Abstract
Latent membrane protein 1 (LMP1), an Epstein-Barr virus (EBV) oncoprotein, mimics a constitutively activated tumor necrosis factor receptor and activates various signaling pathways, including phosphatidylinositol 3-kinase (PI3K)/Akt. LMP1 is essential for EBV-mediated B-cell transformation and is sufficient to transform several cell lines. Cellular transformation has been associated strongly with genomic instability, while DNA repair plays an important role in maintaining genomic stability. Previously, we have shown that LMP1 represses DNA repair by the C-terminal activating region 1 (CTAR1) in human epithelial cells. In the present study, we demonstrate that the PI3K/Akt pathway is required for LMP1-mediated repression of DNA repair. Through the LMP1/PI3K/Akt pathway, FOXO3a, which can induce DNA repair, is inactivated because of phosphorylation and relocalization. Expression of a constitutively active FOXO3a mutant can rescue LMP1-mediated repression of DNA repair. Furthermore, LMP1 can decrease the expression of DNA damage-binding protein 1 (DDB1), which functions in nucleotide excision repair, through the PI3K/Akt/FOXO3a pathway. LMP1-mediated repression of DNA repair is restored by DDB1, although only partially. These results suggest that LMP1 triggers the PI3K/Akt pathway to inactivate FOXO3a and decrease DDB1, which can lead to repression of DNA repair and may contribute to genomic instability in human epithelial cells.
Epstein-Barr virus (EBV), a gammaherpesvirus, infects more than 90% of the human population and is the etiological agent of infectious mononucleosis. It also has been associated with human epithelial and lymphoid malignancies, such as nasopharyngeal carcinoma (NPC), Burkitt's lymphoma, T-cell lymphoma, and Hodgkin's disease (56). In vitro, EBV can infect and immortalize resting B cells to enable permanent growth of lymphoblastoid cell lines. Lymphoblastoid cell lines express nine viral proteins, including the latent membrane protein 1 (LMP1). Furthermore, LMP1 is detected in tumor biopsies from patients with EBV-associated malignancies, including NPC and Hodgkin's disease (16).
LMP1 is regarded as an oncogene because it shows pleiotropic effects; e.g., it transforms several cell lines, is essential for EBV-mediated B-cell transformation, inhibits epithelial cell differentiation, and induces epithelial hyperproliferation and B-cell lymphoma in transgenic mice (4, 5, 7, 10, 17, 29, 34, 67, 68). Recently, it was shown that LMP1 plays a role in genomic instability; expression of LMP1 in human epithelial cells reduces the capacity for DNA repair, enhances micronucleus formation, and represses p53-mediated DNA repair and transcriptional activity (39, 40).
LMP1 mimics a constitutively activated, ligand-independent, tumor necrosis factor receptor (TNFR) (21, 48, 65). It consists of a short N-terminal cytoplasmic tail, six hydrophobic transmembrane segments and a long C-terminal cytoplasmic region containing two critical signaling domains, designated C-terminal activating regions 1 and 2 (CTAR1 and CTAR2). These two domains were identified on the basis of their ability to activate the nuclear factor-κB (NF-κB) transcription factor pathway (24). However, there seems to be some difference between them; CTAR1 is necessary for EBV-mediated B-cell transformation, while CTAR2 is dispensable (26, 30). CTAR1 recruits TNFR-associated factors (TRAFs) directly via its PxQxT motif, but CTAR2 recruits TRAFs indirectly via the TNFR-associated death domain protein (9, 14, 18, 27). CTAR1 activates both canonical and noncanonical NF-κB pathways, and CTAR2 activates the canonical NF-κB pathway (2, 13, 42, 57). However, LMP1 activates the c-Jun N-terminal kinase (JNK) pathway through CTAR2, but not CTAR1 (12, 15, 32). In addition, LMP1 triggers the phosphatidylinositol 3-kinase (PI3K)/Akt pathway in a CTAR1-dependent manner (8, 43).
The PI3K/Akt pathway controls a variety of cellular activities, including proliferation, survival, motility, and morphology. The deregulation of PI3K or Akt activity perturbs these process and has been linked to several human cancers (3). Akt also has been shown to be activated in clinical specimens from EBV-associated NPC and Hodgkin lymphoma (47). The PI3K/Akt pathway has been shown to be activated by EBV LMP1 and LMP2A, which may contribute to several oncogenic effects (8, 58, 62).
The Forkhead box class O (FOXO) transcription factors, including FOXO1, FOXO3a, and FOXO4, are downstream targets of the PI3K/Akt pathway. After PI3K activation, Akt phosphorylates FOXO proteins. These phosphorylated FOXO proteins associate with 14-3-3 proteins, relocate from the nucleus to the cytoplasm, and subsequently lose their ability to regulate target genes. In contrast, unphosphorylated FOXO proteins are located in the nucleus and are active forms regulating their target genes (22). The FOXO target genes are involved in metabolism, cell cycle progression, apoptosis, DNA repair, and protection from oxidative damage. Perturbation of FOXO function leads to uncontrolled cell proliferation and accumulation of DNA damage. Therefore, FOXO proteins play important roles in controlling cell integrity and homeostasis. Several studies have shown that deregulation of FOXO was found in several types of tumor, including breast cancer, prostate cancer, and leukemia (1).
DNA damage-binding protein 1 (DDB1) is associated with DDB2 to form a heterodimeric UV-DDB complex that shows strong and specific binding to UV-irradiated DNA (6, 31). UV-DDB complex recognizes several UV-induced DNA lesions, including pyrimidine-pyrimidone(6-4)photoproducts and cyclobutane pyrimidine dimers, and initiates nucleotide excision repair (NER) (19). UV-DDB complex loses its DNA damage binding activity in repair-deficient disease xeroderma pigmentosum group E patients because of mutations of the DDB2 gene (50). No naturally occurring DDB1 mutation have been reported (69). DDB1 is a 127-kDa protein that is strongly conserved among eukaryotes. The regulation of DDB1 remains to be elucidated; however, the DDB1 gene is induced by active forms of FOXO proteins and is possibly a putative FOXO target gene (55).
Genomic instability has been thought to be associated strongly with tumor formation, while DNA repair plays a role in maintaining genomic stability (23, 51). Previously, the capacity for repair of cellular DNA was shown to be repressed in human epithelial cells by EBV LMP1 (40). However, the molecular mechanisms involved in this process are not well characterized. In the present study, we sought to determine the signaling pathways and mechanisms involved in this LMP1-mediated repression of DNA repair. The PI3K/Akt pathway was found to be necessary for LMP1-mediated repression of DNA repair. In addition, FOXO3a and DDB1, which are known to be important in DNA repair, are linked to this LMP1/PI3K/Akt pathway. Even though DDB1 participates in a minor way, it is evident that LMP1 can repress DNA repair through the PI3K/Akt/FOXO3a pathway.
MATERIALS AND METHODS
Cell culture.
NPC-TW01, an NPC cell line, lost its original infecting EBV after passage (36, 37). It owns a normal p53 protein (25). H1299, a human large cell lung carcinoma cell line, has a deletion of the p53 gene (46). H1299/bcl2 cells were established by transfecting a plasmid expressing bcl-2 into H1299 cells and selected by using G418 (800 μg/ml) (40). All cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum in 5% CO2 at 37°C.
Plasmids.
The plasmids expressing LMP1 and its mutants were described previously (39). The firefly and Renilla luciferase reporter plasmids also were described previously (40). The plasmids expressing the hemagglutinin (HA)-tagged forms of Akt, either constitutively active (myristylated-ΔPH) or inactive (KM mutant), were purchased from Upstate. The plasmids HA-FOXO3a-WT and HA-FOXO3a-TM were obtained from the Addgene plasmid repository (identification numbers 1787 and 1788). The plasmid expressing DDB1 was purchased from the American Type Culture Collection (identification number MBA-126).
Inhibitors.
Wortmannin, LY294002, sodium salicylate, and SB203580 were purchased from Sigma, Cell Signaling, Sigma, and Calbiochem, respectively.
Transfection.
Plasmids were transfected into NPC-TW01 and H1299/bcl2 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
HCR assay.
The host cell reactivation (HCR) assay was described previously (40) and modified partly. Briefly, firefly luciferase reporter plasmid (pCMV-Luc) either UV irradiated (1,500 J/M2) or not, undamaged Renilla luciferase reporter plasmid (pRL-CMV) as the internal control, and various effector plasmids were cotransfected into cells. At 24 h posttransfection, cells were lysed and assayed for firefly and Renilla luciferase activity. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity. To determine the ability of cells to repair damaged DNA, repair conversion and the fold change of HCR were calculated. Repair conversion was calculated by dividing the normalized firefly luciferase activity of cells transfected with UV-irradiated pCMV-Luc by that of nonirradiated pCMV-Luc transfectants. The fold change of HCR was calculated by dividing the repair conversion of effector transfectants by that of vector transfectants. The data from at least five independent experiments were averaged to calculate the means and standard deviations.
Western blotting assay.
Cell extracts were separated by NuPAGE (Invitrogen) and transferred to Hybond-C super membrane (Amersham). The blot was incubated with blocking buffer and reacted with primary antibody for overnight at 4°C. In the present study, various primary antibodies were used: anti-LMP1 (Dako); anti-Flag and anti-β-actin (Sigma); anti-Akt, anti-phospho-Akt (Ser473), anti-phospho-GSK-3α/β (Ser21/9), and anti-phospho-FOXO3a (Ser253) (Cell Signaling); anti-FOXO3a and anti-phospho-FOXO3a (Thr32) (Upstate); and anti-DDB1 and anti-PARP (Santa Cruz). After a washing step, the blots were incubated with horseradish peroxidase-conjugated goat anti-mouse or rabbit immunoglobulin G (Amersham). After development with a freshly prepared substrate (ECL; Amersham), the luminescence was detected by exposure to X-ray film.
Akt kinase assay.
The Akt kinase activity was measured by using an Akt kinase assay kit (kit no. 9840; Cell Signaling Technology) according to the manufacturer's protocol. Briefly, after various treatments, the cells were lysed in cell lysis buffer on ice for 5 min. After centrifugation, the protein concentration of the supernatant was measured by a protein assay (Bio-Rad). A total of 10 μg of protein from each sample was used to detect total Akt by Western blotting. Then, 200 μg of protein from each sample was used for immunoprecipitation with 20 μl of immobilized Akt antibody overnight at 4°C. Next, the immobilized Akt antibody complex was collected, washed, and subjected to Akt kinase assay in kinase buffer, 200 μM ATP, and 1 μg of GSK-3 fusion protein. Phosphorylation of GSK-3 was measured by immunoblotting.
RNA interference.
Silencer Validated small interfering RNA (siRNA) for Akt1 (catalog no. 51321) and Silencer Negative control siRNA (catalog no. 4613) were purchased from Ambion. Transfection of the siRNA was performed with Lipofectamine 2000.
Nuclear and cytosolic protein extraction.
A Qproteome nuclear protein kit (catalog no. 37582; Qiagen) was used. Briefly, a total of 107cells were cultured in 100-mm dishes. Cells were harvested and lysed in lysis buffer. The cell suspension was added 25 μl of detergent solution and then centrifuged. The supernatant was retained as the cytosolic fraction. The remaining pellet was incubated with extraction buffer by gentle agitation at 4°C for 30 min and was then centrifuged. The supernatant was retained as the nuclear fraction.
Immunofluorescence.
Cells were grown on coverslips and transfected with various plasmids. After 24 h of serum starvation, cells were fixed with 4% paraformaldehyde and subsequently were permeabilized with 0.4% Triton X-100. Coverslips were blocked in phosphate-buffered saline containing 4% fetal calf serum, and anti-FOXO3a antibody was added. After a washing step, rhodamine-conjugated anti-rabbit antibody (Jackson Laboratories) was added. Finally, the cells were counterstained with Hoechst 33258 and analyzed on a Leica confocal microscope system.
Cell sorting.
To analyze the successfully transfected LMP1-positive cells, the transfected cells were fixed with 70% ethanol and then permeabilized with 1% Triton X-100. The fixed cells were blocked in phosphate-buffered saline containing 4% fetal calf serum and then incubated with mouse monoclonal anti-LMP1 antibody. After a washing step, the cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody (Jackson Laboratories). The stained cells were sorted by using a FACSAria cell sorter and the FACSDiva software (Becton Dickinson). LMP1-positive cells were sorted based on the intensity of FITC compared to vector control cells.
RESULTS
LMP1 represses DNA repair, and the CTAR1 domain is required.
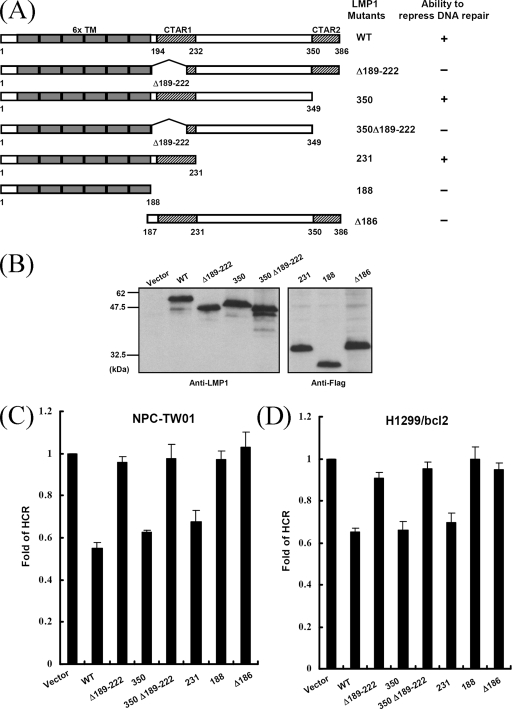
We demonstrated previously that LMP1 could repress DNA repair in a p53-independent manner, and the CTAR1 domain was required for this repression (40). To characterize further this phenomenon, various LMP1 mutants were constructed, as depicted in Fig. 1A: puroLMP1-Δ189-222, puroLMP1-350, puroLMP1-350Δ189-222, puroLMP1-231, puroLMP1-188, and puroLMP1-Δ186 (39). The expression levels of these LMP1 mutants in NPC-TW01 cells were analyzed by immunoblotting (Fig. 1B). We then transfected these plasmids into p53-proficient (NPC-TW01) and p53-deficient (H1299/bcl2) cells and carried out a HCR assay, which determines a cell's ability to repair UV-damaged DNA (54). Similar results were obtained in both cell lines (Fig. 1C and D). The wild-type LMP1-transfected cells lost ca. 40 to 50% of their ability to reactivate a UV-damaged luciferase reporter plasmid compared to cells transfected with the empty vector. Similarly, the repair capacity also was reduced in cells expressing the CTAR2 deletion mutants, LMP1-350 and LMP1-231. However, the repair capacity was almost normal in cells expressing the CTAR1 deletion mutants, LMP1-Δ189-222, LMP1-350Δ189-222, and LMP1-188. Moreover, mutant LMP1-Δ186, which retained only the C terminus, was not able to reduce the capacity for DNA repair. The ability of LMP1 mutants to repress cellular DNA repair capacity is summarized in Fig. 1A. These results are consistent with those of our previous study: LMP1 represses DNA repair in a p53-independent manner. Furthermore, the CTAR1 domain is necessary for LMP1-mediated repression of DNA repair, whereas CTAR2 is dispensable.
FIG. 1.
The CTAR1 domain is required for LMP1 to repress DNA repair. (A) Schematic representations of LMP1 and its mutants used in HCR assays. Six transmembrane (6x TM), CTAR1, and CTAR2 domains are indicated. Also shown is the ability of constructs indicated to repress DNA repair. (B) The expression levels of LMP1 and its mutants. A total of 5 × 105 NPC-TW01 cells were transfected with 2 μg of constructs encoding LMP1 or its mutants. At 24 h posttransfection, equal amounts of protein extracts were analyzed by Western blotting with anti-LMP1 or anti-Flag antibodies. (C and D) Totals of 105 NPC-TW01 (C) and H1299/bcl2 (D) cells per well in 24-well plates were cotransfected with 0.1 μg of an empty vector or constructs encoding LMP1 or its mutants, together with a UV-damaged (1,500 J/M2) or undamaged pCMV-Luc reporter construct (0.1 μg) and an undamaged pRL-CMV reporter construct (0.05 μg) as an internal control. At 24 h after transfection, the firefly and the Renilla luciferase activity were determined. The fold change of HCR represents DNA repair activity, calculated as described in Materials and Methods. All data presented represent the means and the standard deviations of at least five independent experiments conducted in duplicate.
LMP1 induces phosphorylation of Akt and enhances Akt kinase activity through the CTAR1 domain.
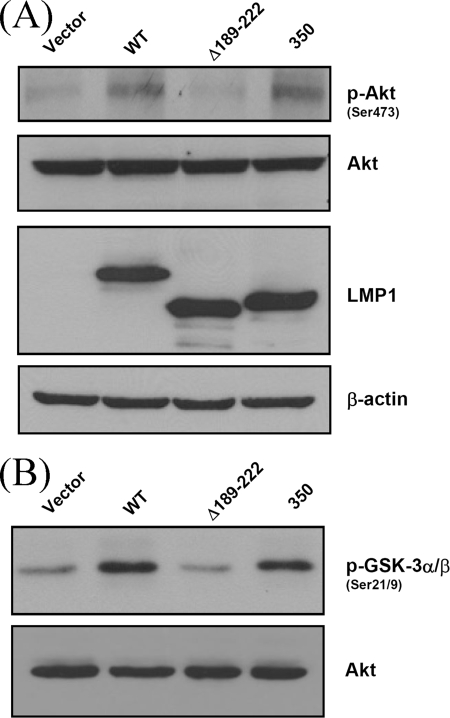
LMP1 is known to activate distinct signaling pathways through the CTAR1 or CTAR2 domain. The signaling pathways triggered by CTAR1 might be involved in LMP1-mediated repression of DNA repair. Earlier findings indicated that the PI3K/Akt pathway was activated by LMP1 CTAR1 but not by CTAR2 (8). To determine whether LMP1 triggers PI3K/Akt signaling by CTAR1 in the NPC cell line, the levels of phosphorylated Akt were determined by Western blotting (Fig. 2A). The levels of phosphorylated Akt in cells expressing LMP1 or the CTAR2 deletion mutant LMP1-350 were clearly higher than in cells expressing the CTAR1 deletion mutant LMP1-Δ189-222 or the empty vector. Furthermore, an Akt kinase assay was carried out with Akt immunoprecipitates and a GSK-3 protein as a substrate (Fig. 2B). There were significantly higher levels of phosphorylated GSK-3 in cells expressing LMP1 or the CTAR2 deletion mutant than in cells expressing the CTAR1 deletion mutant or the empty vector. These data indicate that LMP1 can phosphorylate Akt and enhance Akt kinase activity through the CTAR1 domain.
FIG. 2.
LMP1 induces phosphorylation of Akt and enhances Akt kinase activity through the CTAR1 domain. (A) A total of 5 × 105 NPC-TW01 cells were transfected with 2 μg of an empty vector or constructs encoding LMP1 or its mutants. After 24 h of serum starvation, cell extracts were obtained, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting with anti-phospho-Akt Ser473, anti-Akt, anti-LMP1, and anti-β-actin antibodies. (B) A total of 10 μg of protein lysates as described in panel A were subjected to detection of total Akt by Western blotting. A total of 200 μg of protein lysates as described in panel A were immunoprecipitated using an immobilized anti-Akt antibody, and immunoprecipitates were incubated with a GSK-3 fusion protein as the substrate. Phosphorylation of Ser-21/9 of GSK-3α/β, as a measure of Akt kinase activity, was then detected by Western blotting.
LMP1-mediated repression of DNA repair is dependent on PI3K.
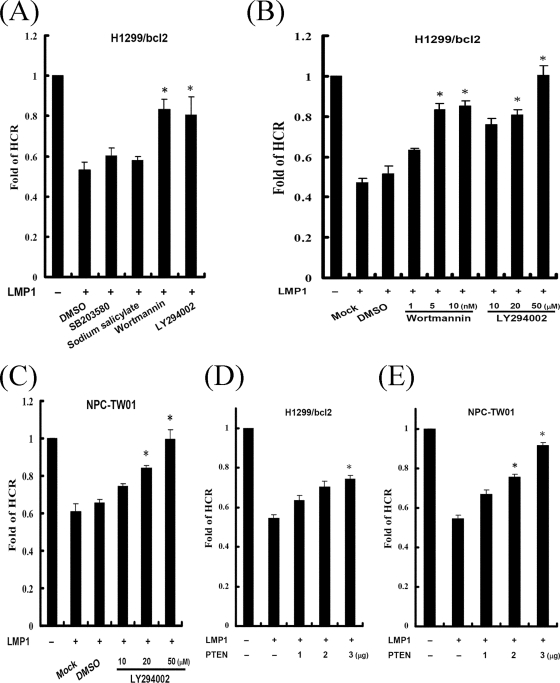
Through the CTAR1 domain, LMP1 not only represses cellular DNA repair but also activates the PI3K/Akt signaling pathway. To understand whether PI3K signaling contributes to LMP1-mediated repression of DNA repair, cells were transfected with the LMP1 vector and then treated with inhibitors of PI3K (wortmannin and LY294002), p38 (SB203580), or NF-κB (sodium salicylate), and the cellular capacity for DNA repair was assessed subsequently by using an HCR assay (Fig. 3A). LMP1 could repress DNA repair even when the p38 or NF-κB pathway was blocked. However, repression by LMP1 was abolished after treatment with PI3K inhibitors, which could rescue LMP1-repressed DNA repair in a dose-dependent manner (Fig. 3B). Similar results were also observed in NPC-TW01 cells (Fig. 3C). PTEN can dephosphorylate the lipid product of PI3K and antagonize PI3K activity (20). Therefore, a PTEN-expressing plasmid was used in the HCR assay to confirm the contribution of the PI3K signaling pathway to LMP1-repressed DNA repair (Fig. 3D and E). LMP1-diminished DNA repair was gradually restored as the levels of PTEN increased. These results suggest that PI3K signaling may be involved in LMP1-mediated repression of DNA repair in human epithelial cells.
FIG. 3.
LMP1-mediated repression of DNA repair is dependent on PI3K. (A) H1299/bcl2 cells were cotransfected with an empty vector or a construct encoding LMP1, together with a UV-damaged or undamaged pCMV-Luc reporter construct and an undamaged pRL-CMV reporter construct. One hour after transfection, cells were incubated in the absence (dimethyl sulfoxide [DMSO]) or presence of various inhibitors, including SB203580 (30 μM), sodium salicylate (10 mM), wortmannin (5 nM), and LY294002 (20 μM). Twenty-three hours later, luciferase assays were performed, and the fold change of HCR was calculated. (B and C) H1299/bcl2 (B) and NPC-TW01 (C) cells were cotransfected with identical plasmids as described in panel A. One hour after transfection, cells were mock treated (DMSO) or treated with various concentrations of signal transduction inhibitors, wortmannin (1, 5, and 10 nM), LY294002 (10, 20, and 50 μM). Twenty-three hour later, luciferase assays were performed, and the fold change of HCR was calculated. (D and E) H1299/bcl2 (D) and NPC-TW01 (E) cells were transfected with various amounts of PTEN expressing vector (1, 2, and 3 μg) and identical plasmids as described in panel A. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. All data presented represent the means and the standard deviations of at least five independent experiments conducted in duplicate. The asterisk indicates statistical significance (P < 0.001, relative to LMP1 alone [Student t test]).
LMP1-mediated repression of DNA repair is dependent on Akt.
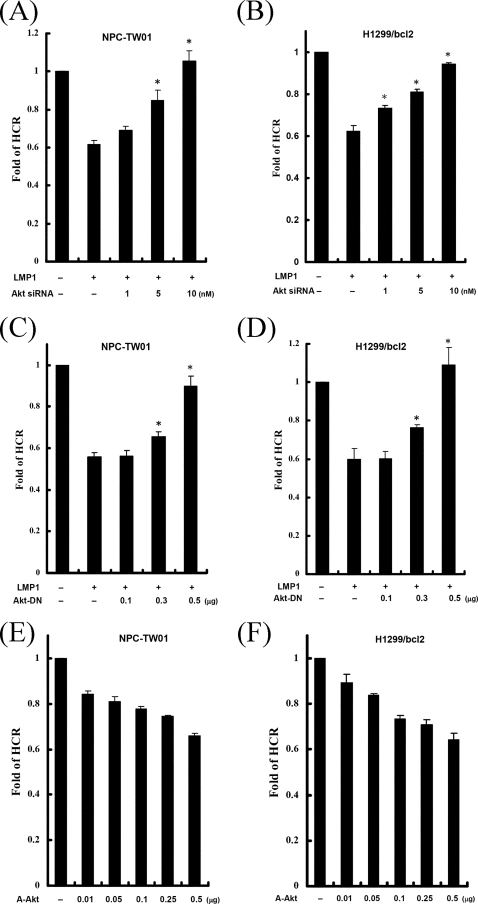
The protein kinase Akt is one of the best-characterized targets of PI3K lipid products. To determine whether Akt is involved in LMP1-repressed DNA repair, an siRNA was used to inhibit Akt1 in the HCR assay. LMP1-mediated repression of DNA repair was recovered by Akt1 siRNA in a dose-dependent manner but not by the scrambled siRNA (Fig. 4A and B). Moreover, we used a dominant-negative Akt1 (Akt-DN) expressing plasmid to perturb the Akt signaling pathway in the HCR assay (Fig. 4C and D). LMP1-diminished DNA repair was gradually rescued to normal with increasing amounts of Akt-DN. To define the role of Akt on DNA repair further, cells were transfected with empty vector or various amounts of the constitutively active Akt (A-Akt) expressing vector and then analyzed by the HCR assay (Fig. 4E and F). Indeed, expression of the constitutively active Akt inhibited cellular DNA repair capacity. Taken together, these data indicate that Akt signaling may be involved in LMP1-mediated repression of DNA repair in human epithelial cells.
FIG. 4.
LMP1-mediated repression of DNA repair is dependent on Akt, and the constitutively active Akt can repress DNA repair. (A and B) NPC-TW01 (A) and H1299/bcl2 (B) cells were cotransfected with an empty vector or a construct encoding LMP1 and negative control siRNA or various concentrations of siRNA for Akt1 (1, 5, and 10 nM), together with a UV-damaged or undamaged pCMV-Luc reporter construct and pRL-CMV reporter construct. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. (C and D) NPC-TW01 (C) and H1299/bcl2 (D) cells were cotransfected with various amounts of Akt-DN expressing vector (0.1, 0.3, and 0.5 μg), together with LMP1 and reporter constructs as described in panel A. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. (E and F) NPC-TW01 (E) and H1299/bcl2 (F) cells were cotransfected with an empty vector or various amounts of a constitutively active myristoylated Akt expressing plasmid (0.01, 0.05, 0.1, 0.25, and 0.5 μg), together with a UV-damaged or undamaged pCMV-Luc reporter construct and pRL-CMV reporter construct. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. All data presented represent the means and the standard deviations of at least five independent experiments conducted in duplicate. The asterisk indicates statistical significance (P < 0.001, relative to LMP1 alone [Student t test]).
The CTAR1 domain of LMP1 induces phosphorylation and relocalization of FOXO3a in a PI3K-dependent manner.
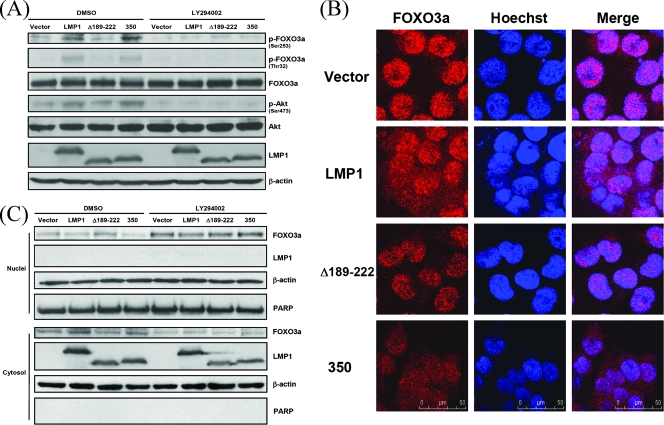
We sought to identify the downstream targets of the PI3K/Akt pathway involved in LMP1-mediated repression of DNA repair. FOXO3a, a member of the FOXO family, was chosen because it has been shown to stimulate DNA repair (63). To confirm that LMP1 influences the activity of FOXO3a, phosphorylated FOXO3a, considered the inactivated form, was detected by immunoblotting. The amount of phosphorylated FOXO3a was higher in cells expressing wild-type LMP1 and LMP1-350 than in control cells but was not increased in cells expressing LMP1-Δ189-222 (Fig. 5A, lanes 1 to 4). Importantly, phosphorylated FOXO3a was suppressed after LY294002 treatment, revealing the specificity of the interaction (Fig. 5A, lanes 5 to 8). Furthermore, the subcellular localization of endogenous FOXO3a in cells expressing LMP1 or its mutants was examined by confocal microscopy (Fig. 5B). Most of the endogenous FOXO3a was localized within the nucleus after overnight serum starvation. However, in the presence of LMP1, some FOXO3a was relocalized from the nucleus to the cytoplasm. Cytoplasmic retention of FOXO3a was also noted in the presence of the LMP1 CTAR2 deletion mutant but not in the presence of the CTAR1 deletion mutant. To analyze further the nuclear and cytosolic distribution of FOXO3a, cells were fractionated into nuclear and cytosolic parts, and the amount of FOXO3a was detected in each by immunoblotting. Compared to cells expressing the empty vector, a decrease of nuclear FOXO3a and an increase of cytosolic FOXO3a were observed in cells expressing LMP1 and LMP1-350, but not in cells expressing LMPΔ189-222 (Fig. 5C, lanes 1 to 4). After treatment with LY294002, an increase of nuclear FOXO3a and a decrease of cytosolic FOXO3a were observed in all cells, suggesting PI3K/Akt specificity (Fig. 5C, lanes 5 to 8). Taken together, it is possible that LMP1 may activate PI3K/Akt signaling by CTAR1 and subsequently cause phosphorylation of FOXO3a, leading to its retention in the cytoplasm and away from the nucleus.
FIG. 5.
LMP1 induces phosphorylation and relocalization of FOXO3a by CTAR1 in a PI3K-dependent manner. (A) A total of 5 × 105 NPC-TW01 cells were transfected with 2 μg of an empty vector or constructs encoding LMP1 or its mutants. After 23 h of serum starvation, cells were incubated without (DMSO) or with LY294002 (50 μM) for 1 h. Protein extracts isolated from the cells were analyzed by Western blotting with anti-phospho-FOXO3a Ser253, anti-phospho-FOXO3a Thr32, anti-FOXO3a, anti-phospho-Akt Ser473, anti-Akt, anti-LMP1, and anti-β-actin antibodies. (B) NPC-TW01 cells were cultured on sterile coverslips and transfected with an empty vector or constructs encoding LMP1 or its mutants. Cells were stained with an anti-FOXO3a antibody, followed by the addition of rhodamine-conjugated anti-rabbit antibody (red), and were analyzed by confocal microscopy. Nuclei were stained with Hoechst 33258 (blue). (C) A total of 107 NPC-TW01 cells were transfected with 4 μg of an empty vector or constructs encoding LMP1 or its mutants. After 23 h of serum starvation, cells were incubated without (DMSO) or with LY294002 (50 μM) for 1 h. Subsequently, nuclear and cytosolic protein extracts isolated from the cells were analyzed by Western blotting with anti-FOXO3a, anti-LMP1, anti-β-actin, and anti-PARP antibodies.
LMP1 represses FOXO3a-induced DNA repair.
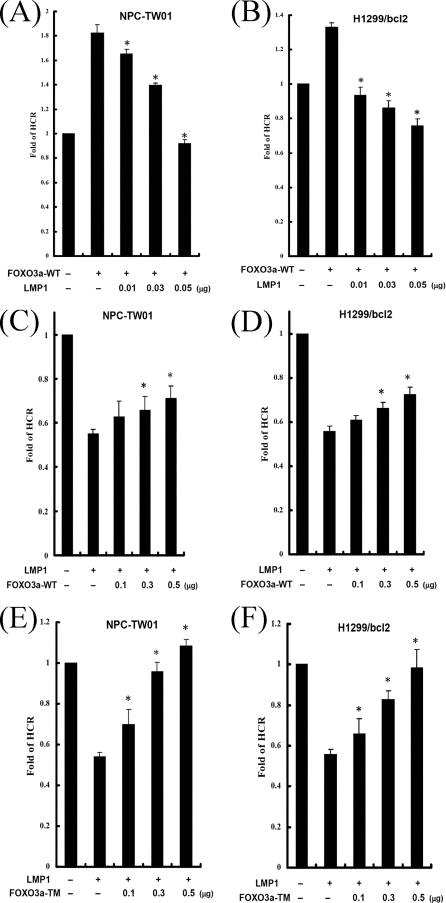
FOXO3a is known to trigger the repair of damaged DNA. Therefore, we sought to determine whether LMP1 influences FOXO3a-mediated DNA repair. Epithelial cells were cotransfected with the vector expressing FOXO3a and various doses of the vector expressing LMP1 and then analyzed by using the HCR assay. Consistent with a previous report, FOXO3a could enhance DNA repair in both cell lines (Fig. 6A and B, lanes 2). However, FOXO3a-induced DNA repair was gradually reduced by increasing LMP1 (Fig. 6A and B, lanes 3 to 5). This suggests that LMP1 can repress FOXO3a-mediated DNA repair.
FIG. 6.
LMP1 represses FOXO3a-mediated DNA repair, and LMP1-diminished DNA repair is partly rescued by FOXO3a-WT and completely rescued by FOXO3a-TM. (A and B) NPC-TW01 (A) and H1299/bcl2 (B) cells were cotransfected with 0.3 μg of an empty vector or wild-type FOXO3a expressing vector and various amounts of LMP1 expressing vector (0.01, 0.03, and 0.05 μg), together with a UV-damaged or undamaged pCMV-Luc reporter construct and pRL-CMV reporter construct as an internal control. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. The asterisk indicates statistical significance (P < 0.001, relative to FOXO3a alone [Student t test]). (C to F) NPC-TW01 (C and E) and H1299/bcl2 (D and F) cells were cotransfected with an empty vector or a construct encoding LMP1 and various amounts of FOXO3a-WT expressing vector (0.1, 0.3, and 0.5 μg) (C and D) or FOXO3a-TM expressing vector (0.1, 0.3, and 0.5 μg) (E and F), together with a UV-damaged or undamaged pCMV-Luc reporter construct and pRL-CMV reporter construct. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. All data presented represent the means and the standard deviations of at least five independent experiments conducted in duplicate. The asterisk indicates statistical significance (P < 0.001, relative to LMP1 alone [Student t test]).
LMP1-diminished DNA repair is rescued partly by FOXO3a-WT and completely by FOXO3a-TM.
Endogenous FOXO3a is known to be inactivated by LMP1/PI3K/Akt signaling, which may lead to repression of DNA repair. To understand whether exogenous FOXO3a rescues the LMP1-mediated repression of DNA repair, cells were transfected with empty vector or LMP1 plasmid and various amounts of the vector expressing wild type-FOXO3a and then analyzed by using the HCR assay (Fig. 6C and D). LMP1-mediated repression of DNA repair was partly restored by an increase of FOXO3a-WT compared to the vector control. To avoid interference by endogenous PI3K/Akt signaling, FOXO3a-TM, which was mutated in three key Akt regulatory sites, was used in the HCR assay (Fig. 6E and F). Without the disturbance of Akt signaling, expression of FOXO3a-TM did indeed restore LMP1-mediated repression of DNA repair. Taken together, LMP1-mediated repression of DNA repair is restored partly by FOXO3a-WT and completely by FOXO3a-TM. It is possible that LMP1 may interfere with endogenous FOXO3a, leading to repression of the cellular DNA repair capacity.
LMP1 represses the expression of DDB1 by CTAR1 in a PI3K-dependent manner.
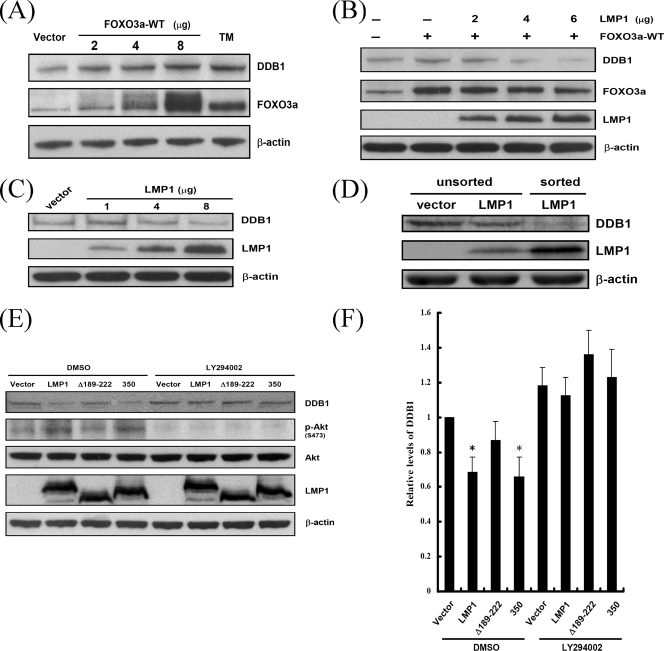
In order to understand how FOXO3a mediates DNA repair functions, we sought to identify the transcriptional targets of FOXO3a. Previously, several DNA damage response genes regulated by FOXO proteins were identified (55). Of these genes, we are especially interested in DDB1 because of its function in NER, which removes most UV damage. To confirm that FOXO3a induces expression of DDB1, cells were transfected with vectors expressing FOXO3a-WT or FOXO3a-TM, and then the expression of DDB1 was analyzed by immunoblotting (Fig. 7A). DDB1 was induced not only by wild-type FOXO3a in a dose-dependent manner but also by the constitutively active FOXO3a. This suggests that DDB1 is a transcriptional target of FOXO3a. However, FOXO3a-induced expression of DDB1 was downregulated by LMP1 (Fig. 7B). In addition, the expression of endogenous DDB1 also was repressed by LMP1 in a dose-dependent manner (Fig. 7C). Nevertheless, the inhibition of DDB1 is relatively modest at low levels of LMP1, which may be due to suboptimal transfection efficiencies. To further confirm the effect of LMP1, the successfully transfected LMP1-positive cells were sorted by fluorescence-activated cell sorting, and the DDB1 protein levels were analyzed by immunoblotting (Fig. 7D). Compared to the unsorted vector control or LMP1-transfected cells, the suppression of DDB1 in the sorted LMP1 expressing cells was more evident. This indicates that LMP1 truly can result in the suppression of DDB1. To determine which domain or triggering pathway of LMP1 is required for the suppression of DDB1, cells were transfected with plasmids expressing LMP1 or its mutant and the expression of DDB1 was analyzed by immunoblotting (Fig. 7E). Accompanied by the increase of phosphorylated Akt, a decrease of DDB1 was detected in cells expressing LMP1 and LMP1-350 compared to vector control cells, but there was no significant decrease in cells expressing LMP1-Δ189-222. The decrease of DDB1 in response to LMP1 and LMP1-350 was blocked by treatment with LY294002, which revealed the specificity of the interaction. The relative levels of DDB1 were given, as shown in Fig. 7F. These results indicate that LMP1 may repress DDB1 by CTAR1 through the PI3K/Akt/FOXO3a signaling pathway.
FIG. 7.
LMP1 decreases the expression of DDB1 by CTAR1 in a PI3K-dependent manner. (A) A total of 107 NPC-TW01 cells were transfected with an empty vector or various amounts of FOXO3a-WT (2, 4, and 8 μg) or FOXO3a-TM (8 μg) expressing plasmid. After 24 h, protein extracts isolated from the cells were analyzed by Western blotting with anti-DDB1, anti-FOXO3a, and anti-β-actin antibodies. (B) A total of 107 NPC-TW01 cells were cotransfected with an empty vector or FOXO3a expressing plasmid (2 μg) and various amounts of LMP1 construct (2, 4, and 6 μg). After 24 h, protein extracts were analyzed by Western blotting with anti-DDB1, anti-FOXO3a, anti-LMP1, and anti-β-actin antibodies. (C) A total of 107 NPC-TW01 cells were transfected with an empty vector or various amounts of LMP1 expressing plasmid (1, 4, and 8 μg). After 24 h, protein extracts were analyzed by Western blotting with anti-DDB1, anti-LMP1, and anti-β-actin antibodies. (D) A total of 107 NPC-TW01 cells were transfected with an empty vector or LMP1 expressing plasmid (2 μg). After 24 h, the cells were fixed and stained with anti-LMP1 antibody followed by the addition of FITC-conjugated antibody. The LMP1-positive cells were sorted based on the intensity of FITC compared to vector control. Protein extracts were analyzed by Western blotting with anti-DDB1, anti-LMP1, and anti-β-actin antibodies. (E) A total of 5 × 105 NPC-TW01 cells were transfected with 2 μg of an empty vector or constructs encoding LMP1 or its mutants. After 23 h of serum starvation, the cells were incubated without (DMSO) or with LY294002 (50 μM) for 1 h. Protein extracts were analyzed by Western blotting with anti-DDB1, anti-phospho-Akt Ser473, anti-Akt, anti-LMP1, and anti-β-actin antibodies. A representative example of four independent experiments is shown. (F) Relative levels of DDB1 as described in panel E. The intensities of the DDB1 bands were quantified by using the AlphaImager computer program. All samples were compared to the vector control. The bars represent the means of four independent experiments, and the error bars indicate standard deviations. The asterisk indicates statistical significance (relative to vector control [Student t test]).
LMP1-mediated repression of DNA repair is partly restored by DDB1.
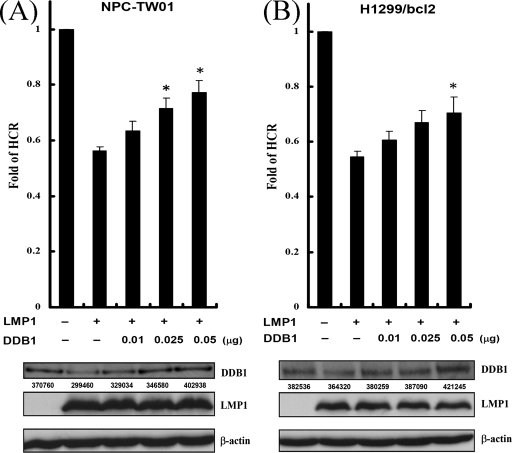
It was likely that DDB1 was inactivated by LMP1 through PI3K/Akt/FOXO3a signaling, which might contribute to the repression of DNA repair. To determine whether exogenous DDB1 rescues the LMP1-mediated repression of DNA repair, cells were transfected with plasmids expressing LMP1 and various amounts of plasmids expressing DDB1, and the HCR assays were carried out (Fig. 8). In parallel, the DDB1 levels were also detected by immunoblotting. Accompanied by the increase of exogenous DDB1, the LMP1-mediated repression of DNA repair was gradually restored. The expression of DDB1 was not sufficient to restore all LMP1-mediated repression of DNA repair, although the DDB1 protein levels were restored completely. These results indicate that LMP1-mediated suppression of DDB1 may partially contribute to the repression of the DNA repair.
FIG. 8.
LMP1-mediated repression of DNA repair is partly restored by DDB1. (A and B) NPC-TW01 (A) and H1299/bcl2 (B) cells were cotransfected with an empty vector or a construct encoding LMP1 and various amounts of DDB1 expressing vector (0.01, 0.025, and 0.05 μg), together with a UV-damaged or undamaged pCMV-Luc reporter construct and pRL-CMV reporter construct as an internal control. At 24 h after transfection, luciferase assays were performed, and the fold change of HCR was calculated. All data presented represent the means and standard deviations of at least five independent experiments conducted in duplicate. The asterisk indicates statistical significance (P < 0.001, relative to LMP1 alone [Student t test]). Western blot analysis was done to show the expression of DDB1 and LMP1. The data are the representative of three experiments. The AlphaImager computer program was used to analyze the DDB1 levels. The integrated density values were listed under the corresponding bands.
DISCUSSION
The major findings of the present study are not only the signaling pathway involved in LMP1-mediated repression of DNA repair but also an elucidation of the oncogenic properties of LMP1. In search of the signaling pathway involved in LMP1-mediated repression of DNA repair, we identified two important proteins that are linked to the LMP1 signaling. One is the FOXO3a transcription factor, which is considered to be a tumor suppressor. The other is the DDB1 protein, which is a subunit of the NER complex. LMP1 can trigger the PI3K/Akt pathway by its CTAR1 domain, in a p53 independent manner, to inactivate FOXO3a and then decrease DDB1 expression, subsequently leading to the repression of DNA repair capacity.
Several gene products of human tumor viruses, such as HTLV-1 Tax, HBV HBx, HPV E6, HCV core protein, and JCV T-antigen, have been reported to induce DNA damage or repress DNA repair, which may result in genomic instability (11, 28, 35, 44, 53, 64, 66). Similarly, the EBV-encoded LMP1 can induce micronucleus formation, repress DNA repair, and enhance cellular sensitivity to DNA-damaging agents in a p53-independent manner (40). Even in the presence of p53, LMP1 can repress p53-mediated DNA repair and transcriptional activity (39). Furthermore, LMP1 can disrupt microtubule structures and induce chromosomal aberrations (45). Taken together, EBV LMP1, in common with the gene products of other tumor viruses but using other mechanisms, can contribute to genomic instability.
In the present study, LMP1 triggered the PI3K/Akt pathway, leading to phosphorylation of FOXO3a and its retention in the cytoplasm. Subsequently, FOXO3a lost its ability to regulate target genes, and cellular DNA repair capacity was diminished. However, DNA repair was partly restored by wild-type FOXO3a and was completely restored by an Akt-resistant FOXO3a mutant. It is possible that endogenous PI3K/Akt signaling disturbs the function of wild-type FOXO3a and has no effect on FOXO3a-TM. However, another possibility that cannot be ruled out is that other FOXO transcription factors, such as FOXO1 and FOXO4, also participate in DNA repair. Thus, the expression of FOXO3a is not sufficient to restore repair capacity completely. However, expression of FOXO3a-TM can rescue the diminished DNA repair completely. This implies that FOXO3a may compensate for the repair functions of other FOXO transcription factors.
In addition to FOXO3a, other FOXO transcription factors, such as FOXO1 and FOXO4, were influenced by LMP1 in our preliminary experiments (data not shown). It still remains to be elucidated whether LMP1 affects these FOXO factors by a similar mechanism. In another study, FOXO1 has been shown to be downregulated in EBV-infected B cells. Two EBV proteins, LMP1 and LMP2A, may contribute to this repression by PI3K-mediated nuclear export, by inhibition of FOXO1 mRNA expression, and by alteration of posttranslational modifications (60). However, as we show here, it seems that the LMP1-mediated PI3K/Akt pathway plays the major role in FOXO3a inactivation because the PI3K inhibitor LY294002 can block LMP1-induced FOXO3a phosphorylation and relocalization. This may be explained by the difference between epithelial and B cells or different regulation of FOXO3a and FOXO1.
In mammals, FOXO transcription factors can induce cell cycle arrest, DNA repair, and apoptosis, which makes them attractive candidates as tumor suppressors (22). Here, we demonstrate that LMP1 can change the subcellular localization of FOXO3a and prevent its transcriptional activity, thereby leading to the repression of DNA repair. Previously, LMP1 was shown to have several oncogenic properties that interrupt normal cellular function and induce uncontrolled cellular growth. Whether LMP1 achieves its oncogenic effects, such as reducing cell cycle arrest or antiapoptosis, to a certain extent by interfering with the function of FOXO3a or other FOXO factors remains to be elucidated. However, it is not unusual for a viral protein to deregulate FOXO factors. Vpr, an associated accessory protein of human immunodeficiency virus type 1, was found to perturb the subcellular localization of FOXO3a (33). Furthermore, a recent report indicated that a product of another gammaherpesvirus, KSHV LANA2, was found to inhibit FOXO3a activity, which may promote tumorigenesis (49).
In addition to its ability to recognize DNA lesions, DDB1 has been shown to be a subunit of the Cullin 4A (CUL4A) E3 ubiquitin ligase. The CUL4A-DDB1 ligase could facilitate global genome and transcription-coupled NER progress by ubiquitinating certain NER factors, such as XPC, DDB2, and CSB. Other than the NER factors, several substrates of CUL4A-DDB1 ligase have been identified: e.g., DNA replication factor CDT1, cell cycle inhibitor p27Kip1, proto-oncogenic transcription factor c-Jun, and the histones H2A, H3, and H4 (52). This suggests that CUL4A-DDB1 ligase may play another role beyond NER in regulating the cell cycle, DNA replication, and genomic stability. In the present study, we demonstrate that EBV LMP1 could decrease the expression of DDB1 through the PI3K/Akt/FOXO3a pathway, leading to impaired DNA repair capacity. It would be interesting to see whether LMP1 also could influence the activity of CUL4A-DDB1 ligase and subsequently affect the stability of its substrates, leading to perturbation of normal biological functions. Interestingly, DDB1 is targeted by the gene products of other viruses, such as HBV HBx, human immunodeficiency virus type 1 Vpr, and simian parainfluenza virus 5 V (38, 59, 61).
Genomic instability often is associated with the progression of cancer but the underlying mechanisms are not completely understood. It has been proposed to involve either the loss of DNA repair capacity or the loss of chromosomal stability. EBV encodes the oncoprotein LMP1 seems to play a role in the development of genomic instability. First, LMP1 represses cellular DNA repair capacity, regardless of p53. Second, LMP1 enhances micronucleus formation, which is related to chromosomal aberration (40). Third, LMP1 disrupts microtubule structures and induces aneuploidy (45). Then, as we show here, LMP1 inactivates FOXO3a and decreases DDB1. FOXO3a can upregulate several DNA repair genes and DDB1 can maintain genome integrity (41, 55). Loss of FOXO3a and DDB1 may lead to genomic instability. Based on these studies, we suggest that, in addition to many other mechanisms, LMP1 may induce genomic instability and subsequently facilitate tumorigenesis.
Acknowledgments
This study was supported by grants from the National Science Council (grants NSC 90-2320-B-002-159, NSC 91-2320-B-002-100, and NSC92-2320-B-002-162) and the National Health Research Institutes, Taiwan.
We thank Tim J. Harrison and S. F. Lin for critical reading of the manuscript and the Core Facilities of National Health Research Institutes for technical support in the confocal microscopy and the fluorescence-activated cell sorting analysis.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Arden, K. C. 2006. Multiple roles of FOXO transcription factors in mammalian cells point to multiple roles in cancer. Exp. Gerontol. 41709-717. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, P. G., H. J. Coope, M. Rowe, and S. C. Ley. 2003. Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-κB2 p100 to p52. J. Biol. Chem. 27851134-51142. [DOI] [PubMed] [Google Scholar]
- 3.Bader, A. G., S. Kang, L. Zhao, and P. K. Vogt. 2005. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer 5921-929. [DOI] [PubMed] [Google Scholar]
- 4.Baichwal, V. R., and B. Sugden. 1988. Transformation of BALB 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2461-467. [PubMed] [Google Scholar]
- 5.Brinkmann, M. M., and T. F. Schulz. 2006. Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae. J. Gen. Virol. 871047-1074. [DOI] [PubMed] [Google Scholar]
- 6.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242564-567. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, C. W., A. B. Rickinson, and L. S. Young. 1990. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 344777-780. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 2783694-3704. [DOI] [PubMed] [Google Scholar]
- 9.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 167098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, and W. Hammerschmidt. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 632982-2989. [PubMed] [Google Scholar]
- 11.Duensing, S., and K. Munger. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 627075-7082. [PubMed] [Google Scholar]
- 12.Eliopoulos, A. G., S. M. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 731023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eliopoulos, A. G., J. H. Caamano, J. Flavell, G. M. Reynolds, P. G. Murray, J. L. Poyet, and L. S. Young. 2003. Epstein-Barr virus-encoded latent infection membrane protein 1 regulates the processing of p100 NF-κB2 to p52 via an IKKγ/NEMO-independent signalling pathway. Oncogene 227557-7569. [DOI] [PubMed] [Google Scholar]
- 14.Eliopoulos, A. G., M. Stack, C. W. Dawson, K. M. Kaye, L. Hodgkin, S. Sihota, M. Rowe, and L. S. Young. 1997. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-κB pathway involving TNF receptor-associated factors. Oncogene 142899-2916. [DOI] [PubMed] [Google Scholar]
- 15.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 161731-1742. [DOI] [PubMed] [Google Scholar]
- 16.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11435-444. [DOI] [PubMed] [Google Scholar]
- 17.Fahraeus, R., L. Rymo, J. S. Rhim, and G. Klein. 1990. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature 345447-449. [DOI] [PubMed] [Google Scholar]
- 18.Franken, M., O. Devergne, M. Rosenzweig, B. Annis, E. Kieff, and F. Wang. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 707819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara, Y., C. Masutani, T. Mizukoshi, J. Kondo, F. Hanaoka, and S. Iwai. 1999. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 27420027-20033. [DOI] [PubMed] [Google Scholar]
- 20.Furnari, F. B., H. J. Huang, and W. K. Cavenee. 1998. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 585002-5008. [PubMed] [Google Scholar]
- 21.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 166131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greer, E. L., and A. Brunet. 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 247410-7425. [DOI] [PubMed] [Google Scholar]
- 23.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411366-374. [DOI] [PubMed] [Google Scholar]
- 24.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10549-560. [PubMed] [Google Scholar]
- 25.Hwang, J. K., and C. T. Lin. 1997. Co-localization of endogenous and exogenous p53 proteins in nasopharyngeal carcinoma cells. J. Histochem Cytochem. 45991-1003. [DOI] [PubMed] [Google Scholar]
- 26.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 941447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 9412592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, S. Y., and S. J. Marriott. 1999. Disruption of nucleotide excision repair by the human T-cell leukemia virus type 1 Tax protein. J. Virol. 734299-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 909150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaye, K. M., K. M. Izumi, H. Li, E. Johannsen, D. Davidson, R. Longnecker, and E. Kieff. 1999. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J. Virol. 7310525-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keeney, S., G. J. Chang, and S. Linn. 1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 26821293-21300. [PubMed] [Google Scholar]
- 32.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 166478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kino, T., M. U. De Martino, E. Charmandari, T. Ichijo, T. Outas, and G. P. Chrousos. 2005. HIV-1 accessory protein Vpr inhibits the effect of insulin on the FOXO subfamily of forkhead transcription factors by interfering with their binding to 14-3-3 proteins: potential clinical implications regarding the insulin resistance of HIV-1-infected patients. Diabetes 5423-31. [DOI] [PubMed] [Google Scholar]
- 34.Kulwichit, W., R. H. Edwards, E. M. Davenport, J. F. Baskar, V. Godfrey, and N. Raab-Traub. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B-cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 9511963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, T. H., S. J. Elledge, and J. S. Butel. 1995. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol. 691107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, C. T., W. Y. Chan, W. Chen, H. M. Huang, H. C. Wu, M. M. Hsu, S. M. Chuang, and C. C. Wang. 1993. Characterization of seven newly established nasopharyngeal carcinoma cell lines. Lab. Investig. 68716-727. [PubMed] [Google Scholar]
- 37.Lin, C. T., C. I. Wong, W. Y. Chan, K. W. Tzung, J. K. Ho, M. M. Hsu, and S. M. Chuang. 1990. Establishment and characterization of two nasopharyngeal carcinoma cell lines. Lab. Investig. 62713-724. [PubMed] [Google Scholar]
- 38.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249189-200. [DOI] [PubMed] [Google Scholar]
- 39.Liu, M. T., Y. T. Chang, S. C. Chen, Y. C. Chuang, Y. R. Chen, C. S. Lin, and J. Y. Chen. 2005. Epstein-Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene 242635-2646. [DOI] [PubMed] [Google Scholar]
- 40.Liu, M. T., Y. R. Chen, S. C. Chen, C. Y. Hu, C. S. Lin, Y. T. Chang, W. B. Wang, and J. Y. Chen. 2004. Epstein-Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene 232531-2539. [DOI] [PubMed] [Google Scholar]
- 41.Lovejoy, C. A., K. Lock, A. Yenamandra, and D. Cortez. 2006. DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 267977-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luftig, M., T. Yasui, V. Soni, M. S. Kang, N. Jacobson, E. Cahir-McFarland, B. Seed, and E. Kieff. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-κB activation. Proc. Natl. Acad. Sci. USA 101141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mainou, B. A., D. N. Everly, Jr., and N. Raab-Traub. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 246917-6924. [DOI] [PubMed] [Google Scholar]
- 44.Majone, F., O. J. Semmes, and K. T. Jeang. 1993. Induction of micronuclei by HTLV-1 Tax: a cellular assay for function. Virology 193456-459. [DOI] [PubMed] [Google Scholar]
- 45.Man, C., J. Rosa, L. T. Lee, V. H. Lee, B. K. Chow, K. W. Lo, S. Doxsey, Z. G. Wu, Y. L. Kwong, D. Y. Jin, A. L. Cheung, and S. W. Tsao. 2007. Latent membrane protein 1 suppresses RASSF1A expression, disrupts microtubule structures and induces chromosomal aberrations in human epithelial cells. Oncogene 263069-3080. [DOI] [PubMed] [Google Scholar]
- 46.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7171-180. [PubMed] [Google Scholar]
- 47.Morrison, J. A., M. L. Gulley, R. Pathmanathan, and N. Raab-Traub. 2004. Differential signaling pathways are activated in the Epstein-Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res. 645251-5260. [DOI] [PubMed] [Google Scholar]
- 48.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80389-399. [DOI] [PubMed] [Google Scholar]
- 49.Munoz-Fontela, C., L. Marcos-Villar, P. Gallego, J. Arroyo, M. Da Costa, K. M. Pomeranz, E. W. Lam, and C. Rivas. 2007. Latent protein LANA2 from Kaposi's sarcoma-associated herpesvirus interacts with 14-3-3 proteins and inhibits FOXO3a transcription factor. J. Virol. 811511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb− phenotype. J. Biol. Chem. 27124317-24320. [DOI] [PubMed] [Google Scholar]
- 51.Nowak, M. A., N. L. Komarova, A. Sengupta, P. V. Jallepalli, M. Shih Ie, B. Vogelstein, and C. Lengauer. 2002. The role of chromosomal instability in tumor initiation. Proc. Natl. Acad. Sci. USA 9916226-16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Connell, B. C., and J. W. Harper. 2007. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 19206-214. [DOI] [PubMed] [Google Scholar]
- 53.Philpott, S. M., and G. C. Buehring. 1999. Defective DNA repair in cells with human T-cell leukemia/bovine leukemia viruses: role of tax gene. J. Natl. Cancer Inst. 91933-942. [DOI] [PubMed] [Google Scholar]
- 54.Protic-Sabljic, M., and K. H. Kraemer. 1985. One pyrimidine dimer inactivates expression of a transfected gene in xeroderma pigmentosum cells. Proc. Natl. Acad. Sci. USA 826622-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramaswamy, S., N. Nakamura, I. Sansal, L. Bergeron, and W. R. Sellers. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 281-91. [DOI] [PubMed] [Google Scholar]
- 56.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 57.Saito, N., G. Courtois, A. Chiba, N. Yamamoto, T. Nitta, N. Hironaka, M. Rowe, and S. Yamaoka. 2003. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-κB through distinct signaling pathways in fibroblast cell lines. J. Biol. Chem. 27846565-46575. [DOI] [PubMed] [Google Scholar]
- 58.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 7410681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrofelbauer, B., Y. Hakata, and N. R. Landau. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. USA 1044130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shore, A. M., P. C. White, R. C. Hui, A. Essafi, E. W. Lam, M. Rowe, and P. Brennan. 2006. Epstein-Barr virus represses the FOXO1 transcription factor through latent membrane protein 1 and latent membrane protein 2A. J. Virol. 8011191-11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sitterlin, D., T. H. Lee, S. Prigent, P. Tiollais, J. S. Butel, and C. Transy. 1997. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 716194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swart, R., I. K. Ruf, J. Sample, and R. Longnecker. 2000. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 7410838-10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran, H., A. Brunet, J. M. Grenier, S. R. Datta, A. J. Fornace, Jr., P. S. DiStefano, L. W. Chiang, and M. E. Greenberg. 2002. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296530-534. [DOI] [PubMed] [Google Scholar]
- 64.Trojanek, J., S. Croul, T. Ho, J. Y. Wang, A. Darbinyan, M. Nowicki, L. D. Valle, T. Skorski, K. Khalili, and K. Reiss. 2006. T-antigen of the human polyomavirus JC attenuates faithful DNA repair by forcing nuclear interaction between IRS-1 and Rad51. J. Cell Physiol. 20635-46. [DOI] [PubMed] [Google Scholar]
- 65.Uchida, J., T. Yasui, Y. Takaoka-Shichijo, M. Muraoka, W. Kulwichit, N. Raab-Traub, and H. Kikutani. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286300-303. [DOI] [PubMed] [Google Scholar]
- 66.van Pelt, J. F., T. Severi, T. Crabbe, A. V. Eetveldt, C. Verslype, T. Roskams, and J. Fevery. 2004. Expression of hepatitis C virus core protein impairs DNA repair in human hepatoma cells. Cancer Lett. 209197-205. [DOI] [PubMed] [Google Scholar]
- 67.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43831-840. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, J. B., W. Weinberg, R. Johnson, S. Yuspa, and A. J. Levine. 1990. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 611315-1327. [DOI] [PubMed] [Google Scholar]
- 69.Wittschieben, B. O., and R. D. Wood. 2003. DDB complexities. DNA Repair 21065-1069. [DOI] [PubMed] [Google Scholar]