Abstract
Sapovirus is a positive-stranded RNA virus with a translational strategy based on processing of a polyprotein precursor by a chymotrypsin-like protease. So far, the molecular mechanisms regulating cleavage specificity of the viral protease are poorly understood. In this study, the catalytic activities and substrate specificities of the predicted forms of the viral protease, the 3C-like protease (NS6) and the 3CD-like protease-polymerase (NS6-7), were examined in vitro. The purified NS6 and NS6-7 were able to cleave synthetic peptides (15 to 17 residues) displaying the cleavage sites of the sapovirus polyprotein, both NS6 and NS6-7 proteins being active forms of the viral protease. High-performance liquid chromatography and subsequent mass spectrometry analysis of digested products showed a specific trans cleavage of peptides bearing Gln-Gly, Gln-Ala, Glu-Gly, Glu-Pro, or Glu-Lys at the scissile bond. In contrast, peptides bearing Glu-Ala or Gln-Asp at the scissile bond (NS4-NS5 and NS5-NS6, or NS6-NS7 junctions, respectively) were resistant to trans cleavage by NS6 or NS6-7 proteins, whereas cis cleavage of the Glu-Ala scissile bond of the NS5-NS6 junction was evidenced. Interestingly, the presence of a Phe at position P4 overruled the resistance to trans cleavage of the Glu-Ala junction (NS5-NS6), whereas substitutions at the P1 and P2′ positions altered the cleavage efficiency. The differential cleavage observed is supported by a model of the substrate-binding site of the sapovirus protease, indicating that the P4, P1, and P2′ positions in the substrate modulate the cleavage specificity and efficiency of the sapovirus chymotrypsin-like protease.
Sapovirus is a RNA virus belonging to Caliciviridae, where it segregates in a separate genus including human pathogenic and nonhuman pathogenic strains. Because of the lack of a cell culture system to propagate human pathogenic sapovirus strains, the translation strategy remains poorly understood. In particular, so far, the active forms of the protease as well as the molecular mechanisms of cleavage by the viral protease remain unclear.
The sapovirus single-stranded positive-oriented RNA genome (about 7.6 kb) encodes structural and nonstructural proteins located in two open reading frames (ORF1 and ORF2). Like other RNA viruses with a positive-oriented genome (i.e., Flaviviridae and Picornaviridae), sapovirus uses a translational strategy based on cotranslational processing of a polyprotein precursor encoded in ORF1. This cotranslational processing is mediated by one of the key players in viral replication, the viral chymotrypsin-like protease. The sapovirus protease (3C-like, NS6, 16 kDa) is predicted to be located in the C-terminal half of the polyprotein precursor, upstream of the viral RNA-dependent RNA polymerase (3D-like, NS7, 57 kDa).
In this study, it was postulated that (i) both NS6 and NS6-7 proteins are active forms of the viral protease, (ii) differential cis/trans processing of the cleavage sites in the polyprotein occurs, and (iii) the specificity of cleavage is regulated by distinct positions in the substrate. Our results show that both NS6 and NS6-7 proteins are able to cleave nearly all the scissile bonds in the polyprotein precursor in trans. Moreover, cleavage specificity and efficiency seem to be modulated by the positions P4, P1, and P2′ of the substrate. Cleavage of peptides displaying the scissile bonds of the sapovirus polyprotein reveals different cleavage efficiencies by NS6 and NS6-7, suggesting a differential release of the nonstructural proteins from the polyprotein precursor.
MATERIALS AND METHODS
Prediction of sapovirus NS6 protein boundaries.
The boundaries of the sapovirus NS6 protein were based on the previous predictions performed on the active domain of the sapovirus NS7 RNA-dependent RNA polymerase (3), indicating a putative cleavage of the protease-polymerase precursor at a Gln1207-Asp1208 scissile bond in the polyprotein precursor of the sapovirus pJG-Sap I clone (GenBank accession no. AY694184).
Expression and purification of recombinant sapovirus NS6 protein.
Production and purification of recombinant sapovirus NS6 were performed as described previously (3, 13-15), with slight modifications. Briefly, a cDNA fragment (459 bp) encompassing the sapovirus 3CL-like protease gene and encoding the NS5-NS6 and NS6-NS7 cleavage sites at its 5′ and 3′ ends, respectively, was generated by PCR from sapovirus clone pJG-SapI (GenBank accession no. AY694184; Fig. 1) using primers VZ-SapNS6-H6C-for (5′-GG GGACAAGTTTGTACAAAAAAGCAGGCTTCAAGGAGATGCCACCATGAAAGCCCCGCAGCAATTGTGGAGTTCACGCAG-3′) and VZ-SapNS6-H6C-rev (5′-GGGGACCACTTTGTACAAGAAAGCTGGGCTTATTAGTGATGGTGATG GTGATGTTGGGTAGTGACCTCCTTGACAACACG-3′). cDNA was cloned into the pDEST14 vector (Gateway; Invitrogen), resulting in expression vector pVZ-SapNS6-H6C, which was sequenced and used to transform Escherichia coli Rosetta(DE3) cells. After induction of cells grown at 37°C in 2YT medium, cultures were incubated at 25°C overnight and harvested by centrifugation at 4,500 rpm for 15 min. The pellet was suspended in buffer A (20 mM Tris, 150 mM NaCl [pH 7.8]) and then incubated on ice for 30 min. Cells were sonicated on ice, and the lysates were centrifuged at 20,000 rpm for 60 min and then resuspended in buffer B (20 mM Tris, 500 mM NaCl, 60 mM imidazole [pH 7.9]) and ultracentrifuged for 1.5 h at 40,000 rpm and 4°C. After purification of the protein by affinity chromatography on Ni-nitrilotriacetic acid (NTA) columns, a final purification step was performed on a GPC HiLoad 16/60 column with 75 pg Superdex (GE Healthcare) in buffer C (50 mM Tris, 100 mM NaCl, 1 mM dithiothreitol [pH 8.0]). Fractions containing proteins were collected and used for the peptide-based cleavage assay. Protein concentration was determined with the bicinchoninic acid protein assay kit (Pierce) based on the Biuret reaction.
FIG. 1.
Organization of the polyprotein precursor encoded in ORF1 of sapovirus clone pJG-SapI (GenBank accession no. [GB acc. No.] AY694184). (A) ORF1 encoding the nonstructural proteins (5,176 nucleotides in length) is shown. Putative cleavage sites are indicated. Both NS6 and NS6-7 proteins and their active-site mutant were expressed in Escherichia coli Rosetta(DE3) cells. The recombinant proteins bear a His6 tag at the C terminus. The nonstructural proteins NS1 to NS7 corresponding to the proteins p11, p28, 2C-like NTPase, p32, VPg, 3C-like protease, and 3D-like polymerase, respectively, are shown. (B) The proteins HA-[NS5/NS6]-NS6-7-His and HA-[NS4/NS5]-NS6-7-His bearing the authentic cleavage sites of the NS5-NS6 and the NS4-NS5 junctions, respectively, were expressed in Escherichia coli Rosetta(DE3) cells. Those constructs bear a His6 tag at the C terminus and an HA tag at the N terminus. The HA tag is underlined.
Gene expression and purification of recombinant sapovirus NS6-7 protein and it active site mutant.
Expression and purification of recombinant sapovirus NS6-7 protein were performed as described above, except that primers VZ2-SapNS6-H6C-for (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGAAGGAGATGCCACCATGAAAGCCCCGACAGCAATTGTGGAGTTCACGCAG-3′) and VZ3-Sap-NS6-H6C-rev (5′-GGGGACCACTTTGTACAAGAAA GCTGGGTCTTATTAGTGATGGTGATGGTGATGCTCCATCTCAAACACTATTTTGTGGGTTCC-3′) were used for generation of the pVZ-SapNS6-7-H6C expression clone. Fractions containing proteins were collected and used for the peptide-based assay. The protein concentration was determined with the bicinchoninic acid protein assay kit (Pierce).
Expression of mutated sapovirus NS6 gene.
Point mutations in the putative active site of sapovirus NS6 protein (GDC116GC) were generated with the Quick Change site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. pVZ-SapNS6-H6C or pVZ-SapNS6-7-H6C templates and primers 84-Sap-μNS6-for (5′-CCAAGAAAGGTGACGCCGGGCTGCCCTATTTC-3′) and 85-Sap-μNS6-rev (5′-GAAATAGGGCAGCCCGGCGTCACCTTTCTGG-3′) were used. This resulted in conversion of the residue C116 in the protein's predicted active site to alanine. Mutant NS6 gene expression was performed as described above, and the resulting proteins were termed μNS6 and μNS6-7 (Fig. 1).
Synthesis and purification of peptides.
Peptides were synthesized by GL Biochem (Shanghai, China) and purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on an RP column. The lyophilized products were characterized by HPLC and fast atom bombardment mass spectrometry (MS).
Analysis of the trans proteolytic activity of sapovirus NS6 and NS6-7 proteins.
Twenty-one different peptides (S1 to S21, 15 to 17 residues) displaying the cleavage sites in the sapovirus ORF1 polyprotein precursor as well as mutations in the P4 to P1 and P1 to P1′ positions were used as substrates (Table 1). Lyophilized peptides were dissolved in Tris buffer (40 mM Tris, 150 mM NaCl [pH 7.2]) at a final concentration of 1 mg/ml. Proteolytic cleavage was examined by HPLC as described previously (15). The cleavage efficiencies of the peptides by the viral protease were reported as percentages of the fraction of substrate that is converted to product based on the integrated peak area (12, 15, 17). Comparative analysis of cleavage efficiencies was performed using a nonparametric Mann-Whitney U test using GraphPad InStat, version 3.0a, for Macintosh (GraphPad Software, San Diego, CA). Statistical significance was set at P < 0.05.
TABLE 1.
Sequence and trans cleavage efficiency of peptides by sapovirus protease
| Peptidea | Cleavage site is sapovirus polyprotein | Mutationb | Sequence (NH2-COOH)c
|
Mean ± SE % cleavage efficiency (n = 4) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | |||||||||||
| SV1 | NS1-NS2 | D | G | Y | T | F | V | E | E | G | L | L | D | M | F | G | T | 100.00 | ||
| SV2 | NS2-NS3 | V | K | T | T | F | T | A | Q | G | P | T | D | L | G | W | A | 13.2 ± 1.8 | ||
| SV3 | NS3-NS4 | M | E | R | R | F | K | E | Q | A | G | P | L | Q | N | L | 43.2 ± 0.6 | |||
| SV4 | NS4-NS5 | T | R | G | S | R | T | E | E | A | K | G | K | T | K | H | G | 0.0 | ||
| SV5 | NS5-NS6 | P | Q | Q | H | Y | E | E | E | A | P | T | A | I | V | E | F | T | 0.0 | |
| SV6 | NS6-NS7 | V | K | E | V | T | T | Q | D | E | F | Q | W | K | G | L | 0.0 | |||
| SV7 | NS7-VP1 | H | K | I | V | F | E | M | E | G | N | G | S | N | P | E | P | K | 94.03 ± 0.5 | |
| SV8 | NS1-NS2 | E68G | D | G | Y | T | F | V | E | G | G | L | L | D | M | F | G | T | 0.0 | |
| SV9 | NS2-NS3 | Q324G | V | K | T | T | F | T | A | G | G | P | T | D | L | G | W | A | 0.0 | |
| SV10 | NS4-NS5 | R936F | T | R | G | S | F | T | E | E | A | K | G | K | T | K | H | G | 0.0 | |
| SV11 | NS4-NS5 | T937K | T | R | G | S | R | K | E | E | A | K | G | K | T | K | H | G | 0.0 | |
| SV12 | NS4-NS5 | R936F, T937K | T | R | G | S | F | K | E | E | A | K | G | K | T | K | H | G | 0.0 | |
| SV13 | NS5-NS6 | E1051K | P | Q | Q | H | Y | K | E | E | A | P | T | A | I | V | E | F | T | 0.0 |
| SV14 | NS5-NS6 | Y1050F | P | Q | Q | H | F | E | E | E | A | P | T | A | I | V | E | F | T | 19.27 ± 1.77 |
| SV15 | NS5-NS6 | E1051D | P | Q | Q | H | Y | D | E | E | A | P | T | A | I | V | E | F | T | 0.0 |
| SV16 | NS5-NS6 | E1051V | P | Q | Q | H | Y | V | E | E | A | P | T | A | I | V | E | F | T | 0.0 |
| SV17 | NS2-NS3 | A232E | V | K | T | T | F | T | E | Q | G | P | T | D | L | G | W | A | 9.48 ± 0.48 | |
| SV18 | NS2-NS3 | A232E, P235N | V | K | T | T | F | T | E | Q | G | N | T | D | L | G | W | A | 33.64 ± 1.14 | |
| SV19 | NS1-NS2 | G69P | D | G | Y | T | F | V | E | E | P | L | L | D | M | F | G | T | 97.95 ± 2.05 | |
| SV20 | NS1-NS2 | G69K | D | G | Y | T | F | V | E | E | K | L | L | D | M | F | G | T | 97.44 ± 2.00 | |
| SV21 | NS5-NS6 | Y1050F, P1055L | P | Q | Q | H | F | E | E | E | A | L | T | A | I | V | E | F | T | 0.0 |
The peptides reflect the authentic or mutated cleavage sites in the sapovirus polyprotein.
The numbering corresponds to the polyprotein of clone pJG-SapI (GenBank accession no. AY694184).
Key positions from P5 to P1 and P1′ to P5′ are noted. The mutated positions are highlighted in boldface.
Molecular characterization of the cleavage products.
MS analysis of HPLC fractions containing the products of limited proteolysis was performed as described previously (15) at the Mass Spectrometry Facility of the Max Planck Institute of Molecular Cell Biology and Genetics (Dresden, Germany). Matrix-assisted laser desorption ionization analysis was carried out on a Reflex IV matrix-assisted laser desorption ionization-time of flight mass spectrometer (Bruker Daltonics, Bremen, Germany) in reflective mode using α-cyano-4-hydroxy-trans-cinnamic acid as the matrix (17). Spectra were externally calibrated using masses of abundant autolysis products of trypsin. Nano-electrospray ionization-MS and tandem-MS (MS/MS) analyses were performed on a hybrid quadrupole time-of-flight mass spectrometer QSTAR Pulsar i (MDS Sciex, Concord, Canada) equipped with an automated chip-based nanoflow ion source (Advion, Ithaca, NJ). Spectra were processed with BioAnalyst QS v.1 software (MDS Sciex, Concord, Canada).
Analysis of the cis proteolytic activity of sapovirus protease.
A time course experiment was performed to investigate the cis cleavage activity of sapovirus NS6-7. Therefore, two constructs bearing the scissile bonds corresponding to the NS4-NS5 or NS5-NS6 cleavage sites fused to a hemagglutinin (HA) tag at the N terminus were designed, resulting in expression vectors pIR-SAP-ΔτNS5/tNS6-7 and pIR-SAP-ΔτNS4NS5/tNS6-7. Expression in Escherichia coli Rosetta(DE3) and purification were performed as described above, yielding proteins HA-[NS5/NS6]-NS6-7-His and HA-[NS4/NS5]-NS6-7-His. The cultures were harvested 8 h after induction, and the pellets were diluted 1:25, 1:50, 1:75, and 1:100. The pellets were suspended in 0.5 ml 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading solution, separated by 12% SDS-PAGE, and transferred onto Hybond nitrocellulose filters (Amersham Biosciences). Filters were blocked overnight in 1% milk powder dissolved in phosphate-buffered saline-0.1% Tween 20. Blotted proteins were detected with the mouse monoclonal anti-His primary antibody (Qiagen) or the mouse monoclonal anti-HA antibody (Sigma) at a 1:500 dilution. A phosphatase-labeled antimouse secondary antibody (Promega) was used at a 1:8,000 dilution. After each incubation step, filters were washed four times with phosphate-buffered saline-0.1% Tween 20. Detection was performed as described previously (15).
Homology model building.
A homology model for the sapovirus 3C-like protease was manually constructed on the basis of the crystal structure of the norovirus 3C-like protease (Protein Data Bank [PDB] identification no. 2FYQ) (19). Sequence alignments and structure modeling were aided by DeepView/SwissPDBViewer, version 3.9b2 (GlaxoSmithKline, 2006).
RESULTS
Expression and purification of recombinant SV NS6 and NS6-7 proteins.
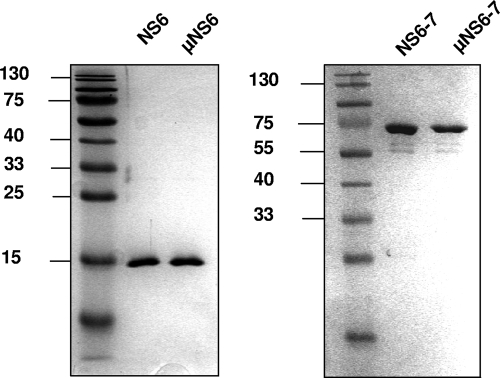
Expression of recombinant SV NS6 and NS6-7 genes as well as their respective active-site mutants was performed in E. coli Rosetta(DE3) cells. After separation of the protein fractions by Ni-NTA affinity chromatography, the eluted protein was further purified by gel filtration, and the fractions containing the highest level of purity were used for further experiments (Fig. 2).
FIG. 2.
Expression and purification of the sapovirus NS6 and NS6-7 proteins and their respective active-site mutants. The cDNA clones pJG-SAP-NS6, pJG-SAP-μNS6, pJG-SAP-NS6-7, and pJG-SAP-μNS6-7 were used for expression in Escherichia coli Rosetta(DE3) cells. The NS6 and NS6-7 proteins as well as their active-site mutants (μNS6 and μNS6-7) were purified by affinity chromatography on a Ni-NTA column followed by gel chromatography. Coomassie staining after SDS-PAGE of eluted fractions is shown.
SV NS6 and NS6-7 display cleavage activities in trans.
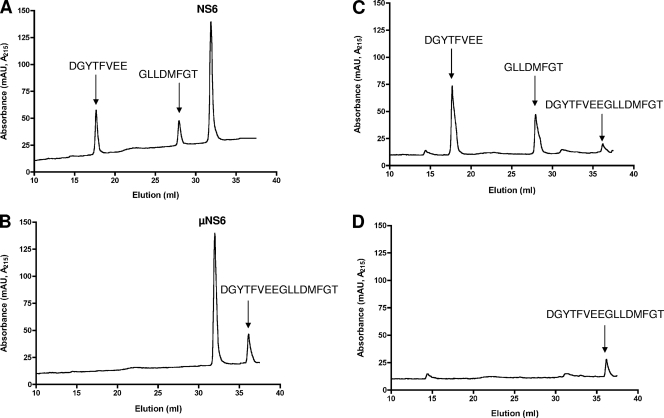
For characterization of the trans cleavage activities of NS6 and NS6-7, a cell-free assay was designed as previously described (15). This assay examines the trans cleavage activity of the purified NS6 and NS6-7 proteins on peptides bearing the scissile bonds from the SV polyprotein. Seven peptides corresponding to the cleavage sites within the sapovirus polyprotein precursor were designed (SV1 to SV7; Table 1). Overnight incubation of the peptides with NS6 or NS6-7 protein followed by analysis of the reaction by RP-HPLC provided evidence of proteolytic cleavage of peptides NS1-NS2, NS2-NS3, NS3-NS4, and NS7-virion protein 1 (VP1). A representative example is shown in Fig. 3, where incubation of peptide NS1-NS2 with NS6 or NS6-7 protein generated cleavage products, as indicated by two RP-HPLC peaks in addition to the substrate peak, presumably resulting from proteolytic processing of this peptide at the Gln-Gly scissile bond. When the active-site mutants were incubated with the peptide, no cleavage was evidenced, indicating that the cleavage is specific for NS6 and NS6-7 proteins. Similarly, incubation of peptides NS2-NS3, NS3-NS4, and NS7-VP1 with purified NS6 or NS6-7 proteins generated two cleavage products, presumably resulting from proteolytic processing of peptides at Glu-Gly, or Glu-Ala or Gln-Ala scissile bonds, respectively (Table 1). In contrast, no trans cleavage products were observed when peptide NS5-NS6, NS6-NS7, or NS4-NS5 was incubated with NS6 or NS6-7 protein, suggesting resistance of these peptides to proteolytic cleavage in trans.
FIG. 3.
trans cleavage of peptides by sapovirus NS6 and NS6-7 proteins. The peptides (30 μM) were incubated with purified NS6 or NS6-7 at 30°C for 17 h. Cleavage products were monitored by RP-HPLC as described in Materials and Methods. As a control, peptides were incubated with the NS6 or NS6-7 active-site mutants (μNS6 and μNS6-7, respectively). The cleavage products were collected and analyzed by MS. (A and B) Representative example of cleavage of the NS1-NS2 peptide by NS6 protein. μNS6 protein was used as a control (B), as indicated. (C and D) Representative example of cleavage of the NS1-NS2 peptide by NS6-7. μNS6-7 protein was used as a control (D). The eluted peaks of the peptide NS1-NS2 (NH2-DGYTFVEEGLLDMFGT-COOH) and its cleavage products (DGYTFVEE and GLLDMFGT) are indicated, according to MS analysis. mAU, milli-absorbance units.
To further characterize the specificities of NS6 and NS6-7 for the predicted cleavage sites, the RP-HPLC fractions containing the products of peptide proteolysis were subjected to MS analysis. Analyzed fractions contained the expected products of proteolysis, allowing the precise identification of the scissile bonds for peptides NS1-NS2, NS2-NS3, NS3-NS4, and NS7-VP1 (Table 1).
Sapovirus peptides NS1-NS2, NS2-NS3, NS3-NS4, NS4-NS5, NS5-NS6, NS6-NS7, and NS7-VP1 are differentially cleaved by NS6 and NS6-7 proteins.
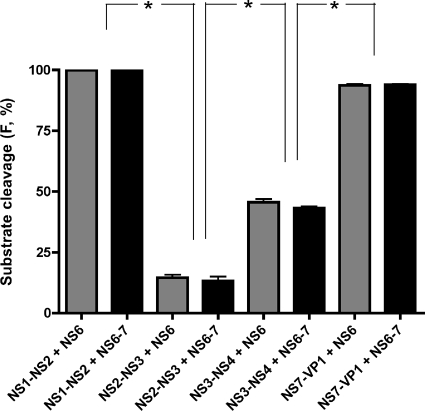
The activity of the sapovirus protease NS6 and NS6-7 was examined in the peptidolytic assay run under standard conditions. As shown in Fig. 4 and Table 1, sapovirus NS6 and NS6-7 proteins displayed similar cleavage efficiencies in trans. However, both proteins cleaved peptides NS1-NS2 and NS7-VP1 more efficiently than NS2-NS3 and NS3-NS4 (P < 0.05 by nonparametric Mann-Whitney U test). Moreover, peptides NS4-NS5, NS5-NS6, and NS6-NS7 were resistant to cleavage in trans. These data further suggest a differential cleavage pattern of the sapovirus polyprotein by NS6 and NS6-7, possibly modulated by the amino acid sequence pattern around the scissile bond.
FIG. 4.
The scissile bonds NS1-NS2, NS2-NS3, NS3-NS4, and NS7-VP1 are differentially cleaved by sapovirus NS6 and NS6-7 proteins. Cleavage efficiency of peptides NS1-NS2, NS2-NS3, NS3-NS4, and NS7-VP1 by NS6 (black bars) or NS6-7 (gray bars) protein is shown. The efficiency is expressed as the percentage of the fraction (F) of substrate converted to product. Means and standard errors of the means of four independent experiments are shown. Comparison of the means was performed by a nonparametric Mann-Whitney U test. The degree of statistical significance is shown: *, significant at P < 0.015.
Investigation of cis cleavage at the NS4-NS5, NS5-NS6, and NS6-NS7 scissile bonds.
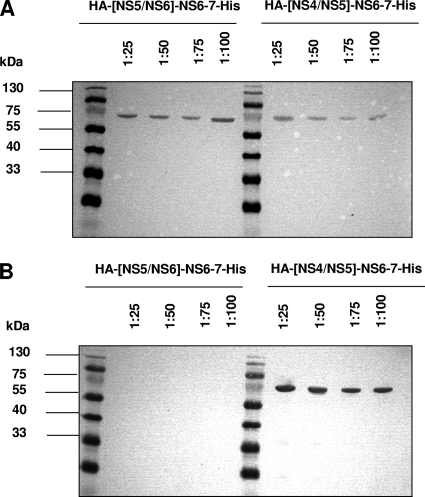
To investigate whether a cis cleavage occurs at the NS4-NS5 and NS5-NS6 junction, two NS6-7 proteins bearing the NS4-NS5 or NS5-NS6 scissile bond fused to an HA tag at the N terminus and a His tag at the C terminus located downstream of the NS6-NS7 cleavage site were expressed in E. coli. The cultures were harvested 8 h after induction, and the pellets were diluted 1:25, 1:50, 1:75, and 1:100, analyzed by SDS-PAGE, and visualized by Western blotting using anti-HA or anti-His antibodies. The NS6-7 protein was not able to cleave the His tag at the C terminus downstream of the NS6-NS7 cleavage site, indicating no cleavage in cis between NS6 and NS7 (Fig. 5A). The NS6-7 protein was able to cleave the HA tag at the N terminus upstream of the NS5-NS6 cleavage site (Fig. 5B) up to a 1:100 dilution, indicating that cis cleavage of the NS5-NS6 scissile bond occurs. In contrast, the NS6-7 protein could not cleave the HA tag at the N terminus upstream of the NS4-NS5 cleavage site scissile bonds in cis.
FIG. 5.
Sapovirus NS6-7 protein processes the cleavage site NS5-NS6 in cis. The cDNA clones pIR-SAP-ΔτNS5NS6/tNS6-7 and pIR-SAP-ΔτNS4NS5/tNS6-7 were used for expression in Escherichia coli Rosetta(DE3) cells, yielding proteins HA-[NS5/NS6]-NS6-7-His and HA-[NS4/NS5]-NS6-7-His, respectively. Protein HA-[NS5/NS6]-NS6-7-His bears at the N terminus the authentic NS5-NS6 cleavage site (PQQHYEEE1053/A) fused to an HA tag, and protein HA-[NS4/NS5]-NS6-7-His bears at the N terminus the authentic NS4-NS5 cleavage site (TRGSRTEE939AKGKTKHG) fused to an HA tag. The proteins were expressed in E. coli, the cultures were harvested 8 h after induction, and the pellets were diluted 1:25, 1:50, 1:75, and 1:100, analyzed by SDS-PAGE, and visualized by Western blotting using anti-HA or anti-His antibodies. Western blot analysis of the eluted proteins diluted to 1:25, 1:50, 1:75, and 1:100 is shown. (A) Detection of proteins HA-[NS5/NS6]-NS6-7-His and HA-[NS4/NS5]-NS6-7-His by anti-His antibody, as indicated. (B) Detection of proteins HA-[NS5/NS6]-NS6-7-His and HA-[NS4/NS5]-NS6-7-His by anti-HA antibody, as indicated.
The amino acid sequence around the scissile bond modulates the trans cleavage by sapovirus protease.
To examine whether the P1 to P4 and P1′ to P2′ positions in the peptide sequence modulate trans cleavage, 14 additional peptides bearing amino acid substitutions at either side of the scissile bond were designed and synthesized. Gln or Glu is present at the P1 position of all cleavage sites of the polyprotein precursor (Table 1). In order to assess the specificity of the protease for either amino acid at this position, substitution of Glu for Gly or Gln for Gly was performed in peptides NS1-NS2 (SV8) or NS2-NS3 (SV9), respectively. This completely abolished processing of the peptides. In contrast, at the P1′position, no effect on cleavage efficiency was observed when Gly was substituted for with a Pro or Lys (SV19 and SV20, respectively), indicating that at the scissile bond (P1-P1′ position), the P1 position modulates the sensitivity to trans cleavage by the sapovirus protease.
At the P2 position, Glu, Ala, Thr, or Met is present. Mutation of Ala to Gln in this position did not result in an increase of cleavage efficiency (SV2 versus SV17). However, by replacing a Pro at the P2′ position with an Asp, cleavage efficiency increased significantly (SV2 versus SV18; P < 0.05). This indicates that the P2′ position also modulates cleavage efficiency in trans by the sapovirus 3C-like protease. This is also observed in peptide SV21, where mutation at the P2′ position (Pro to Leu) completely abolished cleavage of the peptide. The P3 position does not seem to influence cleavage efficiency, as observed in peptides SV11 to SV13, SV15, and SV16. In contrast, position P4 positively modulated cleavage, as observed in peptide SV14 in comparison to peptide SV5. There, replacing Tyr by Phe resulted in cleavage of the peptide NS5-NS6 that was resistant to cleavage in trans. To sum up, the positions P4, P1, and P2′ seem to play an important role in modulating cleavage efficiency of the scissile bonds of the polyprotein precursor by the sapovirus protease.
Modeling of sapovirus NS6 protease active site.
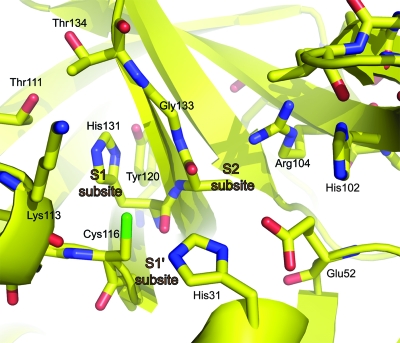
In order to generate a reliable model of the active site of the sapovirus protease, automatic sequence alignment of the sapovirus and norovirus 3C-like protease was used, yielding a sequence identity of 32%. However, as is often the case, this preliminary alignment gave an overestimated value for the sequence identity, and our structure-based alignment yielded a more realistic value of approximately 20%. According to our model (Fig. 6) of the sapovirus protease active site, the walls of the S1 specificity pocket are formed by Ala132mc (“mc” represents main-chain atoms), Gly133, Thr134, Thr111, and Lys113mc. The two threonine residues are at the far end of the pocket (see Fig. 6, upper left). At the bottom, there is His131 residue highly conserved in 3C and 3C-like proteases. As in most of these enzymes, the histidine accepts a hydrogen bond from the hydroxyl of a tyrosine residue: in this case, Tyr120. The structural position of the tyrosine OH group is similar to the one in the coronavirus main proteases (1, 2), although the residue is on a different strand in the β-barrel of domain II. In the S2 subsite, Asn99 at the tip of the bII-cII hairpin forms part of the S2 pocket, and so does the Glu52-His31 pair of the catalytic triad. Interestingly, the guanidinium group of Arg104 is also part of the S2 pocket, according to our model. As to the S3 subsite, and similar to most 3C or 3C-like proteases, there is no evident S3 pocket and a specificity for P3 of the substrate is not recognizable, except that the side chain tends to be hydrophilic since the subsite is open toward bulk solvent. In contrast, there is pronounced specificity for P4 being hydrophobic. Since a part of the S4 pocket involves a segment of the protease that is highly unrelated to norovirus 3C-like protease, we had difficulty completing the model for this subsite. Residues likely involved are Asn99 (side-chain amide), Thr97mc, Ile96, and Val98. The latter residue is Ile109 in norovirus 3C-like protease, but the side chain has to be shorter in the sapovirus protease, otherwise the above-mentioned Arg104 of the S2 pocket would not fit. The S1′ pocket in all 3C and 3C-like proteases is small in size, and the sapovirus NS6 protein is no exception. Thus, the most preferred P1′ residue is glycine. The residues involved are the carbonyl of Lys113, Gly114, His31 of the catalytic triad, and Val32. The main-chain amides of Gly114 and Cys116 are involved in stabilizing the oxanion. The S2′ subsite cannot be modeled reliably, but we note that the P2′ residue will be partly exposed to solvent. Beyond P2′, the substrate residues are not in contact with the enzyme.
FIG. 6.
Homology model for the sapovirus protease catalytic site. The model was manually constructed on the basis of the crystal structure of the norovirus NS6 3C-like protease (PDB identification no. 2FYQ).
DISCUSSION
The sapovirus protease is predicted to play an essential role in the replication strategy of the virus. The molecular mechanisms underlying the catalytic activity and specificity of the sapovirus chymotrypsin-like protease have so far remained enigmatic. Understanding the regulation of polyprotein processing in sapovirus is an important step toward elucidating the sapovirus replication strategy. Such knowledge is also an important prerequisite for the development of antiviral drugs to control sapovirus infection.
In this study, the cleavage efficiency and substrate specificity of the catalytic in-trans and cis activities of sapovirus NS6 and NS6-7 proteins were examined in vitro. The NS6 and NS6-7 proteins were able to cleave peptides containing Gln-Gly, Glu-Gly, or Gln-Ala bonds. However, NS6 and NS6-7 were not able to cleave the NS6-NS7 scissile bond, neither in trans nor in cis, indicating that the NS6-7 protein is the active form of the sapovirus protease. These results are in accordance with previous reports on the processing of sapovirus polyprotein in a cell-free system (9, 10). Indeed, production of the sapovirus ORF1 polyprotein in vitro leads to cotranslational release of the NS1, NS2, NS3, NS4-NS5, and NS6-7 proteins. To characterize the differential cis/trans cleavage pattern of the polyprotein precursor, two constructs featuring the authentic cleavage sites of the NS4-NS5 junction or the NS5-NS6 junction at the N terminus were designed. Expression of the constructs revealed that the NS5-NS6 junction was cleaved in cis, whereas the NS4-NS5 junction was resistant to cis cleavage. Cleavage in cis by the 3C-like protease has already been discussed in the cases of norovirus (16) and poliovirus (6). Our data on the autocatalytic cleavage of the norovirus 3C-like protease from its protease-polymerase precursor (15) support these observations. In our study, the peptide bearing the cleavage site between the NS4 and NS5 proteins was resistant to cleavage in trans by both NS6 and NS6-7 proteins. Discordant results on processing of the sapovirus NS4-NS5 boundary have been reported so far. In the study of Oka et al. (8), processing of the ORF1 polyprotein of strain Mc10 leads to the detection of the NS4 and NS5 proteins putatively released from an N-terminally glutathione S-transferase-labeled NS4-NS5 precursor. The cleavage between the NS4 and NS5 proteins was also observed in cell-free system, where cleavage of the whole polyprotein was investigated (9, 10). However, in a further study using the same system, the NS4-NS5 precursor was also detected (11), indicating no cleavage of the scissile bond, as observed in our study. In order to investigate the reason for this discordance, we have performed a control experiment in which the processing of a so-far cleavage-resistant peptide (SV4 reflecting the putative NS4-NS5 scissile bond) was investigated in the presence of reticulocyte lysates (see the supplemental material). Interestingly, the peptide was completely cleaved in the presence of reticulocyte lysates, whether sapovirus NS6-7, NS6, or their respective active-site mutants were added to the reaction, indicating that a nonspecific cleavage of the NS4-NS5 site occurs in the translation-transcription assay used. This nonspecific cleavage is not due to a cofactor of the NS6 nor NS6-7 present in the reticulocyte lysates, as would have been expected if the active protease incubated with the lysates cleaves the peptide, in comparison to the lack of cleavage observed when the lysates are absent. Those results indicate that processing of the NS4-NS5 scissile bond strongly depends on the expression system used.
Moreover, the cleavage specificity and efficiency of the scissile bonds seem to depend on the sequence of the peptide, more precisely the P4, P1, and P2′ positions. The sapovirus NS6 and NS6-7 proteins displayed a higher cleavage efficiency of NS1-NS2 and NS7-VP1 sites than of the NS2-NS3 and NS3-NS4 sites, indicating that Gln in P1 decreases the cleavage efficiency compared to Glu. Mutation of the P1 Gln to Gly or Glu to Gly abolished completely processing of the peptides. In contrast, mutation at the P1′ position did not affect the cleavage efficiency. These data indicate that at the scissile bond (P1-P1′ position), the P1 position modulates the sensitivity to trans cleavage by the sapovirus protease. The NS5-NS6 scissile bond, consisting of Glu-Ala, was not cleaved by NS6 nor by NS6-7 proteins in trans. Peptide NS6-NS7 that was not cleaved in trans displayed a Gln-Asp scissile bond, thus differing in the P1′ position from the above-mentioned cleavage sites. However, the Glu-Ala scissile bond of the NS5-NS6 junction was cleaved in cis, indicating that this pair of residues may play a role in regulating cis versus trans cleavage. Interestingly, the P2′ position seems to modulate cleavage efficiency in trans by the sapovirus 3C-like protease, as observed in peptide SV21 (NS5-NS6). Moreover, by replacing at the P2′ position of the NS2-NS3 peptide a Pro with an Asn, cleavage efficiency increased significantly (SV2 versus SV18, P < 0.05). Similarly, position P4 positively modulates cleavage, where replacing Tyr with Phe resulted in trans cleavage of the peptide NS5-NS6. The relevance of a Phe at position P4 has also been highlighted in the cases of norovirus (4) and feline calicivirus (11). In summary, the positions P4, P1, and P2′ seem to play an important role in modulating cleavage efficiency of the scissile bonds of the polyprotein precursor by the sapovirus protease.
The differential cleavage observed in this study is supported by our model of the substrate-binding site of the sapovirus protease. In the absence of a crystal structure for an enzyme, three-dimensional homology models have their merits when it comes to interpreting experimental data on substrate specificity (such as in the present case) and even for preliminary steps in drug discovery (5). Of course, the quality of a homology model depends on the overall degree of sequence identity between the template(s) and the model, as well as the quality of the template and the number of homologous sequences available. In this study, we only focus on regions of high similarity between the sapovirus and norovirus 3C-like proteases that allowed reliable construction of a three-dimensional model.
In the SV NS6 3C-like protease, the glutamate residue of the catalytic triad Cys116-His31-Glu52 is in contact with another histidine residue, His102. This residue is not conserved in the norovirus 3C-like protease, where it is replaced by Val114. The reason for the presence of the second histidine in the immediate catalytic site will be discussed below. In order to interpret the cleavage specificity data described above, we will focus on the substrate-binding subsites of the enzyme as revealed by the homology model. At the bottom of the S1 pocket, His131 is absolutely conserved in all calicivirus, picornavirus, and coronavirus 3C-like proteases. It has been shown to interact with the P1 side chain of the substrate in several crystal structures of complexes between substrate-like inhibitors and these enzymes (1, 2, 7, 18). Most of these proteases are absolutely specific for glutamine in the P1 position. In order to reject the binding of glutamate residues in the S1 pocket, the conserved histidine should be uncharged over a wide range of pHs. In the coronavirus 3C-like proteases (main proteases), this is ensured by a tyrosine residue, which donates a hydrogen bond from its hydroxyl group to the histidine (1). A tyrosine is also present in the picornavirus 3C proteases, even though at a very different position in the amino acid sequence (residue 138 in the 3Cpros from human rhinovirus 2, poliovirus, and foot-and-mouth disease virus; as an exception, hepatitis A virus 3Cpro does not have this tyrosine). Interestingly, the hydroxyl group of Tyr138 in the picornavirus 3Cpros occupies exactly the same position as the hydroxyl of Tyr160(161) (numbering for severe acute respiratory syndrome-coronavirus Mpro in brackets) in the coronaviral enzymes and can thus donate an ideal hydrogen bond to the histidine at the bottom of the S1 pocket (1). Sapovirus and norovirus 3C proteases also feature the conserved tyrosine, and yet they accept not only glutamine, but also glutamate in the P1 position of the substrate. As far as the tyrosine is concerned, the calicivirus 3C-like proteases seem to be more similar to the coronavirus main proteases than to the picornavirus 3C enzymes. According to our model, the presence of a guanidinium group of Arg104 in the S2 pocket could explain the observed specificity of the sapovirus protease for a glutamate in P2. In norovirus 3C-like protease, this residue is replaced by Gly116, which obviously cannot interact with a P2-glutamate. Methionine and alanine also occur (once each) at the P2 position of sapovirus cleavage sites. Our model would be compatible with both: methionine is sufficiently flexible to adapt its conformation to the amphiphilic S2 pocket, and alanine does not pose a sterical problem due to its short side chain. The presence of Arg104 at the bottom of the S2 pocket jeopardizes the integrity of the catalytic triad, because according to our model, the guanidinium group of this residue is placed within 4.5 Å from Glu52, which might reorient toward the arginine since the latter is in a largely hydrophobic environment. To prevent this, His102 plays an important role by holding Glu52 in its proper position in the triad. Thus, the triad in fact becomes a Cys116-His31-Glu52-His102 tetrad (Fig. 6). This feature does not exist in norovirus 3C-like protease (which lacks the arginine in the S2 subsite).
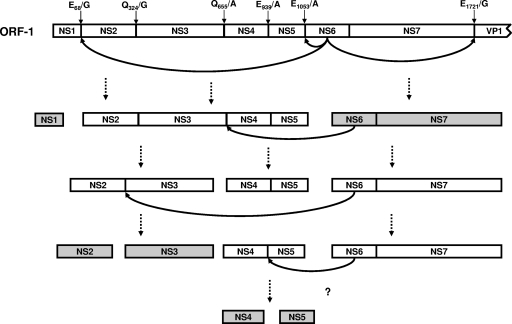
Based on our data, we postulated that differences in susceptibility of the predicted cleavage sites to proteolysis by sapovirus NS6-7 protein may result in a sequential processing of the scissile bonds. According to the observed relative cleavage efficiencies of the peptides by NS6-7 (Fig. 4), a sequential map for processing of the sapovirus polyprotein is proposed (Fig. 7) based on a temporal and/or spatial ranking for release of the active nonstructural proteins from the polyprotein precursor. This ranking may be based on the half-life of the precursor proteins, as observed during the processing of norovirus and poliovirus polyproteins (12, 15), in which a similar mechanism regulating availability of the nonstructural proteins of the replication complex was postulated. Therefore, it is conceivable that polyprotein processing by sapovirus NS6-7 regulates the formation of the replication complex through a ranking order of stability of the nonstructural proteins. It should be emphasized, however, that the proposed model of polyprotein processing cascade is based on data generated in vitro, i.e., in the absence of cellular factors that may play an essential role in modulating the replication of the sapovirus. In this sense, our in vitro model will have to be completed if and when an efficient culture system for sapovirus is available.
FIG. 7.
Proposed model for release of the nonstructural proteins based on the cleavage efficiencies of the peptides. In the first step, NS6-7 cleaves in trans the NS1-NS2 and NS7-VP1 bonds and in cis the NS5-NS6 bonds, releasing NS1, NS2-NS3-NS4-NS5, NS6-7, and VP1 proteins. This is followed by cleavage in trans of the NS2-NS3 and NS3-NS4 scissile bonds by NS6-7 protein, releasing NS3 and NS4-NS5. Cleavage of NS4-NS5 is probably not dependent only on the catalytic activity of the sapovirus protease, but rather on other cellular components. The final stable products are highlighted. The scissile bonds in the sapovirus polyprotein are indicated.
Supplementary Material
Acknowledgments
This study was supported by the European project “VIZIER” (Comparative Structural Genomics of Viral Enzymes Involved in Replication) funded by the 6th Framework Program of the European Commission under reference LSHG-CT-2004-511960.
Footnotes
Published ahead of print on 11 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 213213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 3001763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Fullerton, S. W. B., M. Blaschke, B. Coutard, J. Gebhardt, A. Gorbalenya, B. Canard, P. A. Tucker, and J. Rohayem. 2007. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 811858-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy, M. E. 2005. Norovirus protein structure and function. FEMS Microbiol. Lett. 2531-8. [DOI] [PubMed] [Google Scholar]
- 5.Hillisch, A., L. F. Pineda, and R. Hilgenfeld. 2004. Utility of homology models in the drug discovery process. Drug Discov. Today 9659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan, A. R., N. Khazanovich-Bernstein, E. M. Bergmann, and M. N. James. 1999. Structural aspects of activation pathways of aspartic protease zymogens and viral 3C protease precursors. Proc. Natl. Acad. Sci. USA 9610968-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthews, D. A., W. W. Smith, R. A. Ferre, B. Condon, G. Budahazi, W. Sisson, J. E. Villafranca, C. A. Janson, H. E. McElroy, C. L. Gribskov, et al. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77761-771. [DOI] [PubMed] [Google Scholar]
- 8.Oka, T., K. Katayama, S. Ogawa, G. S. Hansman, T. Kageyama, T. Miyamura, and N. Takeda. 2005. Cleavage activity of the sapovirus 3C-like protease in Escherichia coli. Arch. Virol. 1502539-2548. [DOI] [PubMed] [Google Scholar]
- 9.Oka, T., K. Katayama, S. Ogawa, G. S. Hansman, T. Kageyama, H. Ushijima, T. Miyamura, and N. Takeda. 2005. Proteolytic processing of sapovirus ORF1 polyprotein. J. Virol. 797283-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oka, T., M. Yamamoto, K. Katayama, G. S. Hansman, S. Ogawa, T. Miyamura, and N. Takeda. 2006. Identification of the cleavage sites of sapovirus open reading frame 1 polyprotein. J. Gen. Virol. 873329-3338. [DOI] [PubMed] [Google Scholar]
- 11.Oka, T., M. Yamamoto, M. Yokoyama, S. Ogawa, G. S. Hansman, K. Katayama, K. Miyashita, H. Takagi, Y. Tohya, H. Sato, and N. Takeda. 2007. Highly conserved configuration of catalytic amino acid residues among calicivirus-encoded proteases. J. Virol. 816798-6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pallai, P. V., F. Burkhardt, M. Skoog, K. Schreiner, P. Bax, K. A. Cohen, G. Hansen, D. E. Palladino, K. S. Harris, M. J. Nicklin, et al. 1989. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 2649738-9741. [PubMed] [Google Scholar]
- 13.Rohayem, J., K. Jager, I. Robel, U. Scheffler, A. Temme, and W. Rudolph. 2006. Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J. Gen. Virol. 872621-2630. [DOI] [PubMed] [Google Scholar]
- 14.Rohayem, J., I. Robel, K. Jäger, U. Scheffler, and W. Rudolph. 2006. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 807060-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheffler, U., W. Rudolph, J. Gebhardt, and J. Rohayem. 2007. Differential cleavage of the norovirus polyprotein precursor by two active forms of the viral protease. J. Gen. Virol. 882013-2018. [DOI] [PubMed] [Google Scholar]
- 16.Someya, Y., N. Takeda, and T. Miyamura. 2005. Characterization of the norovirus 3C-like protease. Virus Res. 11091-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas, H., J. Havlis, J. Peychl, and A. Shevchenko. 2004. Dried-droplet probe preparation on AnchorChip targets for navigating the acquisition of matrix-assisted laser desorption/ionization time-of-flight spectra by fluorescence of matrix/analyte crystals. Rapid Commun. Mass Spectrom. 18923-930. [DOI] [PubMed] [Google Scholar]
- 18.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 10013190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeitler, C. E., M. K. Estes, and B. V. Venkataram Prasad. 2006. X-ray crystallographic structure of the Norwalk virus protease at 1.5-Å resolution. J. Virol. 805050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.