Abstract
The extracytoplasmic factor (ECF) sigma factor σE is one of the most studied sigma factors of Mycobacterium tuberculosis. It has been shown to be involved in virulence as well as in survival under conditions of high temperature, alkaline pH, and exposure to detergents and oxidative stress. Unlike many ECF sigma factors, σE does not directly regulate the transcription of its own gene. Two promoters have been identified upstream of the sigE gene; one is regulated by the two-component system MprAB, while the other has been shown to be σH dependent. In this paper, we further characterize the regulation of σE by identifying its anti-sigma factor and a previously unknown promoter. Finally, we show that sigE can be translated from three different translational start codons, depending on the promoter used. Taken together, our data demonstrate that σE not only is subjected to complex transcriptional regulation but is also controlled at the translational and posttranslational levels.
Sigma factors are interchangeable RNA polymerase (RNAP) subunits allowing recognition of specific promoter sequences. They are often subjected to both transcriptional and posttranslational regulation (11, 12). One of the best-characterized Mycobacterium tuberculosis sigma factors, σE (29), has been shown to be induced during growth in human macrophages (8, 30) and in response to various stress conditions, such as detergent-mediated surface stress, alkaline pH, heat shock, and oxidative stress (10, 18, 21, 28). An M. tuberculosis mutant lacking sigE was shown to be more sensitive to several environmental stresses (22), had reduced growth in both macrophages and dendritic cells (7, 22), and was severely attenuated in a mouse model of infection (2, 19). Interestingly, dendritic cells infected with the sigE mutant strain secrete larger amounts of interleukin 10 than those infected with the wild-type parental strain, suggesting that the lack of a functional σE leads to a different interaction of the bacterium with the immune system (7).
Two promoters have been described for sigE transcription. The first one was recently shown to be regulated by the two-component system MprAB in response to surface stress and exposure to alkaline pH (10, 26). Its transcriptional start point (TSP) corresponds to the first nucleotide of the putative σE translation start codon, as annotated in the TubercuList (http://genolist.pasteur.fr/TubercuList/). The second one has a clear σH consensus sequence and is responsible for the σH-dependent induction of sigE after heat shock and exposure to diamide both in M. tuberculosis (28) and in Mycobacterium leprae (34). Surprisingly, it is located 63 bp inside the putative σE coding region, leading some investigators to hypothesize that the actual annotation could be wrong (34). Moreover, the Rv1222 gene, the gene located immediately downstream of sigE, has been proposed to encode a member of the zinc-associated anti-sigma factor (ZAS) family, based on the typical HXXXCXXC motif found in its sequence (25). The members of the ZAS family are proteins able to bind specifically to sigma factors and inhibit their activities and are represented by Streptomyces coelicolor RsrA (15), Rhodobacter sphaeroides ChrR (3), and M. tuberculosis RshA (31) and RslA (9).
In this paper, we show that Rv1222 is indeed a σE-specific anti-sigma factor, and we characterize a third promoter responsible for sigE transcription. Moreover, we show that σE can be translated from three alternative translational start codons, resulting in different isoforms depending on the environmental cues sensed by the bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strains HB101, TOP-10, DH5α, BHT101, and MM294 were grown in Luria broth (Difco) at 37°C or 25°C. When required, antibiotics were added at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml; ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; streptomycin, 20 μg/ml; and hygromycin, 100 μg/ml. In two-hybrid experiments, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) were used at final concentrations of 40 μg/ml and 0.5 mM, respectively.
M. tuberculosis H37Rv was grown in either Middlebrook 7H9 liquid medium or Middlebrook 7H10 solid medium (Difco) supplemented with ADN (2% glucose, 5% bovine serum albumin, 0.85% NaCl) and 0.05% Tween 80. Liquid cultures were grown in roller bottles at 37°C. Plates were incubated at 37°C in sealed plastic bags.
In vitro transcription assay.
In vitro transcription assays were performed as described previously (4, 5). Briefly, 300 fmol of Mycobacterium smegmatis RNAP was mixed with 3 pmol of His-tagged purified M. tuberuculosis sigma factors (14). Anti-sigma factors were added to the reaction mixture when indicated (1.5, 3, 6, and 12 pmol for RseA; 3, 6, and 12 pmol for RshA and RslA). Transcripts were detected using a primer extension assay (4).
DNA manipulation.
All recombinant DNA techniques were performed according to standard procedures using E. coli HB101, TOP-10, and DH5α or E. coli MM294 as the initial host. DNA restriction and modifying enzymes were obtained from New England Biolabs and used according to the manufacturer's recommendations.
GST pull-down assay.
Glutathione S-transferase (GST) pull-down assays were conducted as reported by Beaucher et al. (4). σE was fused to GST, mixed with His6-tagged RseA, and analyzed by immunoblotting using anti-His antibody.
Two-hybrid assay.
DNA fragments coding for σE257 (775 bp) and σE215 (645 bp) were amplified from M. tuberculosis H37Rv chromosomal DNA and cloned into pUT18 to create a 5′ in-frame fusion with the sequence encoding the Bordetella pertussis CyaA fragment T18 (pEL60 and pEL58, respectively). A 482-bp DNA fragment containing rseA was cloned in pKT25 to create a 3′ in-frame fusion with the sequence encoding the CyaA fragment T25 (pEL18) (16). Primers used to obtain these constructs are shown in Table S1 in the supplemental material. Plasmid couples pEL18-pEL60 (RseA-σE257), pEL18-pEL58 (RseA-σE215), and pUT18-pKT25 (empty plasmids) were cotransformed in E. coli BHT101, and transformants were selected on solid medium with appropriate selection. The resulting strains were analyzed by a β-galactosidase assay.
RNA extraction and quantitative RT-PCR.
RNA extraction and quantitative reverse transcription real-time PCR (RT-PCR) were performed using Sybr green master mix (Applied Biosystems) as previously described (17). Results were normalized to the amount of sigA mRNA (18). RNA samples that had not been reverse transcribed were included in all experiments to exclude significant DNA contamination. For each sample, melting curves were used to confirm the purity of the amplification products. Experiments were performed at least twice, starting from independent biological samples. Primers used are shown in Table S1 in the supplemental material.
5′ RACE PCR.
M. tuberculosis cultures grown to the exponential phase were treated with 5 mM diamide or 0.05% sodium dodecyl sulfate (SDS) for 60 or 90 min, respectively. 5′ rapid amplification of cDNA ends (RACE) was performed using the 5′/3′ RACE kit (Roche Molecular Biochemicals) according to the manufacturer's recommendations as previously described (21). Primers used are shown in Table S1 in the supplemental material.
Construction of lacZ translational fusions and single-nucleotide mutagenesis.
Primers for construction of the translational fusions shown in Fig. 6 are reported in Table S1 in the supplemental material. A 775-bp DNA fragment containing the first 240 bp of sigE and the 455 bp upstream of its annotated translational start codon was amplified and cloned into the HindIII site of pMYT131 (a pSM128 derivative) (D. Ghisotti, unpublished data) in order to create a translational fusion with lacZ (see Fig. 6B below). Single-nucleotide mutagenesis to obtain the constructs shown in Fig. 6B was performed by overlap extension PCR according to the procedure described by Ito et al. (13) based on three common primers and a mutagenic primer for each mutation. Common primers are represented by RP277 and RP282 (see Fig. S1 in the supplemental material), able to anneal to the regions flanking the 775-bp sigE region cloned in pMYT131, and RP278, which overlaps one of the two HindIII sites flanking the cloned fragment and contains a mismatch removing the restriction site. RP279, RP280, RP289, and RP281 represent the mutagenic primers. Mutagenesis was achieved in two PCR steps using construct 1 shown in Fig. 6B as a template. In the first step, two different PCRs were performed using the primer couple RP277-RP278 and the primer couple formed by one of the mutagenic primers (RP279, RP281, RP289, or RP280) and RP282. After gel purification, the two PCR products were mixed and used as a template for a second PCR step in which RP277 and RP282 were used. The resulting PCR product was gel purified and digested with HindIII to distinguish the products containing the desired mutations (13) (see Fig. S1 in the supplemental material). The selected DNA fragments containing the desired mutations were then ligated into pMYT131 in frame with lacZ. Finally, the resulting constructs were subjected to sequencing to confirm the introduction of the mutation.
FIG. 6.
Maps of the DNA fragments used to construct lacZ translational fusions. Putative translation start codons with their positions (in parentheses) are indicated. Mutated translation start codons are boxed. Inverted black triangles indicate the positions of frameshift mutations. The results of the β-galactosidase assay performed for M. tuberculosis are shown on the right (expressed in Miller units).
Construct 1 shown in Fig. 6C was obtained by PCR using primers RP120 and RP406, with construct 1 as a template and subsequent cloning in pMYT131. The resulting construct was used as a template for amplification with the upper primer RP120 and lower mutagenic primer RP460, RP461, RP589, RP407, or RP590 to obtain constructs 2 to 6 (see Fig. 6C below). Finally, the resulting constructs were subjected to sequencing to confirm the introduction of the mutation.
Electroporation of M. tuberculosis.
Preparation of electrocompetent bacteria and electroporation were performed as previously described (19).
β-Galactosidase assays.
To perform the two-hybrid assays, E. coli strains were grown at room temperature or at 37°C in overnight culture in the presence of 0.5 mM IPTG and appropriate antibiotics. For evaluation of translational fusions, M. tuberculosis strains were grown at 37°C to an optical density at 600 nm (OD600) of 0.2 to 0.3. β-Galactosidase assays were performed as previously described (23), and the enzymatic activity was expressed in Miller units with the following formula: A = (377.77 × OD420)/(time of incubation at 28°C × 0.3 × protein concentration in mg/ml) (23).
RESULTS
σE posttranslational regulation: Rv1222 (RseA) is a σE anti-sigma factor.
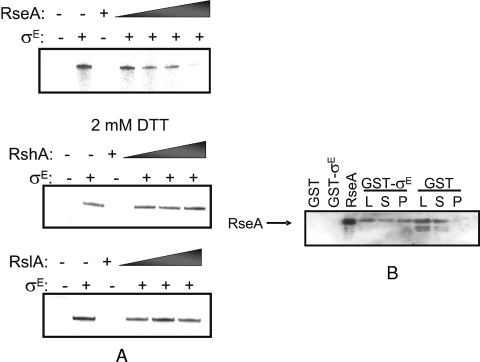
The activities of many M. tuberculosis sigma factors are regulated by antagonist proteins referred to as anti-sigma factors (12). Rv1222 has been proposed to be part of the ZAS family, the members of which all contain an HXXXCXXC motif and are located downstream of their cognate sigma factor-encoding gene (25, 29). In order to test this hypothesis, we conducted in vitro transcription assays with His6-tagged M. tuberculosis σE and RNAP purified from M. smegmatis at the M. tuberculosis sigB promoter, previously demonstrated to be transcribed by σE RNAP (21, 28). Figure 1A shows that increasing amounts of Rv1222 inhibit σE-dependent transcription in a dose-dependent manner. In contrast, the M. tuberculosis anti-sigma factors RshA and RslA (responsible for posttranslational regulation of M. tuberculosis σH and σL, respectively) (5, 31) have no effect on the activity of σE. GST pull-down assays were also conducted to demonstrate that Rv1222 interacts directly with σE (Fig. 1B), while GST alone does not. Taken together, these results show that Rv1222 is indeed an anti-sigma factor negatively regulating the activity of σE in M. tuberculosis, and we will now refer to this gene product as RseA (regulator of sigma E).
FIG. 1.
Rv1222 is a σE anti-sigma factor. (A) Rv1222 inhibits σE-dependent transcription in vitro in a dose-dependent manner. Increasing amounts (1.5, 3, 6, and 12 pmol) of Rv1222 were added to a reaction mixture containing M. smegmatis RNAP and σE. The addition of 3, 6, or 12 pmol of RshA (31) or RslA (5) under conditions where they show activities on their cognate sigma factors had no effect on σE-dependent transcription. DTT, dithiothreitol. (B) Rv1222 interacts with σE. σE was fused to GST, mixed with His-tagged Rv1222, and revealed by immunoblotting with an anti-His antibody. “L” represents a loading control which corresponds to 10% of the reaction mixture prior to any washes, “S” is the supernatant—or unbound Rv1222—recovered upon pelleting the beads immediately before the first wash step, and “P” denotes the bead pellet after all washes. No interaction was detected between GST and Rv1222.
sigE is transcribed from three different promoters.
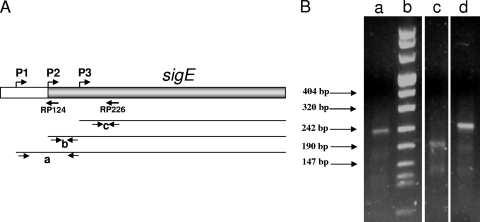
In M. tuberculosis, sigE can be transcribed from two different promoters, one positively regulated by the two-component system MprAB in response to surface stress (10) and one induced in response to heat shock and oxidative stress from the extracytoplasmic factor (ECF) sigma factor σH (28). However, in a previous work, Wu et al. (35) identified two additional sigE TSPs in Mycobacterium bovis BCG, mapping 223 bp and 117 bp upstream of the sigE translational start site. In order to characterize these TSPs in M. tuberculosis, we decided to redefine sigE TSPs in cells subjected to surface stress and oxidative stress as well as in unstressed cells by using the 5′ RACE technique. This technique uses a primer internal to the coding sequence to reverse transcribe the 5′ portion of the mRNA; a poly(A) is then added to the cDNA and is used to anchor a poly(T) primer to allow amplification (6) The amplification product is then sequenced to identify the TSP. In the first experiment, a primer homologous to the sequence immediately downstream of the σH-dependent promoter (RP226) (Fig. 2A) was used for the reverse transcription; under these experimental conditions, we could detect a single band corresponding to the MprAB-regulated promoter TSP (position +1) both in unstressed cells and in cells subjected to surface stress (Fig. 2B). However, when cells were exposed to oxidative stress, this band totally disappeared in favor of a faster-migrating band, corresponding to the TSP of the σH-dependent promoter (Fig. 2B). The first TSP corresponds to the first nucleotide of the putative translational start codon of σE, while the second one is internal to its putative open reading frame (position +63).
FIG. 2.
sigE promoters characterization. (A) Map of the sigE region. The sigE open reading frame is shown in gray, and the positions of the transcription start points of the three promoters (P1, P2, and P3) as well as of the two primers used for reverse transcription in the 5′ RACE experiments (RP124 and RP226) are shown. The three thin lines represent the transcripts originating from each of the three promoters. Arrow pairs below each transcript indicate the primer couples used in quantitative RT-PCR experiments. Couple “a” detects only transcripts originating from P1, couple “b” detects transcripts from P1 and P2, and couple “c” detects transcripts originating from all promoters. (B) Gel electrophoresis of the amplicons obtained by 5′ RACE (reverse transcription from RT226) from unstressed cells (lane a), cells subjected to diamide-induced oxidative stress (lane c), and cells subjected to SDS-induced surface stress (lane d). The molecular weight marker (marker 8; Roche) is shown in lane b.
In order to map other upstream TSPs, a second experiment was performed using unstressed cells. In this case, the mRNA was reverse transcribed starting from a different primer (RP124) whose sequence was located upstream of the one used to detect the other two promoters (Fig. 2B). In this case, we were able to detect a band, corresponding to a third TSP, located 55 bp upstream of the sigE translation start site (data not shown). However, we could not detect the TSPs at −223 bp and −117 bp, previously identified in M. bovis BCG (35). RT-PCR experiments also suggested that the region upstream of the −55 TSP is not transcribed in M. tuberculosis under this growth condition (data not shown). We propose to name the promoter located at −55 bp “P1,” the MprAB-regulated promoter “P2,” and the σH-dependent promoter “P3.” The positions of the three sigE TSPs are shown in Fig. 3.
FIG. 3.
Sequence of the sigE upstream region containing its three promoters. The gray box indicates the sigE coding region. Transcription start points are shown in capital letters, and positions −10 and −35 are underlined. MprA binding sites are boxed. Gray letters indicate residues protected by MprA binding (10).
sigE transcriptional regulation in response to stress.
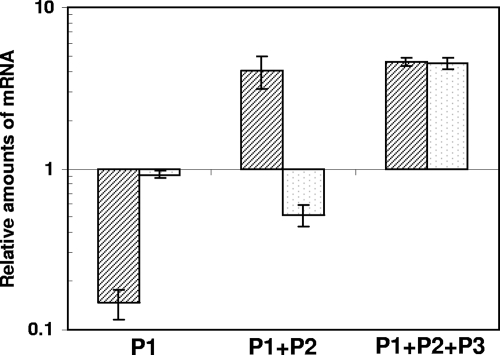
A quantitative RT-PCR assay was implemented to determine the contribution of each of the three promoters to sigE transcription under physiologic conditions and following stress. Three couples of primers were designed, the first able to detect cDNA derived from mRNA transcribed by all of the three promoters, the second able to detect cDNA derived from mRNA that started at both P1 and P2, and the third able to detect only cDNA derived from mRNA that started at P1 (Fig. 2A). Figure 4 shows the variation of the mRNA levels detected with these three couples of primers and normalized to the levels of sigA-specific mRNA, used as an internal invariant control (18). RNA was obtained from cells stressed with either SDS (to induce surface stress) or diamide (to induce oxidative stress) and from unstressed cells.
FIG. 4.
Changes in sigma factor mRNA levels after exposure to SDS-induced surface stress (black stripes) or diamide-induced oxidative stress (gray dots). Three different primer couples were used to evaluate the variation of transcripts initiated at P1, P1, and P2, or from all of the three promoters (P1+P2+P3) (Fig. 1A). Values are expressed as the ratio between the number of cDNA copies detected in samples obtained from the stressed cultures and the number of cDNA copies detected in samples obtained from unstressed bacteria. The values were normalized to the level of sigA cDNA, which represented the internal invariant control.
As expected, the total amount of transcripts due to the cumulative transcription from the three promoters increased under both stress conditions. However, the amount of mRNA transcribed from P1 and P2 increased following surface stress, but slightly decreased following oxidative stress, suggesting that induction in response to oxidative stress is due only to P3. However, the level of P1 transcripts strongly decreased following surface stress, suggesting that P1 and P2 are inversely regulated under these conditions. MprA has two binding sites (10); interestingly, one of them overlaps the P1 TSP, while the other falls immediately before P2 (Fig. 3). Since it was previously demonstrated that MprA is an activator of P2 (10, 26), it seems reasonable to think that the binding of MprA to this operator, while activating P2, represses the transcription of P1.
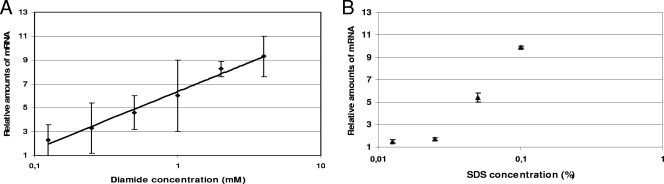
In order to better characterize the induction of sigE, we also performed two dose-response experiments measuring the sigE mRNA increase in response to different amounts of SDS and diamide. The amounts of sigE mRNA increased linearly with the logarithm of diamide concentration (Fig. 5A). However, when the same experiment was performed with SDS, the induction was almost absent up to a concentration of 0.025% and then increased linearly with the logarithm of SDS concentration (Fig. 5B).
FIG. 5.
Sigma factor mRNA levels after exposure to different diamide (A) or SDS (B) concentrations. Values are expressed as the ratio between the number of cDNA copies detected in samples obtained from the stressed cultures and the number of cDNA copies detected in samples obtained from unstressed bacteria. The values were normalized to the level of sigA cDNA, which represented the internal invariant control.
σE is translated from one canonical and two noncanonical initiation codons.
The positions of the two previously characterized TSPs (+1 bp and +63 bp of the annotated sigE putative open reading frame, respectively) led others to hypothesize that the annotated initiation codon was incorrect and that the translation of σE rather starts downstream of P3 (34). We have amplified the sigE upstream region and constructed a translational fusion between the annotated translational start codon of sigE and lacZ (deprived of its own translation start codon) (Fig. 6A). The construct was subcloned into an integrative vector and introduced into M. tuberculosis H37Rv. β-Galactosidase assays were next performed using cells either treated or not treated with SDS. As shown in Fig. 6A, β-galactosidase activity was detected in both cases but was higher with the SDS treatment, demonstrating that this translation start codon is functional and that the MprAB-dependent promoter produces a leaderless, yet translated, mRNA.
Although the mRNA molecules transcribed from the two upstream promoters can generate an open reading frame beginning at the annotated start codon, the transcription of P3 occurs downstream of this site and should result in a smaller σE isoform. In order to test if alternative translation start codons exist, the region upstream of sigE and the first 260 bp of the sigE coding region were amplified and fused in frame to lacZ (again, deprived of its own translation start codon) (Fig. 6B). This region was subjected to single-nucleotide mutagenesis to change the annotated translation start codon from ATG to ATC and introduce a frameshift mutation in order to eliminate translation from this site (Fig. 6B). The two resulting translational fusions were once again integrated into the M. tuberculosis genome, and β-galactosidase assays were performed using unstressed cells. As shown in Fig. 6B, β-galactosidase activity was detected in both cases, confirming the presence of an alternative translation start site downstream of P3.
The region downstream of P3 contains two possible alternative translation start codons in frame with sigE at positions +142 bp and +240 bp relative to the start codon of the annotated coding sequence (Fig. 6B). Following the previous scheme, single-nucleotide mutagenesis was used to abolish translation initiation from any of these codons. When the resulting constructs were introduced into M. tuberculosis, no change in β-galactosidase activity was observed, suggesting that none of the two potential translation initiation codons was functional in vivo (Fig. 6B). However, a frameshift mutation immediately downstream of the putative translation start codon at +142 bp totally abolished β-galactosidase activity (Fig. 6B), suggesting that the alternative translation start codon should be located between position +63 bp (P3) and position +142 bp.
Analyzing this sequence, we found two rare alternative start codons at positions +118 bp and +127 bp (ATC and TTG, respectively). Interestingly, a ribosome binding site (GGAGG) that was very well conserved was found 18 bp and 27 bp upstream of these codons, respectively.
A region of 628 bp including the sigE upstream region and its coding region up to the two alternative start codons, in which the translation start codon at position +1 bp was inactivated, was amplified and used to obtain translational fusion with lacZ (Fig. 6C). In order to study the role of the first alternative translation start codon, this construct was further mutagenized to obtain two other constructs where a frameshift mutation was introduced either immediately before or immediately after the ATC codon (+118 bp) (Fig. 6C). The resulting translational fusions were integrated into the M. tuberculosis genome, and β-galactosidase assays were performed. The construct with the wild-type sequence and the construct where the frameshift mutation was introduced before the ATC codon (Fig. 6C) had comparable activities. However, the construct with the frameshift downstream of the ATC codon showed an activity of about 50% of that obtained from the other two (Fig. 6C), strongly suggesting that this codon is only partially involved in the initiation of translation of the smaller isoform of σE.
To study the role of the second rare translational initiation codon (TTG), we constructed two additional mutants. In the first one, a frameshift mutation was introduced after the codon, while in the second one, the codon was changed from TTG to TTA but no frameshift was introduced (Fig. 6C). The first construct did not show any β-galactosidase activity, indicating that no translational initiation occurs downstream of this codon. However, the second construct showed a level of activity similar to that of construct 3, where a frameshift mutation was placed after the ATC codon and had about 50% of the activity of the wild-type sequence (Fig. 6C). Taken together, these data strongly suggest that both translational start sites are functional and that they participate equally in the translation of two smaller isoforms of σE. To confirm this hypothesis, we designed another construct, where both codons were mutated (Fig. 6C). In this case, the β-galactosidase activity decreased by approximately 90% with respect to the wild-type construct. Taken together, these data indicate that σE can exist in the following three isoforms: a larger one of 257 amino acids (aa) (σE257), and two smaller isoforms of 218 and 215 aa (σE218 and σE215, respectively).
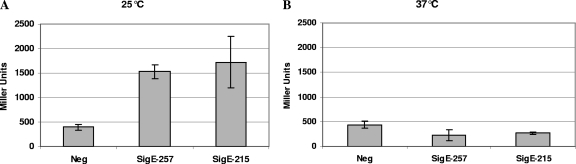
σE215 binds to the anti-sigma factor RseA as well as σE257.
We have shown that RseA is able to regulate the activity of σE257 (Fig. 1). In order to determine whether σE215 is also posttranslationally regulated by RseA, we performed a bacterial two-hybrid experiment based on the reconstitution in E. coli of the catalytic domain of B. pertussis adenylate cyclase CyaA formed by the two independently folding domains, T18 and T25 (16). As shown in Fig. 7A, when bacteria were grown at 25°C, almost the same amount of β-galactosidase activity (due to reconstitution of the CyaA catalytic domain) was detected when the plasmid encoding a T25-RseA fusion was cotransformed in E. coli BTH101 with either one of the two plasmids encoding the T18-σE257 or T18-σE215 fusion, suggesting that both isoforms are able to bind to RseA. Interestingly, when bacteria were grown at 37°C, β-galactosidase activity remained below the background level (Fig. 7B), which could suggest that the interaction between RseA and both of the two σE isoforms is sensitive to temperature.
FIG. 7.
Results of a β-galactosidase assay measuring the interaction of σE257 (SigE-257) and σE215 (SigE-215) with the anti-sigma factor RseA in a bacterial two-hybrid system based on the reconstruction of B. pertussis adenylate cyclase. (A) bacteria grown at 25°C; (B) bacteria grown at 37°C. Results are expressed in Miller units. Negative control (Neg) represents the background activity of E. coli cotransformed, with the two plasmids encoding the two fragments of adenylate cyclase.
DISCUSSION
The transcriptional regulation of sigE has been extensively studied in M. tuberculosis, M. bovis BCG, M. smegmatis, and M. leprae (20, 29, 34). The results of these studies indicate that sigE is transcribed from at least two promoters. The first one, whose TSP maps at the first codon of the putative sigE coding sequence, is under the control of the two-component system MprAB and is induced following surface stress and alkaline pH (10). The second one, whose TSP maps 63 bp inside of the putative sigE coding sequence, is transcribed from a σH RNAP promoter and is induced under conditions of oxidative stress and heat shock (28). In M. bovis BCG and in M. smegmatis, two other TSPs were mapped at 223 bp and 117 bp (M. bovis BCG) or at 157 bp or 85 bp (M. smegmatis) upstream of the sigE putative translational start site (35). These findings prompted us to verify if those promoters are also found in the region upstream of sigE in M. tuberculosis. Surprisingly, and in spite of the fact that the DNA sequences upstream of sigE are identical in M. tuberculosis and M. bovis BCG, we were unable to detect those promoters. However, we identified a TSP 55 bp upstream of the sigE putative coding region, preceded by a sequence with some similarity to a σE or σH consensus promoter sequence (Fig. 3). Interestingly, this TSP was previously identified by Wu et al. (35) in an in vitro transcription assay using reconstituted σE RNAP, even if the authors ascribed it to an artifact because the result could not be confirmed by primer extension in M. bovis BCG. These data stress once again that, despite the similarity between M. tuberculosis and M. bovis BCG genomes, extrapolation of gene regulation data between these two organisms is often misleading (20). Since the mRNA initiated at this TSP strongly decreased following SDS-mediated surface stress (which leads to the induction of sigE) (Fig. 4), it is unlikely that P1 is recognized by σE in vivo. This hypothesis is also supported by the fact that the P1 promoter sequence is distant (even if somewhat related because of degenerate bases) from the reported σE consensus promoter (32) and by the observation that the SDS-mediated induction of sigE is maintained in a sigE mutant strain (22). Since ECF sigma factors usually recognize similar promoters, it is possible that P1 is indeed recognized in vivo by an ECF sigma factor that is different from σE, which could be able to recognize it under less-stringent in vitro conditions. The fact that, unlike most of the ECF sigma factor genes, sigE is not directly autoregulated underscores the peculiarity of its transcriptional regulation (22).
The finding that following SDS-mediated surface stress, P1 is repressed while P2 is induced suggests that these two promoters are subjected to inverse regulation. Observing the sequence of this region, we found that one of the MprA binding boxes overlaps the P1 TSP, suggesting that MprA binding, while activating P2, inhibits transcription from P1 (Fig. 3). Interestingly, the same genome structure is conserved in other mycobacteria, such as M. bovis, Mycobacterium avium, M. leprae, and partially in M. smegmatis, suggesting that this regulatory mechanism evolved very early in the evolutionary history of mycobacteria (see Fig. S2 in the supplemental material).
The presence of an additional promoter (P3) inside the predicted sigE coding region suggests that either the real translation start codon is downstream of the predicted one, as hypothesized by other authors (34), or two σE isoforms are translated depending on which promoter is used for sigE transcription. The fact that translational fusions of lacZ with the predicted translation start codon were active and SDS inducible (Fig. 6A) strongly suggests that the latter hypothesis was correct and that a σE isoform of 257 aa can be translated from a larger mRNA starting at P1 during growth under physiologic conditions or from a shorter, leaderless mRNA starting at P2, following surface stress.
It has previously been shown that leaderless mRNAs bind preferentially to 70S ribosomes (compared to 30S subunits) and can be efficiently translated, starting with the A of the initiation codon (24, 33). For this reason, it has been proposed that they could be preferentially translated under adverse conditions in which 70S ribosomes are prevalent, such as carbon source downshifts, a stationary phase, and slow growth (24). The finding that several leaderless mRNAs in Streptomyces spp. encode proteins that confer resistance to antibiotics, which have the ribosome as a target, reinforces this hypothesis (24). Since several of the adverse conditions known to prevent 70S ribosome dissociation are probably experienced by M. tuberculosis during latency, the finding that sigE could be translated from a leaderless mRNA opens the interesting hypothesis that σE257 could be preferentially translated under these conditions.
In order to identify the translation start codon of the smaller σE isoform, we analyzed the P3 downstream region by single-nucleotide mutagenesis. Surprisingly, none of the two ATG codons present in the region was involved in σE translation (Fig. 6B). However, we identified two rare initiation codons immediately downstream of a well-conserved putative ribosome binding site able to initiate translation with almost the same activity (Fig. 6C). The σE isoforms translated from these start codons measure 218 and 215 aa, respectively. Several attempts to reveal the three σE isoforms in M. tuberculosis protein extracts by Western blotting and immunoprecipitation from cells growing under physiologic conditions or after stress were unsuccessful, probably due to their low levels of expression (data not shown).
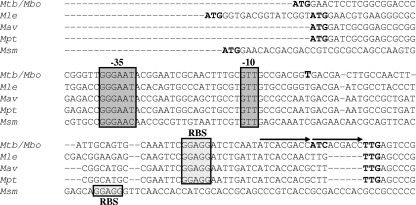
In order to evaluate if the presence of three σE isoforms was a peculiarity of M. tuberculosis or if it was conserved in other mycobacterial species, we compared the sigE coding regions of M. tuberculosis, M. leprae, M. avium, M. paratuberculosis, and M. smegmatis. It appears that an MprAB-regulated P2-like promoter and a downstream P3-like promoter are conserved in all of them (Fig. 8; see Fig. S1 in the supplemental material), suggesting that the presence of at least two σE isoforms is a common prerogative among mycobacteria. While the presence of the TTG rare initiation codon was conserved in all species, with the exception of M. smegmatis, the other rare initiation codon (ATC) was found only in M. tuberculosis. Interestingly, this region in M. tuberculosis and in M. bovis contains a 9-bp duplication, moving the TTG codon 6 bp apart from the ribosome binding site (Fig. 8). It is possible that before the occurrence of the duplication, coincident with M. tuberculosis speciation, the TTG codon was the only translational start codon of a single σE smaller isoform, and translational initiation from the ATC codon began only after it was moved at the critical distance from the ribosome binding site by the duplication.
FIG. 8.
Sequence of the first part of the sigE coding region in M. tuberculosis (Mtb), M. bovis (Mbo), M. leprae (Mle), M. avium (Mav), M. paratuberculosis (Mpt), and M. smegmatis (Msm). Putative translation start codons and P3 transcription start points are bold. P3 positions −35 and −10 are boxed, as well as the putative ribosome binding site (RBS). The two arrows show the 9-bp duplication (ATCACGACC) found in the M. tuberculosis and M. bovis sequences.
Sigma factors are often regulated at the posttranslational level by anti-sigma factors whose genes are usually located next to the gene encoding their cognate sigma factor. Since the gene downstream of sigE (Rv1222) shows an HXXXCXXC motif found in anti-sigma factors from the ZAS family, we designed in vitro transcription and GST pull-down experiments to determine if these two proteins were able to interact. As shown in Fig. 1, we demonstrate that Rv1222 could specifically bind and inhibit the transcription activity of σE257 in a dose-dependent manner, demonstrating that it is the σE257-specific anti-sigma factor, which we named RseA. In order to determine if also the smallest isoform σE215 was subjected to RseA posttranslational regulation, we performed a bacterial two-hybrid assay based on the reconstitution of the catalytic domain of B. pertussis adenylate cyclase CyaA (16). Two σE isoforms were fused at their C terminus to one fragment of the catalytic domain of CyaA, while RseA was fused at its N terminus to the other fragment. Both σE257 and σE215 were shown to be able to interact with RseA, indicating that both isoforms are posttranslationally regulated by this anti-sigma factor. Interestingly, the interaction was detected at 25°C and not at 37°C (Fig. 7), suggesting that RseA could respond to temperature. This finding is consistent with the fact that sigE plays a very important role in the heat shock response (14, 18). The other major cue sensed by SigE is surface stress (22). In proteobacteria, the ECF sigma factor responsible for the response to surface stress is posttranslationally regulated by an anti-sigma factor, whose activity is modulated by intramembrane proteolysis (1). Interestingly, the gene encoding M. tuberculosis RseA is followed by two cotranscribed genes. The first encodes a membrane serine protease (HtrA) similar to that involved in intramembrane proteolysis in proteobacteria, while the second encodes an integral membrane protein belonging to the twin-arginine translocation (Tat) system (TatB). The presence of two arginine residues in the RseA N terminus suggests that this protein could be targeted to the bacterial membrane through the Tat system to allow its degradation by HtrA (27). Experiments to address this hypothesis are actually in progress in our laboratory. Based on the fact that the role of σE in oxidative stress response seems to be minor (22), it appears less likely that RseA could directly sense redox potential, despite the fact that some ZAS anti-sigma factors do.
The translation of different σE isoforms in response to specific environmental conditions could suggest that these isoforms have different biological activities. Since domains involved in promoter recognition (regions 2 and 4) are entirely present in all of the three σE isoforms, it seems unlikely that they could recognize different promoters, although the possibility that conformational changes between the σE isoforms could lead to a different promoter specificity cannot be excluded. Interestingly, the N-terminal region of σE257 does not share homology with any other sigma factor and could be involved in protein stability and/or in the interaction with other regulatory proteins. Complementation experiments, where the sequences encoding the different σE isoforms will be reintroduced in a sigE mutant strain, will help to investigate the difference in their physiological functions.
Taken together, our data demonstrate that σE is subjected to complex regulation at transcriptional, translational, and posttranslational levels, demonstrating once again its fundamental importance in M. tuberculosis physiology.
Supplementary Material
Acknowledgments
This work was supported by MIUR-COFIN 2006 grant no. 2006064583 (awarded to R.M.). S.R. was supported by fellowships from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Fonds de la recherche en santé du Québec (FRSQ).
We thank Issar Smith for carefully reading the manuscript, Angela De Iuliis for helpful collaboration, Daniel Ladant for the two-hybrid system, Daniela Ghisotti for pMYT131, and Ida Rosenkrands for the anti-σE serum.
Footnotes
Published ahead of print on 7 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E. 2004. Control of the alternative sigma factor SigE in Escherichia coli. Curr. Opin. Microbiol. 7157-162. [DOI] [PubMed] [Google Scholar]
- 2.Ando, M., T. Yoshimatsu, C. Ko, P. J. Converse, and W. R. Bishai. 2003. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect. Immun. 717170-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, J. R., J. D. Newman, and T. J. Donohue. 2004. Interactions between the Rhodobacter sphaeroides ECF sigma factor, SigE, and its anti-sigma factor, ChrR. J. Mol. Biol. 341345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control SigF activity by distinct mechanisms. Mol. Microbiol. 451527-1540. [DOI] [PubMed] [Google Scholar]
- 5.Dainese, E., S. Rodrigue, G. Delogu, R. Provvedi, L. Laflamme, R. Brzezinski, G. Fadda, I. Smith, L. Gaudreau, G. Palu, and R. Manganelli. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL and roles in virulence and in global regulation of gene expression. Infect. Immun. 742457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frohman, M. A. 1994. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 4S40-S58. [DOI] [PubMed] [Google Scholar]
- 7.Giacomini, E., A. Sotolongo, E. Iona, M. Severa, M. E. Remoli, V. Gafa, R. Lande, L. Fattorini, I. Smith, R. Manganelli, and E. M. Coccia. 2006. Infection of human dendritic cells with a Mycobacterium tuberculosis sigE mutant stimulates production of high levels of interleukin-10 but low levels of CXCL10: impact on the T-cell response. Infect. Immun. 743296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 9611554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn, M. Y., S. Raman, M. Anaya, and R. N. Husson. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 1877062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, H., R. Hovey, J. Kane, V. Singh, and T. C. Zahrt. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 1882134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 4647-110. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52231-286. [DOI] [PubMed] [Google Scholar]
- 13.Ito, W., H. Ishiguro, and Y. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 10267-70. [DOI] [PubMed] [Google Scholar]
- 14.Jacques, J. F., S. Rodrigue, R. Brzezinski, and L. Gaudreau. 2006. A recombinant Mycobacterium tuberculosis in vitro transcription system. FEMS Microbiol. Lett. 255140-147. [DOI] [PubMed] [Google Scholar]
- 15.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 184292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 955752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31715-724. [DOI] [PubMed] [Google Scholar]
- 19.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extra cytoplasmic function sigma factor SigE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 723038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor SigH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45365-374. [DOI] [PubMed] [Google Scholar]
- 22.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor SigE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41423-437. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Moll, I., G. Hirokawa, M. C. Kiel, A. Kaji, and U. Blasi. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 323354-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paget, M. S., J. B. Bae, M. Y. Hahn, W. Li, C. Kleanthous, J. H. Roe, and M. J. Buttner. 2001. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol. 391036-1047. [DOI] [PubMed] [Google Scholar]
- 26.Pang, X., P. Vu, T. F. Byrd, S. Ghanny, P. Soteropoulos, G. V. Mukamolova, S. Wu, B. Samten, and S. T. Howard. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 1531229-1242. [DOI] [PubMed] [Google Scholar]
- 27.Raman, S., A. Cascioferro, R. Husson, and R. Manganelli. 2008. Mycobacterial sigma factors and surface biology, p. 223-234. In M. Daffe and J. M. Reyrat (ed.), The mycobacterial cell envelope. ASM Press, Washington, DC.
- 28.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 1836119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30926-941. [DOI] [PubMed] [Google Scholar]
- 30.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50949-959. [DOI] [PubMed] [Google Scholar]
- 32.Song, T., S.-E. Song, S. Raman, M. Anaya, and R. N. Husson. 2008. Critical role of a single position in the −35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J. Bacteriol. 1902227-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udagawa, T., Y. Shimizu, and T. Ueda. 2004. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 2798539-8546. [DOI] [PubMed] [Google Scholar]
- 34.Williams, D. L., T. L. Pittman, M. Deshotel, S. Oby-Robinson, I. Smith, and R. Husson. 2007. Molecular basis of the defective heat stress response in Mycobacterium leprae. J. Bacteriol. 1898818-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 1792922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.