. Sci. USA 952723-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas, J. A., S. C. Hardies, M. Rolando, S. J. Hayes, K. Lieman, C. A. Carroll, S. T. Weintraub, and P. Serwer. 2007. Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305varphi8-36. Virology 368405-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchiyama, J., M. Rashel, I. Takemura, H. Wakiguchi, and S. Matsuzaki. 2 May 2008. In silico and in vivo evaluation of bacteriophage phiEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl. Environ. Microbiol. doi: 10.1128/AEM.02371-07. [DOI] [PMC free article] [PubMed]
- 61.van der Mee-Marquet, N., M. Loessner, and A. Audurier. 1997. Evaluation of seven experimental phages for inclusion in the international phage set for the epidemiological typing of Listeria monocytogenes. Appl. Environ. Microbiol. 633374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Wang, J., Y. Jiang, M. Vincent, Y. Sun, H. Yu, J. Wang, Q. Bao, H. Kong, and S. Hu. 2005. Complete genome sequence of bacteriophage T5. Virology 33245-65. [DOI] [PubMed] [Google Scholar]
- 65.Webb, V., E. Leduc, and G. B. Spiegelman. 1982. Burst size of bacteriophage SP82 as a function of growth rate of its host Bacillus subtilis. Can. J. Microbiol. 281277-1280. [DOI] [PubMed] [Google Scholar]
- 66.Weis, J., and H. P. Seeliger. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 3029-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wendlinger, G., M. J. Loessner, and S. Scherer. 1996. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 142985-992. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, D. L., and L. P. Gage. 1971. Certain aspects of SPO1 development. J. Mol. Biol. 57297-300. [DOI] [PubMed] [Google Scholar]
- 69.Reference deleted.
- 70.Reference deleted.
- 71.Zimmer, M., E. Sattelberger, R. B. Inman, R. Calendar, and M. J. Loessner. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50303-317. [DOI] [PubMed] [Google Scholar]
- 72.Zink, R., and M. J. Loessner. 1992. Classification of virulent and temperate bacteriophages of Listeria spp. on the basis of morphology and protein analysis. Appl. Environ. Microbiol. 58296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.