Abstract
The La antigen (SS-B) associates with a wide variety of cellular and viral RNAs to affect gene expression in multiple systems. We show that La is the major cellular protein found to be associated with the abundant 44-nucleotide viral leader RNA (leRNA) early after infection with respiratory syncytial virus (RSV), a nonsegmented negative-strand RNA virus. Consistent with this, La redistributes from the nucleus to the cytoplasm in RSV-infected cells. Upon RNA interference knockdown of La, leRNA is redirected to associate with the RNA-binding protein RIG-I, a known activator of interferon (IFN) gene expression, and this is accompanied by the early induction of IFN mRNA. These results suggest that La shields leRNA from RIG-I, abrogating the early viral activation of type I IFN. We mapped the leRNA binding function to RNA recognition motif 1 of La and showed that while wild-type La greatly enhanced RSV growth, a La mutant defective in RSV leRNA binding also did not support RSV growth. Comparative studies of RSV and Sendai virus and the use of IFN-negative Vero cells indicated that La supports the growth of nonsegmented negative-strand RNA viruses by both IFN suppression and a potentially novel IFN-independent mechanism.
RNA and RNA-binding proteins are increasingly being recognized as activators of interferon (IFN) that play a profound role in innate immunity (70). Essentially all viruses activate type I IFNs, alpha IFN (IFN-α) and IFN-β, which signal through the common type I IFN receptor as part of the host defense system (53). Due to their critical role in host-virus relationships, type I IFN induction has been studied in detail (7, 32, 60). The current dogma is that all viruses make double-stranded RNAs that act as pathogen-associated molecular patterns that are recognized by pattern recognition receptors such as retinoic acid-inducible gene I (RIG-I) and MDA5 (35, 59, 71, 72). Both pattern recognition receptors contain DExD/H-box helicase and caspase recruitment domains and recruit adaptors that trigger specific kinases. These kinases phosphorylate IFN regulatory factor 3 and IFN regulatory factor 7, transcription factors that activate type I IFNs (32, 70). Recent results have shown that RIG-I and MDA5 distinguish between different RNA viruses (35, 71, 72). While MDA5 appears to be important for picornaviruses, RIG-I is important for negative-strand RNA viruses including influenza virus and paramyxoviruses such as respiratory syncytial virus (RSV), parainfluenza virus, and Sendai virus (SeV), all of which constitute significant human and animal health hazards. MDA5 is inhibited by the direct binding of paramyxoviral V proteins that share sequence motifs (3, 20, 68). In contrast, the RIG-I pathway is inhibited by apparently diverse viral proteins such as SeV C (63), influenza virus NS1 (49), and hepatitis C virus nonstructural proteins (65); however, the exact mechanisms of their interaction with RIG-I remain unknown.
Although RNA is a suspected ligand of RIG-I in infected cells, its identity has been speculative. Transcription and replication in nonsegmented negative-strand RNA viruses are catalyzed by viral RNA-dependent RNA polymerase in the cytoplasm. Infected cells contain three types of viral RNA, mRNAs, genomic (and antigenomic) RNAs, and viral leader RNA (leRNA) (4, 25), with no evidence of extensive double-stranded RNA (4). Because RIG-I prefers 5′-phosphorylated rather than 5′-m7G-capped RNA (33, 54), the capped viral mRNAs (6) are not expected to be efficient ligands for RIG-I. The full-length genomic (and antigenomic) RNAs are single stranded, encapsidated with viral nucleocapsid protein (1, 30), and resistant to RNase or small interfering RNA (siRNA) (11); it is unclear whether they will serve as efficient ligands for RIG-I. Recent studies indicate that uncapped, 5′-triphosphate-containing RNA (5′-ppp-RNA) activates type I IFN production (33, 54). RIG-I specifically bound 5′-ppp-RNA but not RNA with an m7G cap or other 5′ modifications (33).
The third type of viral RNA is short leRNA containing an unmodified 5′-ppp and an unmodified 3′ end (21, 41). It was recently shown that naked leRNA with 5′-ppp is indeed a potent activator of RIG-I and IFN using a measles virus-derived system (55). The leRNA gene is the most promoter-proximal transcription unit in the viral genome, and accordingly, leRNA is an early and abundant viral transcript generated in vitro. In more extensively studied viruses such as vesicular stomatitis virus (VSV), leRNA is detectable as a ∼44-nucleotide (nt) RNA species in infected cells (4, 14, 41). The N gene is the second transcription unit in all nonsegmented negative-strand viral genomes (except in RSV, where it is fourth), and thus, N is an abundant viral protein in infected cells (5). After a substantial amount of de novo N protein accumulates, it begins to encapsidate leRNA, promoting a transcription-to-replication switch (14, 15, 16, 52). However, until this happens, the leRNA should be available to associate with cellular RNA-binding proteins. Our experiments uncovered the cellular La antigen, an abundant RNA-binding phosphoprotein involved in the metabolism of many cellular and viral RNAs, as being a leRNA-interacting factor that serves to attenuate RIG-I activation in RSV-infected cells and promote virus growth.
The La antigen is a member of the ribonucleoprotein (RNP) family of RNA recognition motif (RRM) proteins with diverse roles in the metabolism of a large variety of cellular and viral RNAs (18, 42, 46). Binding studies, crystal structures, and in vivo analyses indicate that La recognizes the unmodified UUU-3′-OH termini of RNA polymerase III transcripts (66) and functions in the processing of abundant small noncoding RNAs such as tRNAs, ribosomal 5S rRNA, and adenovirus VA1 RNA (23, 46). La is also found to be associated with selected polymerase II transcripts, such as snRNA precursors that end with UUU-3′-OH (46). La binds with highest affinity to nascent 5′-ppp-containing RNAs with two to three U residues and a free-OH group at the 3′ end (10).
Pioneering and later studies have shown an association of La with the leRNA of nonsegmented negative-strand RNA viruses such as VSV (39, 69), human parainfluenza virus type 3 (24), Rinderpest virus (56), and rabies virus (40) and with nonleader RNA species of influenza virus (51) and human immunodeficiency virus (19, 64). It was also postulated that La stabilizes hepatitis B virus mRNA (31). For several viruses, La enhances internal ribosome entry site function (2, 9, 22, 48, 58, 61). However, none of these viral leRNAs, including that of RSV, contains a UU-3′-OH end, suggesting a noncanonical interaction with La. Unfortunately, the biological significance of these La-leRNA interactions remained unknown. Here, we examined the encapsidation status of leRNA in RSV-infected cells. Our results document that RSV leRNA is not naked in infected cells and does not associate with RIG-I. Rather, we find that RSV leRNA is bound and sequestered by La antigen early in infection. Only when La levels were decreased by siRNA was leRNA found to be associated with RIG-I, and this was accompanied by the early induction of IFN mRNA. These results provide the first evidence that a virus uses La to inhibit IFN activation by shielding viral leRNA from RIG-I. We map the domains of La involved in leRNA binding and show that La is also essential for viral growth in an IFN-independent pathway.
MATERIALS AND METHODS
Virus growth.
All cell cultures were done using Dulbecco's modified Eagle's medium with 10% fetal calf serum. Wild-type RSV strain Long of the A serotype was routinely grown on HEp-2 cells, whereas SeV strain Enders was grown in Vero cells. For SeV, the medium additionally contained acetylated trypsin (1 μg/ml; Sigma).
Isolation and use of leRNA and leader RNP (leRNP).
The 44-nt positive-strand leRNA of RSV, 5′-ACGCGAAAAAAUGCGUACAACAAACUUGCAUAAACCAAAAAAAU-3′ (25, 50), with biotin at either the 5′ or 3′ end, was purchased from Dharmacon (Lafayette, CO) and attached to streptavidin agarose beads. In vitro-transcribed leRNA (with 5′-ppp) was synthesized by reconstituted RSV as described previously (5, 25), purified after denaturing 25% polyacrylamide gel electrophoresis, ethanol precipitated, and dissolved in Tris-EDTA. For electrophoretic mobility shift assay (EMSA), unbiotinylated leRNA was phosphorylated by T4 polynucleotide kinase using [γ-32P]ATP.
The in vivo leRNA/leRNP was isolated from RSV-infected A549 cells at 8 h postinfection (p.i.). Cells from two 10-cm dishes were lysed as described previously (14), with some modifications. The monolayers were washed with phosphate-buffered saline (PBS) and lysed by gentle pipetting in a total volume of 1 ml buffer A (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 10 mM MgCl2) plus 0.5% NP-40. The lysate was clarified by centrifugation at 13,000 × g for 5 min. CsCl stock solution (1.2 ml at a concentration of 1.55 g/ml) was made fresh in buffer A containing 0.5 mM each of phenylmethylsulfonyl fluoride and dithiothreitol and mixed with 0.8 ml of the clarified lysate. Tubes were centrifuged at 90,000 rpm for 20 h at 10°C in a Beckman TLX tabletop ultracentrifuge using a TLV 100 rotor. A control gradient contained the exact density positions as determined by weighing 100-μl portions of its fractions.
Following collection, the RNP or RNA fraction was dialyzed at an 8-kDa cutoff (BRL/Invitrogen, Carlsbad, CA) in cold for 2 h against buffer B (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4-KH2PO4 [pH 7.6], 25 mM HEPES [pH 7.6], 5 mM MgCl2) (67). The dialyzed material was used for three purposes: electroporation, immunoprecipitation (IP), and analysis of RNA. For electroporation, RSV nucleocapsid was transcribed in vitro, and the 44-nt leRNA was isolated by denaturing gel electrophoresis and purified (25).
Electroporation with leRNA or leRNP was done as described previously for influenza virus RNP (43), with buffer modifications. Trypsinized A549 cells (300 μl) were resuspended at 2 × 107 cells/ml in buffer B and mixed with the leRNP or leRNA (100 or 200 nmol RNA equivalent, determined by UV after deproteinization) in 100 μl of buffer B. The mixture was electroporated at 1.5 keV and at 25 μF (Gene Pulser; Bio-Rad, Hercules, CA) in a sterile cuvette with a 2-mm gap, was left undisturbed for 15 min, and was then overlaid onto a culture dish. To detect the internalized RNA (Fig. 1B, inset), total RNA at 10 h p.i. was purified by RNeasy Plus (Qiagen, Valencia, CA).
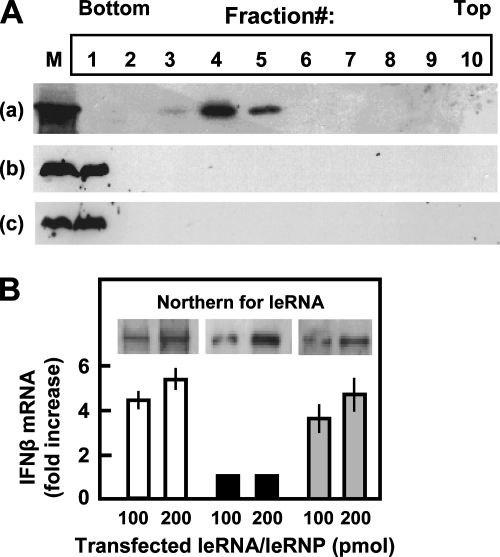
FIG. 1.
RSV leRNA is encapsidated and does not activate type I IFN. (A) Cytosolic extracts from RSV-infected A549 cells at 8 h p.i. without any treatment (a) or after digestion with protease K and 0.1% SDS (b) and pure, synthetic leRNA (c) were subjected to CsCl centrifugation, and fractions were collected and analyzed for leRNA content by Northern analysis as described in Materials and Methods. Autoradiographs of Northern panels are shown. In lane M, synthetic leRNA was electrophoresed to serve as a molecular weight marker. (B) A549 cells were electroporated with 100 or 200 pmol of the following: in vitro RSV-transcribed leRNA (white bars), CsCl-banded leRNP from A (black bars), and leRNA deproteinized from CsCl leRNP (gray bars). Northern analysis (inset) shows leRNA and leRNP from electroporated cells. IFN-β mRNA was quantified by qRT-PCR and plotted as the increase relative to the amount of 18S rRNA in the same samples.
Reverse transcription and real-time PCR (quantitative reverse transcription [qRT]-PCR) using total RNA as a template were done as described previously (13) using primers listed in Table 1.
TABLE 1.
qRT-PCR primersa
| Gene | Primer sequence |
|---|---|
| NS1 | GAATGGCATTGTGTTTGTGC |
| TGGCATTGTTGTGAAATTGG | |
| N | TGCAGGGCAAGTGATGTTAC |
| TTCCATTTCTGCTTGCACAC | |
| P | GGCAAGACTCAGGAATGAGG |
| TCCCTTCCAACAGGTTGTTC | |
| M | AATGCCCAGCAAATTTACCA |
| GCCTTGATTTCACAGGGTGT | |
| SH | CCAATCTGATGGCACAAAAC |
| GCTTGCATGGTGAGATGTTG | |
| G | AAGTCAACCCTGCAATCCAC |
| TTTGTTTTGGCGTTGTTTTG | |
| F | TAGGAGCCATTGTGTCATGC |
| ATCGCACCCGTTAGAAAATG | |
| M2 | CCCATGCACTGCTTGTAAGA |
| CCAACTCTGCAGCTCCACTT | |
| L | TCCTGCTACAGATGCAACCA |
| ACAGGCAATTCAGCATCACA | |
| GAPDH | GGCCTCCAAGGAGTAAGACC |
| AGGGGAGATTCAGTGTGGTG | |
| IFN-β | TGCTCTGGCACAACAGGTAG |
| TGGAGAAGCACAACAGGAGA |
The GenBank accession numbers for RSV, GAPDH, and IFN-β sequences are M74568, NM_002046, and NM_002176, respectively. For each gene, the top sequence is the sense strand and the bottom sequence is the antisense strand. All primers are written 5′ to 3′.
IP was done according to standard procedures (38). The leRNP (50 pmol, equivalent to ∼8 μg N) in buffer B was mixed with an excess (40 μg) of either mouse monoclonal anti-RSV N (Biodesign International), rabbit anti-La (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit RIG-I, or anti-MDA5 (ProSci, Poway, CA) antibody. The complexes were bound to protein A-Sepharose beads by mixing in cold for 2 h. The bound proteins in one-third of the IP pellet were solubilized in sodium dodecyl sulfate (SDS) sample buffer and analyzed by immunoblotting (Western) using the following goat antibodies: anti-RSV N from Chemicon, Billerica, MA, and anti-La, anti-RIG-I, and anti-MDA5 from Santa Cruz. The blot was developed with horseradish peroxidase-conjugated anti-goat secondary antibody, followed by ECL detection (Pierce, Rockford, IL). leRNA in the remaining two-thirds of the IP pellet was detected by Northern analysis.
Northern analysis of leRNA.
Where mentioned, RNP was deproteinized by phenol-chloroform, followed by ethanol precipitation with glycogen carrier, and dissolved in Tris-EDTA. Protein-free leRNA was fractionated on 1.8% agarose morpholinepropanesulfonic acid-formaldehyde gels (56), transferred onto a Hybond-N membrane, UV cross-linked, and probed by 32P-labeled antisense DNA using stringent conditions as described previously (56). The washed membrane was autoradiographed.
Immunofluorescence studies.
Cell monolayers at various times p.i. were rinsed in PBS; fixed with ice-cold 10% trichloroacetic acid for 15 min; washed successively in cold 70%, 90%, and 100% ethanol (3 min each); permeabilized with 0.2% Triton X-100 in cold PBS for 10 min; and then incubated with mouse antibody against La (BD Transduction Laboratories, San Jose, CA) and goat antibody against RSV (Chemicon) for 2 h. Cells were then washed with PBS (three times for 5 min) and incubated with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G and tetramethyl rhodamine isothiocyanate-conjugated anti-goat immunoglobulin G. Finally, the cells were washed three times in PBS, mounted with DAPI (4′,6′-diamidino-2-phenylindole)-DABCO (1,4=diazabicyclo[2.2.2]octane), and analyzed using Nikon TE2000E2 Perfect Focus fluorescence imaging. For best contrast in the merged pictures, red and green were digitally switched such that La was red and RSV was green (see Fig. 5).
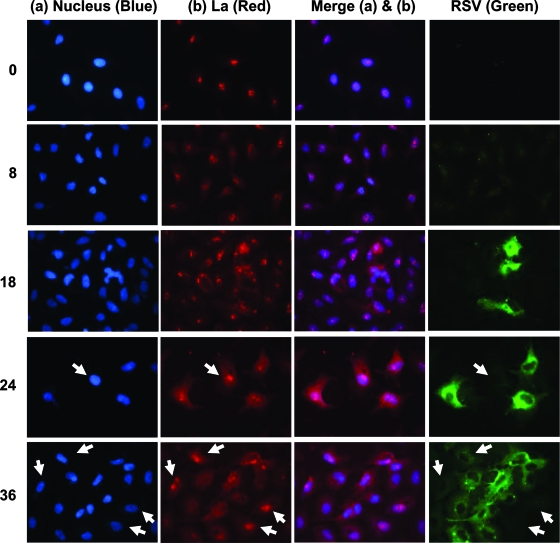
FIG. 5.
Nucleus-to-cytoplasm translocation of La in RSV-infected cells. A549 cells were infected with RSV and at times thereafter (hours on the left) and were fixed and stained with DAPI (nuclei) (blue), anti-La (red), and anti-RSV (green), as indicated above the columns. A few representative nuclei (marked with white arrowheads) show higher levels of nuclear La (red), even at 24 and 36 h p.i., in cells that are poorly infected with RSV (green) (see the text).
Cloning and RNA interference.
cDNAs corresponding to the full-length human La protein or amino acids 1 to 187 of human La protein (hLa1-187) were cloned into the EcoRI/BglII sites of vector pCAGGS using primers that contained EcoRI and BamHI sites (La has an internal BglII site). The forward primer is ATAGCCAgaattcGCCGCCACCATG(CATCATCATCATCATCAC)GCTGAAAATGGTGATAATGAAAAGATG (the EcoRI site is in lowercase type, the Kozak sequence is underlined, and six-His codons are in parentheses), followed by 27 nt of La after its initiator ATG (initiation is upstream of His codons). The recombinant proteins contained a N-terminal His6 tag and were detected by immunoblotting with anti-La or an anti-His antibody.
Transfections with siRNA were performed as described previously (11). Briefly, siRNAs (Dharmacon) were transfected with siPORT NeoFX reagent (Ambion, Austin, TX). Where mentioned, the cells were infected 24 h later. The sense strands of the siRNA, corresponding to the (N)19 core (26), were as follows, and a dTdT extension was added to the 3′ ends of both strands of the siRNA (11, 26): GACCUGUAGAGAAGAUUUA (La), GCCCATTTAAACCAAGAAA (RIG-I), and GGCCTTACCAAATGGAAGT (MDA5). Negative control luciferase siRNA was described previously (12). All siRNAs were purchased from Dharmacon and Ambion.
To create recombinant His-tagged La containing wild-type amino acids but resistant to the siRNA described above, the siRNA target region was changed to alternate but synonymous codons using QuikChange mutagenesis (Stratagene, La Jolla, CA). The mutagenic sense primer had the sequence 5′-CTTGCTGAAATTTTCGGGTGATTTAGATGATCAGACGTGCAGGGAGGACCTCCACATACTTTTCTCAAATCATGGTGAAATAAAATGG-3′ (mutated nucleotides are underlined).
Statistical analysis.
Changes were analyzed by one-way analysis of variance and by Student's t test with Bonferroni correction. All numerical data were collected from at least three separate experiments. Results were expressed as means ± standard errors of the means (error bars in graphs). Differences were considered to be significant at a P value of <0.05.
RESULTS
Viral leader RNA in the early-infected cell is fully complexed with protein.
Classical studies have shown that the nonsegmented negative-strand RNA viral (Mononegavirales) leRNA binds viral N protein and that this is a prerequisite to the replication of the viral genome (4, 16, 52). Since the innate immune response is an early process in infection, we focused on leRNA, the earliest transcriptional product of the incoming RNA virus. For VSV and SeV at 16 h p.i., the leRNA was found exclusively as RNP particles sedimenting at a 1.33-g/ml buoyant density in CsCl (14), which is typical of RNPs. The complexes contained leRNAs and N proteins, although the possible presence of other proteins was not examined. To examine the RNP at early times of infection, we gently lysed RSV-infected cells at 8 h p.i. and performed centrifugation in CsCl. In a parallel experiment, in vitro-synthesized leRNA was subjected to a similar centrifugation. A total of 10 fractions were collected from each gradient, and an equal portion of each was deproteinized and analyzed for leRNA by Northern analysis. The CsCl concentration of each fraction from a parallel tube was determined by weighing it. Results (Fig. 1A) show that essentially all of the leRNA was in a single peak corresponding to about 1.33 g/ml (mainly fraction 4 from the bottom) (Fig. 1Aa), which is characteristic of RNP. In clear contrast, the synthetic naked leRNA sedimented to the bottom (Fig. 1Ab). When the infected cell extract was digested with protease and then subjected to CsCl centrifugation, the majority of leRNA sedimented to the bottom, indicating that the CsCl banding was due to encapsidation by a protein. Thus, even as early as 8 h p.i., RSV leRNA was sequestered as an leRNP.
We then tested if this leRNP is proficient in activating IFN by transfecting it into A549 cells. Control cells were transfected with a comparable quantity of in vitro-transcribed naked leRNA, and transfection efficiency was monitored by Northern analysis (Fig. 1B, inset). IFN-β mRNA levels, measured 18 h p.i., revealed that while naked leRNA induced IFN-β mRNA (Fig. 1B, white bars), leRNP did not (black bars). As another control, the isolated leRNP was deproteinized, and its leRNA, when transfected, induced IFN-β mRNA (Fig. 1B, gray bars). Taken together, these results show that RSV leRNA, as found in its natural state in infected cells, is sequestered as an RNP and is largely inactive for inducing IFN-β.
Viral leader RNA in the infected cell is associated with either N or La but not RIG-I.
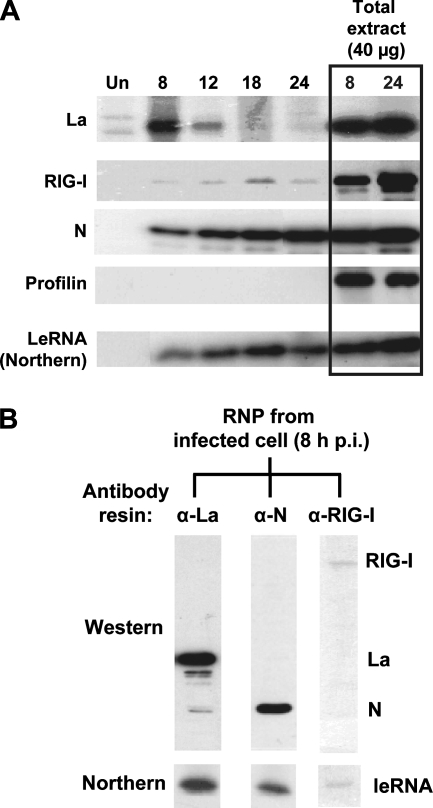
To obtain an estimate of the relative contributions of N, La, and RIG-I in constituting the leRNP, we obtained the CsCl-banded infected cell viral RNPs (corresponding to fraction 4) (Fig. 1A) and analyzed them by two mutually complementary approaches. First, RNPs obtained at various times of infection (8 to 24 h) were subjected to Western analysis using antibodies against La, RIG-I, and N to obtain a dynamic picture of the RNP protein content. As can be seen in Fig. 2A, the RNP at 8 h contained primarily La and some N protein. As N accumulated with time, the RNP protein profile shifted from La to N and was essentially all N by 24 h of infection. RIG-I was notably absent from RNP at all times, which could not be due to the lack of RIG-I in the cell because appreciable amounts of RIG-I could be detected in the total cell extract (the two rightmost lanes in Fig. 2A). Northern analysis showed gradually increasing amounts of leRNA synthesized by the replicating virus, as expected.
FIG. 2.
Protein composition of intracellular viral leRNP. (A) Western analysis of the total pool of infected cell RNP at various times postinfection. Equal amounts of uninfected (Un) or RSV-infected A549 cell extracts (at 8, 12, 18, and 24 h p.i.) were subjected to CsCl buoyant density centrifugation, and the materials from the RNP band region (i.e., fraction 4 in Fig. 1A) were subjected to Western analysis to detect La, RIG-I, or N. In parallel lanes (boxed), total cell extracts (equivalent to 40 μg protein) were directly subjected to Western analysis. Profilin serves as a negative control for leRNA binding and as evidence for equal loading in the total extract lanes. (Bottom) Northern analysis of the corresponding fractions to determine their leRNA contents. Note the high level of content of La in early-stage RNP, which is replaced by increasing N content of the RNP over time. (B) Protein-based selection and analysis of infected cell RNP. Equal amounts of CsCl-banded RSV-infected cell leRNP were incubated with protein A-Sepharose beads conjugated to antibodies against La, N, or RIG-I, as indicated above the lanes. Equal portions of the bound material from each column were analyzed by immunoblotting (Western) using a mixture of antibodies to La, N, and RIG-I. The remainder of the bound material was deproteinized, and the leRNA content was determined by Northern analysis (bottom).
In the second approach, we affinity purified RNP species with specific bound proteins using Sepharose beads coupled to antibody against La, N, or RIG-I. The RNP from each affinity column was examined by immunoblotting (Western) using a mixture of all three antibodies and by Northern analysis for leRNA content. Results (Fig. 2B) with the infected cell RNP at 8 h show that RNPs from the anti-La and anti-N columns contained leRNA (Fig. 2B, bottom), with the former making a greater contribution. In contrast, the anti-RIG-I column bound little RNP such that only trace amounts of leRNA were detected. A simple interpretation of these data is that infected cells contain mainly two protein-bound leRNA species, La-leRNA and N-leRNA, with little or no RIG-I bound, which agrees with our Western data for the unfractionated RNP (Fig. 2A).
In addition, we noticed that leRNP, affinity selected by anti-La or anti-N, contained the corresponding single protein but not both proteins. This suggests that each molecule of leRNA is encapsidated by either N or La but not a mixture of the two, the reason for which is currently unclear.
The loss of La protein leads to an increased association of leader RNA with RIG-I.
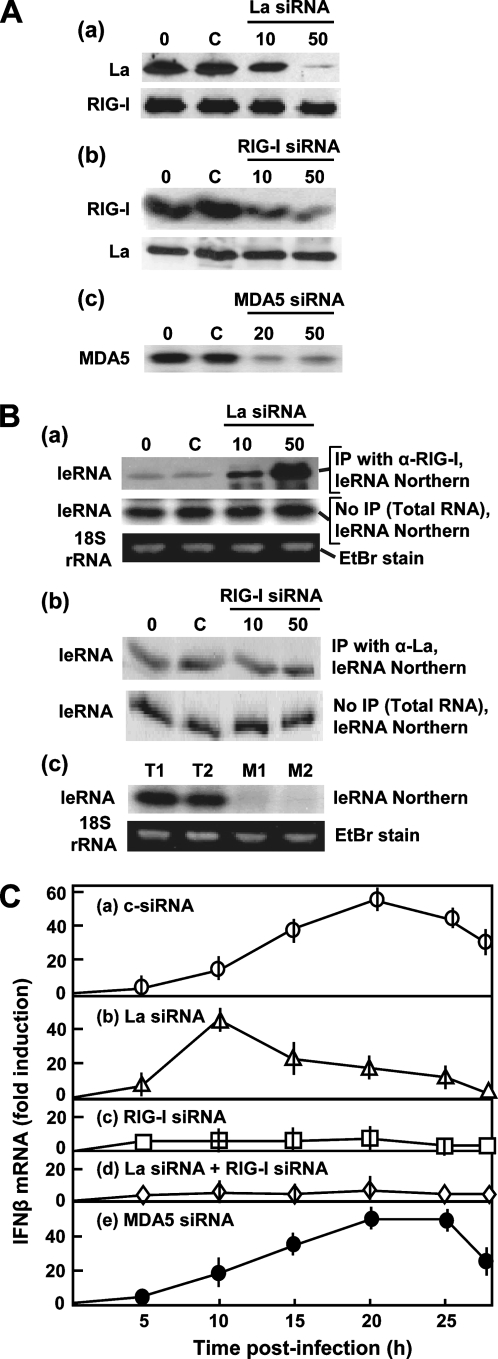
If La binds leRNA early in infection, it may serve to exclude RIG-I from leRNA, potentially moderating IFN gene induction. Accordingly, we hypothesized that the loss of the La protein may free leRNA to bind RIG-I and activate IFN expression. To test this, we knocked down La and RIG-I using siRNAs. We first confirmed that the siRNAs indeed specifically silenced the respective target and not the other protein (Fig. 3Aa and b). Another control siRNA, designed against luciferase, also had no effect on La and RIG-I (not shown).
FIG. 3.
Relationship of La with the RIG-I-mediated IFN response. A549 cells were infected with RSV at 24 h post-siRNA addition, and IP was done 8 h thereafter. (A) Immunoblots show the silencing efficacies of La (a), RIG-I (b), and MDA5 (c) siRNA at concentrations (in nM) indicated above the lanes. Lane 0, no siRNA; lane C, 50 nM of control, anti-luciferase siRNA. For La and RIG-I siRNA, the reciprocal proteins also serve as controls for specificity. (B) Northern analysis of leRNA. Panels correspond to those in A. IP was done with anti-RIG-I when La was silenced by siRNA (a) and vice versa (b). Equal amounts of total RNA were also analyzed (without prior IP), as indicated to the right of each panel, for an internal control. For MDA5, fewer samples are presented because results were negative. T1 and T2, total RNA; M1 and M2, after IP with anti-MDA5; T1 and M1, cells transfected with no siRNA; T2 and M2, transfected with 40 nM MDA5 siRNA. In a and c, where the leRNA levels were different between lanes, an ethidium bromide (EtBr)-stained 18S RNA band is shown to confirm equal loading. (C) Kinetics of IFN-β gene expression following RSV infection of A549 cells in which the indicated genes were knocked down by siRNA (50 nM).
To test the effect of the knockdown on leRNPs, we performed reciprocal IPs of the leRNP at 8 h p.i. and determined the amount of leRNA in the precipitate. Results (Fig. 3B) indeed revealed that with an increasing reduction of the La protein by siRNA, progressively more leRNA bound to RIG-I, providing support for the idea that La sequesters leRNA from RIG-I. In contrast, the loss of RIG-I did not increase the La association with leRNA. MDA5, the other prominent member of the DExD/H-box caspase recruitment domain helicase family, did not associate with leRNA (Fig. 3Bc, lanes M1 and M2), ruling out a significant role of MDA5 in constituting the leRNP. These data show that RIG-I is capable of binding leRNA (in the absence of La) and support the hypothesis that La outcompetes RIG-I for the association with leRNA.
To see if La prevents IFN activation by RIG-I, we determined the kinetics and magnitude of IFN-β gene expression by infecting the La- and/or RIG-I-knocked-down cells with RSV. IFN-β levels peaked at about 20 h p.i. in the control siRNA-treated cells (Fig. 3Ca), confirming previously reported kinetics (44, 55). In La-deficient cells (Fig. 3Cb), higher IFN-β mRNA levels could be seen at much earlier times, peaking at about 10 h p.i. If this shift in the IFN-β response is dependent on RIG-I, the loss of RIG-I should abrogate IFN-β induction. Indeed, in RIG-I knockdown cells with (Fig. 3Cc) or without (d) La, RSV infection failed to induce IFN-β. The loss of MDA5 had no effect on either the kinetics or magnitude of IFN induction by RSV (Fig. 3Ce). We conclude that RIG-I is required for IFN-β induction at all stages of RSV infection and that early induction is blocked by La. The cumulative data provide strong support for the model that La sequesters or shields leRNA from RIG-I, thereby blocking the RIG-I-mediated early induction of IFN.
La greatly enhances RSV replication.
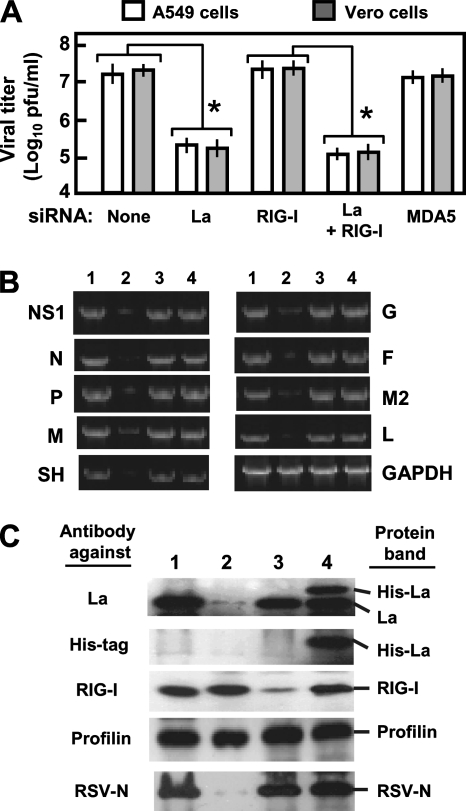
Next, we wanted to assess if La is important for RSV growth. For this, we knocked down La, RIG-I, or both in A549 cells by siRNA, infected the cells with RSV, and measured progeny viral titer at 40 h p.i. The knockdown of La decreased RSV growth by about 100-fold, with or without the concomitant knockdown of RIG-I, suggesting that this strong positive effect of La on RSV replication occurs independently of IFN, which is consistent with the fact that RSV is resistant to type I IFN (45, 57, 62). We show below (see Fig. 8) that this is also true for an IFN-sensitive virus, namely, SeV. Control siRNA or siRNA that did not silence La had no effect on virus growth (Fig. 4A and data not shown). The knockdown of RIG-I, for example, did not significantly affect RSV growth relative to that with no siRNA in A549 cells. In line with this finding, the double knockdown of La and RIG-I was no more inhibitory than the knockdown of La alone. Finally, the knockdown of MDA5 had no effect, in agreement with a lack of its role in leRNA binding, as described above. As shown in Fig. 7, RSV titers can be fully rescued by an La expression construct engineered to be resistant to this La siRNA, excluding any nonspecific effects of the La siRNA on RSV growth.
FIG. 8.
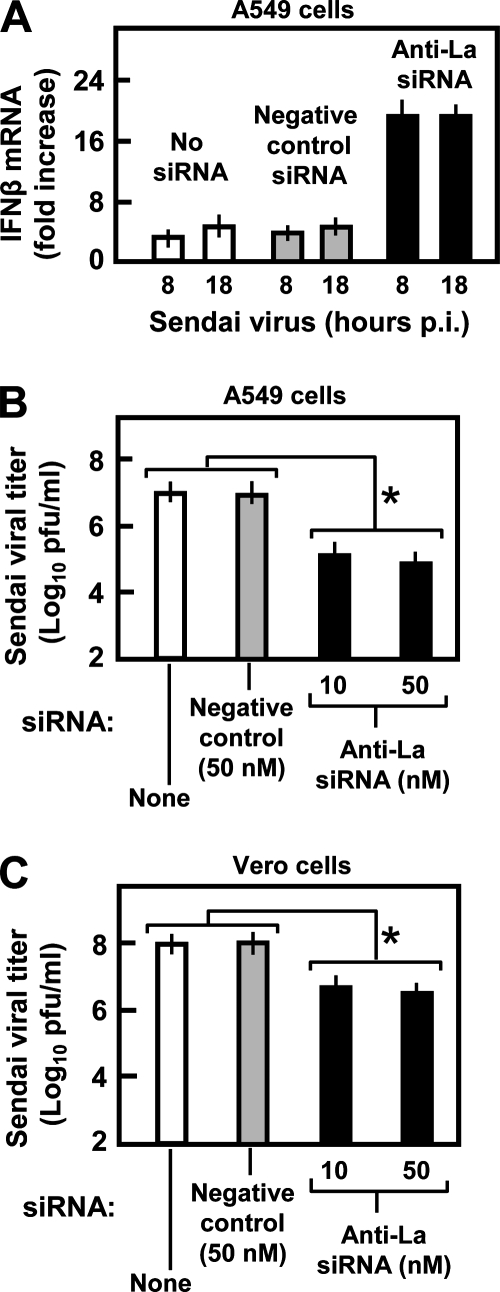
Inhibition of SeV replication in La-depleted cells. (A) A549 cells were transfected with no siRNA, negative control luciferase siRNA, or La siRNA (all 50 nM). Cells were infected with SeV at 3 multiplicities of infection. Thirty-six hours later, and after another 8 h and 18 h, RNA was harvested, and IFN-β mRNA levels were determined by qRT-PCR. Amounts were expressed as the change over uninfected A549 cells. (B and C) A549 and Vero cells, respectively, were transfected with siRNA and infected with SeV as described above (A), and at 72 h p.i., the viral titer in the extracellular medium was measured by standard plaque assay on Vero cells. Asterisks indicate highly significant changes (P < 0.05).
FIG. 4.
La is needed for optimal RSV growth. (A) Progeny viral titer (PFU/ml) in medium collected at 48 h p.i. from RSV-infected A549 and Vero cells in which the indicated genes were knocked down by siRNA (same as in Fig. 3C). An asterisk indicates significant reduction (P < 0.05). (B) Inhibition of RSV transcription in La-silenced cells. A549 cells were transfected with luciferase siRNA (lane 1), La siRNA (lane 2), RIG-I siRNA (lane 3), and plasmid pCAGGS-His-La (lane 4) and infected with RSV 36 h later. Total RNA was isolated after another 12 h and subjected to qRT-PCR as described in Materials and Methods. Transcript levels of RSV genes (NS1, N, P, M, SH, G, F, M2, and L) and control cellular GAPDH were determined by qRT-PCR as described in Materials and Methods. For visual presentation, 10 μl of the PCR was analyzed in a 2% agarose gel and stained with ethidium bromide. For a given gene, samples were taken after the same number of cycles from lanes 1 through 5. However, because of the polarity of viral gene transcription, samples from progressively greater numbers of cycles were taken for more promoter-distal genes so that relatively similar band intensities could be presented for photographic equality. For example, all NS1 samples are shown after 24 cycles, all N samples are shown after 25 cycles, and all P samples are shown after 26 cycles, etc. (C) Western blot of the cell lysates corresponding to the same treatments as above (B) (with the same lane numbers) using the indicated antibodies (left). The detected bands are marked (right). Profilin is an unaffected control.
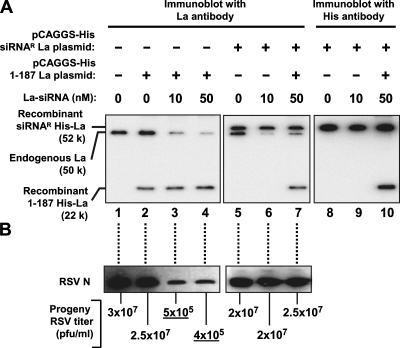
FIG. 7.
An La deletion mutant defective in leRNA binding does not support RSV growth. A549 cells were transfected with anti-La siRNA (10 nM or 50 nM) or no siRNA (0) and, where indicated (+), also with pCAGGS plasmids expressing full-length La encoded by an siRNA-resistant (siRNAR) gene (lanes 5 to 10) or the deletion mutant La1-187 (lanes 2 to 4 and 7), each containing a His tag. (A) The proteins were detected by Western analysis with anti-La or anti-His antibody. (B) RSV growth was monitored by immunoblotting detecting viral N protein and by extracellular RSV titers at 40 h p.i., as indicated below the lanes. Underlined titers were substantially lower than those of the no-siRNA control. Note that the siRNA targets La mRNA sequence encoding amino acids 243 to 250 and hence does not silence the recombinant at residues 1 to 187. By design, the siRNA-resistant La was also unaffected by the siRNA (see Materials and Methods).
To more directly rule out the involvement of IFN, we repeated the knockdown experiments in Vero cells that are deleted of type I IFN genes (27). RSV production was greatly reduced when La was silenced in Vero cells, and again, the loss of RIG-I had no effect (Fig. 4A). In fact, note that the gray (Vero) and white (A549) bars in Fig. 4A have essentially equal heights, showing an IFN-independent role of La. The data indicate that the ∼100-fold positive effect of the La protein on RSV growth occurs by an IFN-independent mechanism.
To initiate studies on this mechanism, we tested if the effect is on viral RNA synthesis and, if so, whether specific viral genes are affected. We determined the levels of individual RSV gene mRNAs by qRT-PCR using primers described in Table 1. These results are presented in Fig. 4B. Clearly, a global reduction of all RSV transcription was observed in La-silenced A549 cells (Fig. 4B, lane 2). To test whether La levels in the cell are rate limiting for RSV growth, we determined the RSV mRNA levels following the recombinant expression of La by transient transfection with a pCAGGS-His-La plasmid. This had no effect on RSV transcription (Fig. 4B, lane 4). The silencing of RIG-I by siRNA (Fig. 4B, lane 3) also had no discernible effect. The control cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was not affected by any treatment, and the irrelevant luciferase siRNA (Fig. 4B, lane 1) also had no effect on any gene. We confirmed the expression of His-La and the specificity and efficacy of the siRNAs by Western blotting using appropriate antibodies (Fig. 4C). Finally, RSV protein expression, judged by viral N protein levels, correlated with the mRNA levels (Fig. 4C, bottom).
Together, these results show an essential IFN-independent role of La in RSV gene expression and progeny RSV production.
RSV infection leads to the redistribution of La protein from the nucleus to the cytoplasm.
Although La is mostly nuclear, it shuttles between the nucleus and the cytoplasm, and redistribution has been observed after infection by some viruses, during apoptosis, and after granzyme B cleavage (8, 47). We examined the subcellular localization of La by immunostaining at various times after infection by RSV (Fig. 5, red) and counterstained with DAPI to visualize nuclei (blue) and with anti-RSV (green). Prior to infection, La was most concentrated in the nucleus, with little La detected in the cytoplasm (0 h) (Fig. 5, top). However, La began to appear in the cytoplasm as early as 8 h p.i., when RSV proteins were first detectable. Although the response was not synchronized in all cells, there was general correlation in the cytoplasmic RSV stain and a paucity of nuclear La as time progressed. For example, by 24 and 36 h, individual cells with the lowest RSV staining (arrows) had the greatest amount of nuclear La. Conversely, individual cells with the highest level of RSV staining showed the lowest concentration of La remaining in the nucleus, and in these cells, the La protein appeared distributed throughout the cell.
RRM1 and surrounding regions of the La protein are important for leRNA binding.
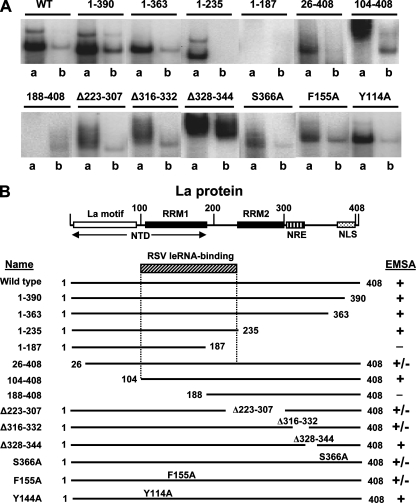
We next sought to map the determinants of the La protein that are important for leRNA interactions. Various nested deletions of the La protein, each with a C-terminal His tag, were purified and used in the EMSA as described previously (8, 29). The EMSA employed 5′-32P, 3′-OH RSV leRNA synthesized in vitro. Competitions were done with nonradioactive 5′-P-leRNA. Since our goal was to identify mutations that decreased binding relative to that of wild-type La, binding constants were not determined. The representative autoradiograms in Fig. 6A and summary in Fig. 6B reveal that removing some or all of the N-terminal domain, as in hLa26-408 and hLa104-408, or parts of the C terminus, as in hLa1-390, hLa1-363, and hLaΔ316-332, had little or no effect on leRNA binding. Mutants with the greatest loss of leRNA binding were hLa188-408 and hLa1-187. While these results indicate that multiple regions of La may contribute to optimal leRNA binding, they point to RRM1 and the linker region between RRM1 and RRM2 (compare regions at amino acids 1 to 235 and 1 to 187) as being the most important.
FIG. 6.
Mapping of RSV leRNA-binding regions of the La antigen. (A) EMSA was performed using 5′-32P-labeled synthetic RSV leRNA. Lanes from a representative autoradiogram are shown, and only the shifted band(s) is presented to conserve space. Each protein (indicated above the lanes) was used at 6 nM, and concentration was confirmed by SDS-polyacrylamide gel electrophoresis (29). Lanes “a” and “b,” indicated below the lanes, represent EMSA with no competitor and a 20-fold excess of unlabeled leRNA, respectively. WT, wild type. (B) The La protein is depicted in schematic form, and binding activity was graded as described previously (29): +, strong binding; +/−, weak binding; −, little or no binding. NTD, N-terminal domain; NRE, nuclear retention element; NLS, nuclear localization signal. Based on the EMSA data, the approximate region (residues 104 to 235) most responsible for RSV leRNA binding is indicated (angularly striped box).
An leRNA binding-defective La mutant does not support RSV growth.
We next expressed mutant hLa1-187 in A549 cells in which endogenous wild-type La was knocked down by the siRNA used previously (in Fig. 3 and 4), which targeted a sequence downstream of the region at positions 1 to 187. In control cells that were mock treated with 0 nM siRNA, the expression of hLa1-187 had little if any negative effect on the replication of RSV, measured by the RSV N protein level and viral titer (Fig. 7, compare lanes 1 and 2). However, as levels of endogenous La decreased with siRNA, levels of RSV protein and titer decreased despite the expression of hLa1-187 (Fig. 7, lanes 3 and 4).
To examine if the siRNA effect was due to the loss of the La protein rather than a nonspecific effect of siRNA, we engineered an La mutant in which the siRNA target sequence was mutated to decrease complementarity to the siRNA while maintaining the same amino acid coding specificity. The resulting clone should be resistant to siRNA directed to endogenous La and yet produce a His-tagged La protein with wild-type amino acid sequence (siRNAR-His-La) (Fig. 7). This clone produced the His-La protein (Mr, ∼52,000) and supported viral growth even in the presence of siRNA that decreased levels of endogenous La protein (Fig. 7, lanes 5 to 7). The recombinant La proteins were further distinguished by their reaction with anti-His antibody (Fig. 7, lanes 8 to 10). A 100-fold decrease in virus production by the La-knocked-down cells and rescue with siRNAR-His-La allows the conclusion that La specifically and greatly enhances RSV growth. The finding that the leRNA binding-defective protein hLa1-187 failed to rescue viral growth (Fig. 7, lanes 3 and 4) is consistent with the idea that the ability of La to bind leRNA is important for virus growth.
Loss of La inhibits an IFN-sensitive virus by both IFN-dependent and -independent mechanisms.
As RSV growth is IFN resistant, one can argue that La's ability to suppress IFN activation may not be particularly relevant for RSV infection. To extend these finding to a relevant virus, we initiated studies in an SeV strain that is known to be IFN sensitive (17, 63). Indeed, as shown in Fig. 8, the basic observations were all validated. First, in IFN-positive A549 cells, the knockdown of La by siRNA resulted in the activation of the IFN-β gene in the SeV-infected cell (Fig. 8A). Second, this resulted in a nearly 100-fold reduction in progeny SeV measured in the extracellular medium at 72 h p.i. (Fig. 8B). Third, although less severe, the inhibition (10-fold) of SeV yield was also observed in IFN-negative Vero cells upon the silencing of La (Fig. 8C). Note that the IFN-sensitive nature of SeV is also reflected in the 10-fold-lower titer in A549 cells than in Vero cells, confirming previously reported findings (17, 37). Thus, these SeV results reveal that when an IFN-sensitive virus is tested in an IFN-positive cell, two roles of the La protein can be appreciated, both of which are needed to support optimal viral growth: the suppression of IFN and a potentially novel IFN-independent stimulatory function.
DISCUSSION
The major finding of this work is that La binds RSV leRNA, which likely prevents RIG-I binding early in infection, thereby preventing the activation of IFN. For IFN-sensitive viruses such as SeV, this should constitute an important mechanism to promote optimal virus growth. In addition, La enhances viral growth by a second mechanism that appears to be independent of its ability to suppress IFN and hence is important for both RSV and SeV. As an RNA-binding protein, La possibly regulates an “RNA regulon” (36) consisting of a set of cellular and/or viral mRNAs that are important for virus growth.
In suppressing IFN, La is critically needed early in infection, when the de novo N protein is in short supply. According to the polarity of nonsegmented negative-strand RNA viral gene expression, viral leRNA should be transcribed at high levels before enough N protein is translated to encapsidate it. However, essentially all leRNA, even at early time points, was found in a protein-bound state (Fig. 1A). We propose a kinetic model in which La is the major protein to sequester leRNA shortly after infection, and as the viral protein levels build up with time, the N protein assumes an increasingly greater role. In support of this model, more RSV leRNA was found to be associated with La than with N at 8 h p.i., and much less, if any, leRNA was ever associated with RIG-I (Fig. 2). Moreover, siRNA knockdown of La led to an association of leRNA with RIG-I (Fig. 3B) with concomitant premature RIG-I-mediated induction of IFN mRNA (Fig. 3C), providing evidence that La functionally sequesters leRNA from RIG-I. The subcellular redistribution of La presumably facilitates its interaction with leRNA, since naked leRNA transfected into uninfected cells in which La is nuclear could activate IFN (Fig. 1B) (55). Clearly, the recruitment of La would afford the virus the ability to accumulate leRNA with a reduced risk of RIG-I-mediated IFN induction.
A substantial portion of La appeared in the cytoplasm after infection with the RNA viruses VSV, parainfluenza virus, poliovirus, and Rinderpest virus (24, 39, 48, 55), all of which, like RSV, replicate in the cytoplasm. Our time course (Fig. 5) shows that this translocation begins very early in RSV infection and is highly efficient. La is a nucleocytoplasmic shuttling protein with a nuclear localization signal, a nuclear retention element, and a nuclear export sequence (8, 28). The early effects of RSV suggest that it activates a signaling pathway that regulates one or more of these domains of La.
The 44-nt RSV leRNA used here for RNA-binding experiments ends with AAU and is not expected to be a high-affinity substrate for the La motif-mediated binding observed in the crystal structure, since it lacks U in the penultimate position, the most critical of the three U residues (71). This is consistent with the disparity in binding to La26-408 and hLa1-187 compared to UUU-containing RNAs (29) and with the possibility that leRNA uses an RNA-binding mode different from UUU-containing RNAs, as is suspected for mRNAs with which La associates (46). Our data are consistent with the possibility that La makes multiple contacts to leRNA, as expected of La as a modular protein that uses multiple surfaces to bind and protect RNA (34). Studies of binding of La with the leRNA of other nonsegmented negative-strand RNA viruses including SeV should illuminate the mechanism of the La-leRNA interaction. This is in progress.
Lastly, the exact viral trigger(s) of RIG-I needs to be identified, but we offer the following possible candidates: (i) small amounts of leftover naked leRNA that escape La and N may suffice to activate enough RIG-I; (ii) the 5′ capping of viral mRNAs may be an inefficient process, and a small fraction of mRNAs that remains uncapped binds and activates RIG-I; and (iii) the 5′ untranslated region or other internal sequences of viral mRNAs may bind and activate RIG-I through novel mechanisms.
Acknowledgments
This research was supported in part by an NIH grant to S.B. (AI059267). R.J.M. and M.A.B. were supported by the Intramural Research Program of the NICHD, NIH. R.J.M. is a Commissioned Officer in the U.S. Public Health Service.
S.B. is also a member of Lions-USA Eye Research group and is indebted to the Lions Club International Fund and the local Lions clubs for the gift of the Nikon TE2000E2 imaging station.
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313360-363. [DOI] [PubMed] [Google Scholar]
- 2.Ali, N., G. J. Pruijn, D. J. Kenan, J. D. Keene, and A. Siddiqui. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 27527531-27540. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, A. K., and S. Barik. 1992. Gene expression of vesicular stomatitis virus genome RNA. Virology 18817-428. [DOI] [PubMed] [Google Scholar]
- 5.Barik, S. 1992. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s). J. Virol. 666813-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barik, S. 1993. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 74485-490. [DOI] [PubMed] [Google Scholar]
- 7.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Mühlberger, M. Bray, H. D. Klenk, P. Palese, and A. García-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 777945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayfield, M. A., T. E. Kaiser, R. V. Intine, and R. J. Maraia. 2007. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol. Cell. Biol. 273303-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsham, G. J., N. Sonenberg, and Y. V. Svitkin. 1995. The role of the La autoantigen in internal initiation. Curr. Top. Microbiol. Immunol. 20385-98. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya, R., K. Perumal, K. Sinha, R. Maraia, and R. Reddy. 2002. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 10243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 1150-55. [DOI] [PubMed] [Google Scholar]
- 13.Bitko, V., N. E. Garmon, T. Cao, B. Estrada, J. E. Oakes, R. N. Lausch, and S. Barik. 2004. Activation of cytokines and NF-kappa B in corneal epithelial cells infected by respiratory syncytial virus: potential relevance in ocular inflammation and respiratory infection. BMC Microbiol. 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumberg, B. M., and D. Kolakofsky. 1981. Intracellular vesicular stomatitis virus leader RNAs are found in nucleocapsid structures. J. Virol. 40568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blumberg, B. M., C. Giorgi, and D. Kolakofsky. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32559-567. [DOI] [PubMed] [Google Scholar]
- 16.Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23837-845. [DOI] [PubMed] [Google Scholar]
- 17.Bousse, T., R. L. Chambers, R. A. Scroggs, A. Portner, and T. Takimoto. 2006. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 12123-32. [DOI] [PubMed] [Google Scholar]
- 18.Chambers, J. C., and J. D. Keene. 1985. Isolation and analysis of cDNA clones expressing human lupus La antigen. Proc. Natl. Acad. Sci. USA 822115-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang, Y. N., D. J. Kenan, J. D. Keene, A. Gatignol, and K. T. Jeang. 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 687008-7020. (Erratum, 69:618-619, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359190-200. [DOI] [PubMed] [Google Scholar]
- 21.Colonno, R. J., and A. K. Banerjee. 1978. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell 1593-101. [DOI] [PubMed] [Google Scholar]
- 22.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 246861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curry, S., and M. R. Conte. 2006. A terminal affair: 3′-end recognition by the human La protein. Trends Biochem. Sci. 31303-305. [DOI] [PubMed] [Google Scholar]
- 24.De, B. P., S. Gupta, H. Zhao, J. A. Drazba, and A. K. Banerjee. 1996. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and La protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 27124728-24735. [DOI] [PubMed] [Google Scholar]
- 25.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase; phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 738384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411494-498. [DOI] [PubMed] [Google Scholar]
- 27.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 28.Fok, V., K. Friend, and J. A. Steitz. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 173319-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier, J. L., H. Fan, and R. J. Maraia. 1997. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol. Cell. Biol. 175823-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313357-360. [DOI] [PubMed] [Google Scholar]
- 31.Heise, T., L. G. Guidotti, and F. V. Chisari. 2001. Characterization of nuclear RNases that cleave hepatitis B virus RNA near the La protein binding site. J. Virol. 756874-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiscott, J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 28215325-15329. [DOI] [PubMed] [Google Scholar]
- 33.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 34.Huang, Y., M. A. Bayfield, R. V. Intine, and R. J. Maraia. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 13611-618. [DOI] [PubMed] [Google Scholar]
- 35.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 36.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8533-543. [DOI] [PubMed] [Google Scholar]
- 37.Koyama, A. H., H. Irie, A. Kato, Y. Nagai, and A. Adachi. 2003. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5373-378. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, R., A. Musiyenko, and S. Barik. 2003. The heat shock protein 90 of Plasmodium falciparum and antimalarial activity of its inhibitor, geldanamycin. Malar. J. 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurilla, M. G., and J. D. Keene. 1983. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell 34837-845. [DOI] [PubMed] [Google Scholar]
- 40.Kurilla, M. G., C. D. Cabradilla, B. P. Holloway, and J. D. Keene. 1984. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J. Virol. 50773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leppert, M., L. Rittenhouse, J. Perrault, D. F. Summers, and D. Kolakofsky. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18735-747. [DOI] [PubMed] [Google Scholar]
- 42.Lerner, M. R., J. A. Boyle, J. A. Hardin, and J. A. Steitz. 1981. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211400-402. [DOI] [PubMed] [Google Scholar]
- 43.Li, S., M. Xu, and K. Coelingh. 1995. Electroporation of influenza virus ribonucleoprotein complexes for rescue of the nucleoprotein and matrix genes. Virus Res. 37153-161. [DOI] [PubMed] [Google Scholar]
- 44.Liu, P., M. Jamaluddin, K. Li, R. P. Garofalo, A. Casola, and A. R. Brasier. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 811401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo, M. S., R. M. Brazas, and M. J. Holtzman. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 799315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maraia, R. J., and M. A. Bayfield. 2006. The La protein-RNA complex surfaces. Mol. Cell 21149-152. [DOI] [PubMed] [Google Scholar]
- 47.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 21367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 673798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mibayashi, M., L. Martínez-Sobrido, Y. M. Loo, W. B. Cárdenas, M. Gale, Jr., and A. García-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mink, M. A., D. S. Stec, and P. L. Collins. 1991. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology 185615-624. [DOI] [PubMed] [Google Scholar]
- 51.Park, Y. W., and M. G. Katze. 1995. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 27028433-28439. [DOI] [PubMed] [Google Scholar]
- 52.Patton, J. T., N. L. Davis, and G. W. Wertz. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J. Virol. 49303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pestka, S. 2007. The interferons: 50 years after their discovery, there is much more to learn. J. Biol. Chem. 28220047-20051. [DOI] [PubMed] [Google Scholar]
- 54.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 55.Plumet, S., F. Herschke, J. M. Bourhis, H. Valentin, S. Longhi, and D. Gerlier. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE 2e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raha, T., R. Pudi, S. Das, and M. S. Shaila. 2004. Leader RNA of Rinderpest virus binds specifically with cellular La protein: a possible role in virus replication. Virus Res. 104101-109. [DOI] [PubMed] [Google Scholar]
- 57.Ramaswamy, M., L. Shi, S. M. Varga, S. Barik, M. A. Behlke, and D. C. Look. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344328-339. [DOI] [PubMed] [Google Scholar]
- 58.Ray, P. S., and S. Das. 2002. La autoantigen is required for the internal ribosome entry site-mediated translation of coxsackievirus B3 RNA. Nucleic Acids Res. 304500-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 1755260-5268. [DOI] [PubMed] [Google Scholar]
- 60.Sen, G. C., and S. N. Sarkar. 2005. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytok. Growth Factor Rev. 161-14. [DOI] [PubMed] [Google Scholar]
- 61.Spangberg, K., L. Wiklund, and S. Schwartz. 2001. Binding of the La autoantigen to the hepatitis C virus 3′ untranslated region protects the RNA from rapid degradation in vitro. J. Gen. Virol. 82113-120. [DOI] [PubMed] [Google Scholar]
- 62.Spann, K. M., K.-C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 784363-4369. (Erratum, 78:6705.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strähle, L., J. B. Marq, A. Brini, S. Hausmann, D. Kolakofsky, and D. Garcin. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 8112227-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svitkin, Y. V., A. Pause, and N. Sonenberg. 1994. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 687001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tasaka, M., N. Sakamoto, Y. Itakura, M. Nakagawa, Y. Itsui, Y. Sekine-Osajima, Y. Nishimura-Sakurai, C. H. Chen, M. Yoneyama, T. Fujita, T. Wakita, S. Maekawa, N. Enomoto, and M. Watanabe. 2007. Hepatitis C virus non-structural proteins responsible for suppression of the RIG-I/Cardif-induced interferon response. J. Gen. Virol. 883323-3333. [DOI] [PubMed] [Google Scholar]
- 66.Teplova, M., Y. R. Yuan, A. T. Phan, L. Malinina, S. Ilin, A. Teplov, and D. J. Patel. 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 2175-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 202902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber, F., and O. Haller. 2007. Viral suppression of the interferon system. Biochimie 89836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilusz, J., and J. D. Keene. 1984. Interactions of plus and minus strand leader RNAs of the New Jersey serotype of vesicular stomatitis virus with the cellular La protein. Virology 13565-73. [DOI] [PubMed] [Google Scholar]
- 70.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 28215315-15318. [DOI] [PubMed] [Google Scholar]
- 71.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 72.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]