Abstract
Protective immunity after resolved hepatitis C virus (HCV) infection has been reported. However, the breadth of this immunity has remained controversial, and the role of neutralizing antibodies has not been well-defined. In the present study, two chimpanzees (CH96A008 and CH1494) with resolved monoclonal H77C (genotype 1a) infection were rechallenged with low-dose homologous H77C virus about 12 months after viral clearance; CH96A008 became persistently infected, and CH1494 had transient viremia lasting 2 weeks. CH1494 was subsequently either partially or completely protected following five homologous rechallenges with monoclonal H77C or polyclonal H77 and after six heterologous rechallenges with HC-J4 (genotype 1b) or HC-J6 (genotype 2a) viruses. Subsequently, a final challenge with H77C resulted in persistent HCV infection. In both chimpanzees, serum neutralizing antibodies against retroviral pseudoparticles bearing the H77C envelope proteins were not detected during the initial infection or during rechallenge. However, anamnestic cellular immune responses developed during the initial homologous rechallenge, in particular in CH96A008, which developed a persistent infection. Polyprotein sequences of viruses recovered from CH1494 after the two homologous rechallenges that resulted in transient viremia were identical with the H77C virus. In contrast, the polyprotein sequences of viruses recovered from both chimpanzees after homologous rechallenge resulting in persistent infection had numerous changes. These findings have important implications for our understanding of immunity against HCV; even in the best-case scenario with autologous rechallenge, low-level viral persistence was seen in the presence of primed T-cell responses.
Worldwide, 3 to 4 million people are infected with hepatitis C virus (HCV) each year, and persistent infection develops in 70 to 80% of these individuals. Thus, about 180 million people are persistently infected with HCV, and this virus is an important cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Although the virus was discovered in 1989, there is still no commercial HCV vaccine, and to further the development of such a vaccine it is important to fully understand the immunological correlates of protective immunity (reviewed in references 3 and 24). HCV has been classified as a member of the Flaviviridae virus family. Its positive-strand RNA genome exhibits significant genetic heterogeneity, with multiple serotypes, which represents a great challenge for developing broadly reactive vaccines. Viral isolates from around the world have been classified into six major genotypes and multiple subtypes (genotypes 1a, 1b, 2a, etc.). Furthermore, HCV circulates as a quasispecies of closely related genomes in an infected individual. Finally, HCV evolves rapidly and has a great potential to escape host immune responses, which represents another challenge for vaccine development.
For the future perspective of developing an effective vaccine against HCV, it is encouraging that 20 to 30% of infected individuals are able to mount an immune response that can control this infection. Therefore, understanding the components of this response might yield important clues to what is needed for effective control (reviewed in reference 3). On the other hand, it appears that the immune response detected in resolving infections does not prevent reinfection and also might not even prevent viral persistence following such reexposure (22). This scenario might be explained by strain- or genotype-specific protective immunity.
The chimpanzee is the only recognized animal model for the study of HCV (5, 7), and this model has been used in previous studies to address the issue of protective immunity. It was originally demonstrated that chimpanzees that resolved their initial experimental HCV infection were not protected against challenge with a quasispecies even of the same strain (9, 26). Also, by repeated heterologous rechallenge some animals became persistently infected. However, in a more recent study, rapid viral clearance was observed following homologous and heterologous genotype 1 rechallenge in chimpanzees that had recovered from their initial infection 1.5 to 16 years earlier (2). The same group subsequently reported that this protective immunity extended to challenge with viruses of other major genotypes (20). Other investigators reported rapid control of HCV in chimpanzees that were rechallenged with homologous monoclonal or polyclonal virus (17, 21, 25, 29, 33). Taken together, these studies found that viral clearance was associated with early anamnestic HCV-specific CD4+ and CD8+ T-cell responses, including memory CD4+ responses, and with intrahepatic induction of gamma interferon. Except for the first study these studies suggested that immunity following a resolved infection in chimpanzees would prevent persistence following challenge with viruses of the same genotype and even with viruses of other genotypes. However, a more recent study suggested that challenge with virus belonging to a different genotype than the virus originally infecting an animal would frequently lead to viral persistence (27). Of note, the role of neutralizing antibodies was not addressed in any of these prior studies on protective immunity.
The role of cellular immunity in protective immunity was further examined in studies in which chimpanzees with resolved genotype 1a infection were depleted of either CD4+ or CD8+ cells before homologous rechallenge (17, 25, 29). In all cases, depletion resulted in prolonged viremia compared with a previous rechallenge in the same animal, and the CD4+ depletion appeared to promote the development of viral persistence through a CD8+ T-cell immune escape mechanism. Thus, these studies shed new light on the role of CD4+ and CD8+ cells in protective immunity.
In the present study, we examined protective immunity against HCV by performing true homologous rechallenge of two chimpanzees with resolved monoclonal genotype 1a infection. We examined the role of neutralizing antibodies and peripheral and intrahepatic T-cell responses. Furthermore, we performed a comprehensive genome analysis of infecting virus in order to determine whether immune escape mutants could potentially have caused reinfection or viral persistence.
MATERIALS AND METHODS
Experimental infections of chimpanzees.
The housing, maintenance, and care of the chimpanzees were in compliance with all relevant guidelines and requirements. The animals were housed in facilities that are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. CH1494 was initially inoculated intrahepatically, in a percutaneous procedure, with RNA transcripts from 20 μg of XbaI-digested pCV-H77C (8). Subsequently, the animal was inoculated intravenously with known infectious virus doses of H77C (15), H77 (12), HC-J4 (36), and HC-J6 (35), respectively. CH96A008 was inoculated intravenously with plasma collected at week 4 from a chimpanzee (CH1590) inoculated intrahepatically with RNA transcripts from a pCV-H77C mutant lacking E2 HVR1 (16). Subsequently, the animal was inoculated intravenously with acute-phase plasma collected at week 7 from CH96A008. Serum samples, plasmapheresis units, peripheral blood mononuclear cells (PBMC), and liver biopsy samples were collected weekly from both chimpanzees. Serum samples were tested for HCV RNA (in-house reverse transcription [RT] nested PCR [6]; Monitor 2.0, Roche Diagnostics, Indianapolis, IN), HCV antibodies (enzyme-linked immunosorbent assay [ELISA] 2.0; Abbott, Chicago, IL), and alanine aminotransferase (ALT; Anilytics, Gaithersburg, MD). Selected biopsy specimens were tested for HCV RNA in the RT-nested PCR assay (16).
In vitro neutralization test.
The neutralization test was performed as described previously for similar HCV neutralization experiments performed in our laboratory (10, 11). A “hyperimmune” pool of serum collected at weeks 1, 2, and 3 following the third rechallenge with polyclonal H77 was heat inactivated at 56°C for 30 min and diluted 1:5 in phosphate-buffered saline (PBS), pH 7.4. The virus pool H77C was diluted in ice-cold PBS immediately before the neutralization experiment. For the in vitro neutralization experiment 1 ml of CH1494 hyperimmune serum (1:5) was mixed with 1 ml of a dilution of the H77C challenge pool containing approximately 64 50% chimpanzee infectious doses (CID50) of H77C. The neutralization mixtures were incubated overnight at 4°C. Thereafter, the entire content was inoculated intravenously into CH1573. Serum samples were collected weekly and tested as described above.
H77C pseudoparticle assay to detect neutralizing antibodies.
The percent neutralization in postchallenge serum, compared to a preinfection sample, was determined in a retroviral HCV pseudoparticle assay using ppH77C, as described in detail previously (23). To generate pseudotyped particles, HEK-293T cells were transfected with a total of 4 mg of plasmid DNA, including 1.5 mg of CMV-Gag-Pol packaging construct, 1.5 mg of MLV-GFP plasmid, and 1.0 mg of HCV E1E2 vector expressing the H77C envelope proteins, using Lipofectamine Plus reagents (Invitrogen, Carlsbad, CA) as per the supplied protocol. The DNA-Lipofectamine mixture was added to the cells and incubated at 37°C. After 3 hours cells were washed once and cultured in 7.5 ml of complete medium for 2 days. Transfection efficiency was consistently over 80%. Culture supernatants were harvested and passed through a 0.45-μm filter to remove cells and debris.
HCVpp infections of Huh7 cells were performed on the same day. Approximately 4 × 104 cells/well were added to 24-well plates and cultured overnight. Neutralization of ppH77C was performed in the presence of 5 to 10% fetal bovine serum. A 1:25 dilution of test antibody samples was incubated for 1 h at room temperature with ppH77C, added to Huh-7 cells, and incubated at 37°C. Supernatants were removed after 8 h, and the cells were incubated in Dulbecco's modified Eagle's medium-10% fetal calf serum for 72 h at 37°C. For green fluorescent protein (GFP) expression analysis, detached cells were washed with 1 ml of PBS and resuspended in 300 ml of PBS. Twenty thousand cells were analyzed for GFP expression using a FACScan (BD Biosciences, Rockville, MD). Percentage neutralization for each sample was calculated by comparison with the presample. Significant neutralization was defined as a 50% reduction of the number of GFP-positive cells.
Assays for peripheral and intrahepatic T-cell responses.
The details of protocols and HCV antigens used to detect cellular immune responses have been published previously (31). PBMC were isolated from 40 ml of blood. Liver-infiltrating lymphocytes were isolated from liver tissue obtained by needle biopsy. Cell suspensions were incubated with magnetic beads coupled to anti-CD4 or anti-CD8, and bound cells were isolated using a magnetic particle concentrator, and cells from liver tissue were next expanded for 2 weeks. All subsequent analyses were performed on freshly isolated cells without prior cryopreservation. In CH1494 and CH96A008, PBMC CD4+ T cells or polyclonally expanded intrahepatic CD4+ T cells were tested for HCV-specific proliferative capacity after 6 days of culture with HCV-1 proteins (C22, C33-c, c100, and NS5; provided by Michael Houghton, Chiron, Emeryville, CA). [3H]thymidine was added for 16 h, and the mean level of thymidine incorporation in the HCV protein-stimulated and control cultures was used to calculate the stimulation index (SI); values of >2.0 were considered positive. In CH96A008, the PBMC CD8+ T-cell responses were tested against a panel of HCV peptides and expressed as the percentage of CD8+ T cells that produced gamma interferon. In CH96A008, polyclonally expanded intrahepatic CD8+ T cells were tested by intracellular gamma interferon staining after 5 h of stimulation with autologous Epstein-Barr virus-immortalized B-cell lines that were infected with recombinant HCV H77-encoding vaccinia virus vHCV 1-1488 or vHCV 827-3011 together with VTF7 provided by C. M. Rice (Rockefeller University, New York, NY) or with VTF7 alone. The frequency of HCV-specific CD8+ T cells was defined as the percentage of CD8+ T cells that produced gamma interferon in response to stimulation by B-cell lines coinfected by vHCV and VTF7 after subtraction of the gamma interferon-positive CD8+ T cells detected after stimulation in the absence of vHCV.
Sequence analysis of viruses recovered from infected chimpanzees.
To determine the consensus sequence of the entire open reading frame (ORF) of H77C virus recovered from CH1494 and CH1573, or of H77C virus lacking the E2 HVR1 recovered from CH96A008, we used two procedures. In serum samples with relatively high HCV RNA titers, we performed long RT-PCR followed by nested PCR with 1a-specific primers of 10 fragments (34) using the primers listed in Table S1 of the supplemental material. In samples with relatively low titers that did not permit amplification with this procedure, we performed RT-nested PCR of 19 fragments (28) using the 1a-specific primers listed in Table S2 of the supplemental material. PCR amplicons were sequenced directly to obtain the ORF consensus sequence. In addition, to analyze the envelope (E2) HVR1 sequences of viruses recovered in CH1494 following rechallenge with H77C, HC-J4, and HC-J6, respectively, E1-E2 fragments were amplified in RT-nested PCR using the primers listed in Table S3 of the supplemental material. Obtained amplicons were cloned and sequenced using standard procedures (28).
RESULTS
Protective immunity in CH1494 following monoclonal homologous genotype 1a (strain H77) rechallenge.
We previously constructed an infectious cDNA clone of strain H77 of HCV (pCV-H77C) (34). The infectivity of this genotype 1a clone was demonstrated by direct intrahepatic injection of RNA transcripts into two chimpanzees (8, 34). Even though the two chimpanzees were infected with the same monoclonal HCV, they had a different outcome. Viremia persisted in one chimpanzee (CH1530). However, in the other chimpanzee (CH1494) the infection resolved during week 24 (Fig. 1). During the primary acute infection, CH1494 had peak HCV RNA titers of 105 to 106 genome equivalents (GE)/ml. Anti-HCV (second-generation ELISA) was detected from week 15. The chimpanzee had evidence of acute viral hepatitis with elevated liver enzyme values. The ORF sequence of virus recovered at week 9 had a single nucleotide change (G3946A) (Table 1), which resulted in an amino acid change (G1202E) in NS3 (Table 2). This mutation also evolved in CH1530 (34) and was thus contained in the monoclonal H77C challenge virus used in this study (15). In the virus recovered from CH1494 at week 21, after the virus reemerged following a fourfold decrease in titers during weeks 12 to 15, we detected a number of additional mutations (Tables 1 and 2).
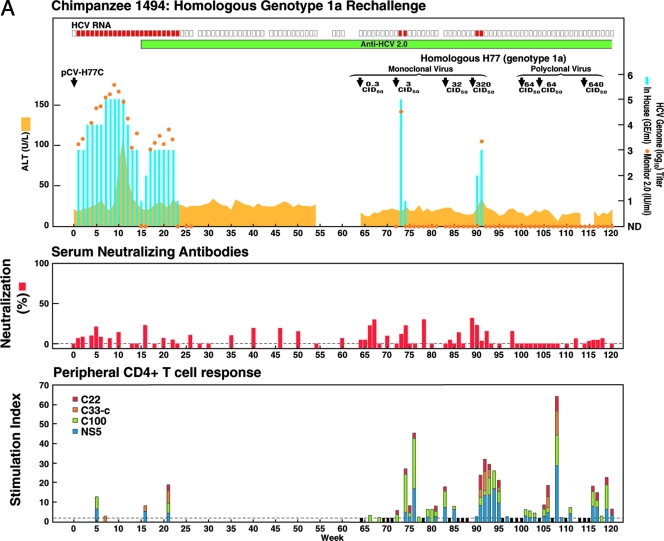
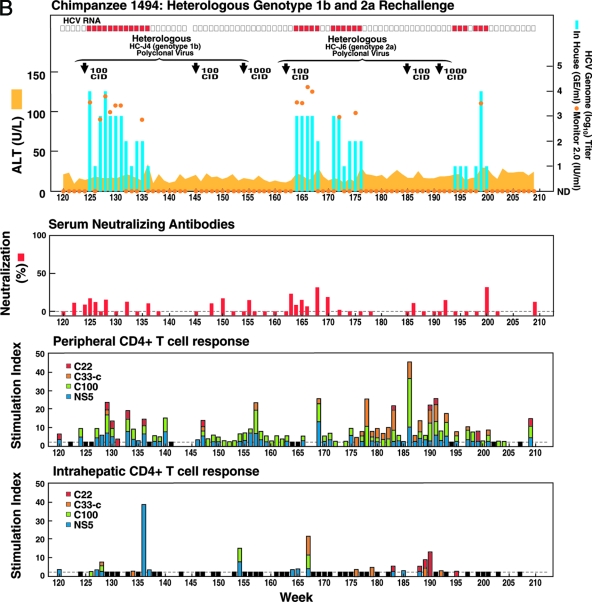
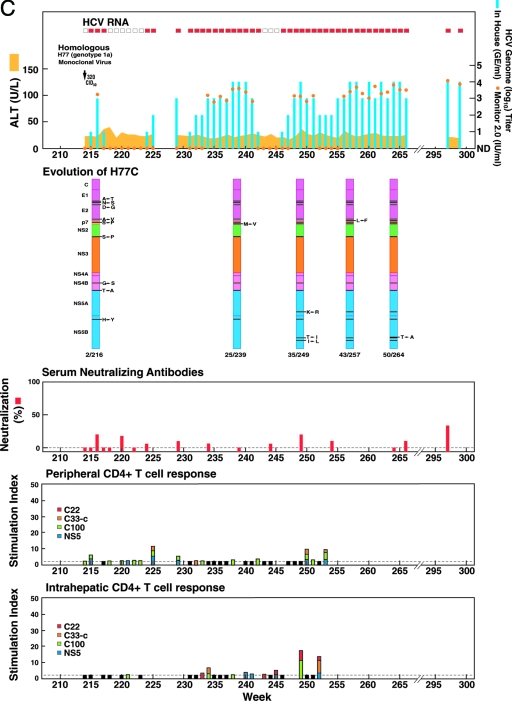
FIG. 1.
Homologous and heterologous rechallenge experiments in CH1494 with resolved monoclonal HCV infection following intrahepatic transfection with RNA transcripts of pCV-H77C (genotype 1a). The CID of viruses used for each rechallenge are indicated. (A) Weeks 0 to 120; about 12 months after viral clearance, CH1494 was rechallenged with homologous monoclonal H77C virus and polyclonal H77 virus. (B) Weeks 120 to 210; heterologous rechallenges with HC-J4 (genotype 1b) and HC-J6 (genotype 2a). (C) Weeks 210 to 300; final challenge with monoclonal H77C, which resulted in persistent infection. The course of infection was evaluated as follows: serum samples collected weekly were tested for HCV RNA by in-house RT-nested PCR with 5′-untranslated region primers and/or with the Roche Monitor Test 2.0. Red rectangle, positive by RT-nested PCR and/or by Monitor; empty rectangle, negative by RT-nested PCR; blue bars, HCV titers determined by performing the RT-nested PCR on 10-fold serial dilutions of extracted RNA; orange dots, HCV Monitor titers. Samples below the detection limit of 600 IU/ml are shown as not detected. Green horizontal bar, seroconversion in the second-generation ELISA; yellow-orange shaded area, serum ALT. For determination of neutralizing antibodies, the percent neutralization of retroviral pseudoparticles bearing the H77C envelope proteins was measured (>50% considered significant). For peripheral and intrahepatic CD4+ T-cell responses, the CD4+ T-cell responses to core (red), NS3 (orange), NS3-NS4 (green), and NS5 (blue) were detected against HCV-1 (genotype 1a) antigens, which are shown according to the specific SI. A specific SI of >2 was considered significant. At the weeks tested, in which the SI was ≤2 against all four antigens, the negative result is indicated by a black bar (with a value of 2). For the evolution of H77C, the H77C genome is shown as a vertical bar with core at the top and NS5B at the bottom. Black lines with capital letters indicate new amino acid changes that were identified when a sequence was compared with the sequence obtained at the previous time point. Solid lines without capital letters represent amino acid changes that persisted. The week analyzed is indicated at the bottom of each genome.
TABLE 1.
Nucleotide changes of consensus ORF sequence of HCV recovered from chimpanzee 1494 following transfection with pCV-H77Ca
| Sample | Nucleotide at indicated position of HCV genome region
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Core
|
E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
||||||
| 593 | 1729 | 2544 | 2628 | 2706 | 2916 | 3946 | 4403 | 5723 | 6231 | 7177 | 7434 | 7788 | |
| H77C | C | C | T | C | G | G | G | C | A | A | G | A | C |
| Wks | |||||||||||||
| 2 | • | ||||||||||||
| 4 | G/a | ||||||||||||
| 9 | • | • | • | • | • | • | A/g | • | • | • | • | • | • |
| 21 | T | T | T/G | T | A | A | A | T | G | T | A | G | T |
The sequence of H77C is shown on the top. Nucleotides identical with H77C are indicated with dots. Dominant changes are indicated by capital letters. Minor sequences are indicated with lowercase letters. Only a short sequence, including nucleotide 3946, was analyzed for weeks 2 and 4.
TABLE 2.
Amino acid changes of HCV variants recovered from chimpanzee 1494 after transfection with pCV-H77Ca
| Sample | Amino acid encoded by HCV genome region at indicated polyprotein position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
||||
| 463 | 735 | 763 | 789 | 859 | 1202 | 1964 | 2279 | 2365 | 2483 | |
| H77C | T | L | H | A | V | G | I | R | T | H |
| HCV recovered at wk: | ||||||||||
| 2 | • | |||||||||
| 4 | G/e | |||||||||
| 9 | • | • | • | • | • | E/g | • | • | • | • |
| 21 | I | L/V | Y | T | M | E | L | Q | A | Y |
The sequence of H77C is shown on the top. Amino acids identical with H77C are indicated with dots. Dominant changes are indicated by capital letters. Minor sequences are indicated with lowercase letters. Only a short sequence encoding amino acid 1202 was analyzed for weeks 2 and 4.
We experimentally rechallenged CH1494 after it had been negative for HCV RNA in the serum for about 12 months when tested with a highly sensitive RT-nested PCR assay (Fig. 1). As challenge virus we used homologous monoclonal H77C virus collected during the early acute phase (weeks 7 to 9) from chimpanzee 1530 (8, 15). The challenge with approximately 3 CID50 produced a viremia lasting only 2 weeks with the viral titer at week 1 as high as the peak titers seen during the primary infection. After a rechallenge with 32 CID50, infection was not detected. Subsequent rechallenge with 320 CID50 again resulted in a transient infection with viremia at weeks 1 and 2 after challenge. Analysis of the entire polyprotein sequence of viruses recovered from the chimpanzee during both episodes of transient viremia demonstrated that they did not represent immune escape variants, since their sequence was identical with that of viruses recovered at week 9 of the original acute infection.
Sterilizing immunity in CH1494 following subsequent challenges with polyclonal homologous genotype 1a virus (strain H77).
The chimpanzee CH1494 was next challenged with the homologous polyclonal virus pool (strain H77) collected from acute-phase infection in patient H (10, 12). This challenge pool is known to contain a quasispecies virus population with significant heterogeneity, in particular within the envelope proteins (11, 34). The chimpanzee was not infected following each of three consecutive challenges with 64 CID50, 64 CID50, and 640 CID50 of this virus pool (Fig. 1A). Apparently, sterilizing immunity against the homologous HCV strain was obtained in this chimpanzee.
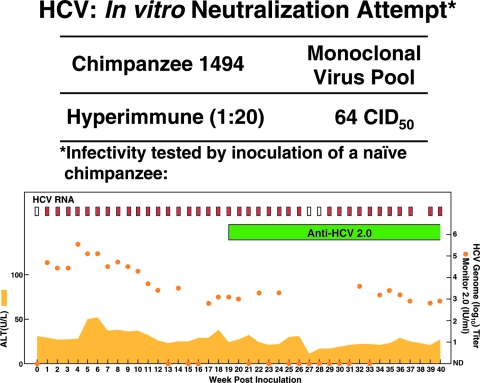
The sterilizing immunity to HCV observed in CH1494 did not appear to be associated with the development of neutralizing antibodies. We attempted to neutralize HCV H77C in vitro with “hyperimmune” serum obtained from the chimpanzee after the third challenge with the polyclonal H77 virus. If this immunity resulted from the development of neutralizing antibodies, these would be expected to increase to high titers following seven virus challenges. Neutralization was attempted with a 1:20 dilution of a serum pool collected from CH1494 at weeks 1 to 3 following the final rechallenge with polyclonal H77 virus (Fig. 1A). As challenge virus we used 64 CID50 of the monoclonal H77C virus pool, the same virus as was used for the initial homologous rechallenges. The mixtures of immune serum and virus were incubated overnight at 4°C, and residual infectivity was tested by inoculating the mixture intravenously into a naïve chimpanzee (CH1573). CH1573 became infected with a wild-type-like course of infection (Fig. 2). Furthermore, we found that HCV viruses recovered at week 4 from CH1573 had the polyprotein sequence of the monoclonal virus pool. Thus, the infection observed in CH1573 was not caused by a preexisting minor variant in the virus pool which could not be neutralized but probably represented true failure of neutralization. These data are in agreement with the lack of neutralizing antibodies previously reported for this hyperimmune sera in an in vitro test using H77 pseudoparticles (1).
FIG. 2.
Attempt to neutralize H77C virus in vitro with serum collected from CH1494 following multiple rechallenges with monoclonal H77C or polyclonal H77 virus. Residual infectivity was tested in CH1573. See also the legend to Fig. 1.
No detectable neutralizing antibody responses were found, but anamnestic cellular immune responses occurred after homologous rechallenges in CH1494.
The availability of an in vitro neutralization assay using pseudoparticles bearing E1 and E2 envelope glycoproteins of HCV enabled us to further evaluate the status of neutralizing antibodies (1). In the present study, we used ppH77C that had the same envelope glycoprotein sequence as the H77C challenge virus. Weekly samples were tested during the initial acute infection and following each rechallenge. However, at the 1:25 serum dilution tested we did not observe evidence of the development of significant autologous neutralizing antibodies, since all samples tested resulted in <50% neutralization of ppH77C infectivity in Huh7 cells (Fig. 1A).
To study the cellular immune responses in CH1494 CD4+ T cells isolated from PBMC or polyclonally expanded CD4+ T cells from liver biopsy specimens were tested for HCV-specific proliferative responses with a panel of recombinant HCV genotype 1a proteins (from strain HCV-1), including core, NS3, NS4, and NS5 (31). Unfortunately, with the exception of samples taken after the last polyclonal challenge, the intrahepatic CD4+ responses were studied too infrequently to adequately address their significance. Also, we were not able to study CD8+ T-cell responses, since we failed most times to expand these cells from the liver and we also did not identify CD8+ T-cell epitopes in this animal. A peripheral HCV-specific CD4+ T-cell response was detected during the initial acute infection, and an anamnestic response occurred immediately following each rechallenge (Fig. 1A). In addition, intrahepatic CD4+ T-cell responses were detected after the challenge with 640 CID50 of the polyclonal H77 virus (data not shown). Overall, our results indicated that effective immunity to HCV did not depend on the development of neutralizing antibodies. However, even in the absence of detectable viremia, rechallenge initiated robust anamnestic peripheral and intrahepatic T-cell responses that might be responsible for the observed homologous immunity.
Sterilizing immunity to HCV observed in CH1494 following repeated genotype 1a challenge did not prevent transient infection following subsequent challenge with genotype 1b or 2a viruses.
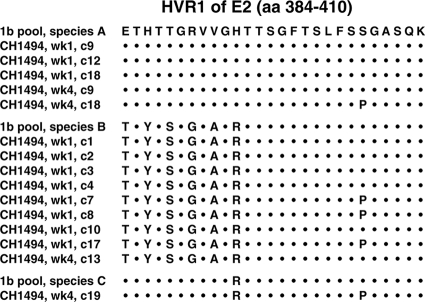
After a subsequent challenge with 100 CID of a heterologous strain (HC-J4), belonging to genotype 1b (36), the chimpanzee developed a transient infection with viremia during weeks 1 to 12 postinfection (Fig. 1B). The animal had peak titers of ∼104 GE/ml at weeks 1 and 4 postchallenge. However, at week 2 the titers were only ∼10 GE/ml, indicative of an anamnestic immune response. Cloning of the E1-E2 region, including HVR1, of virus recovered at weeks 1 and 4 demonstrated a polyclonal virus population, which represented the population found in the 1b challenge pool (36) (Fig. 3). The chimpanzee was not infected following two subsequent challenges with 100 and 1,000 CID of this 1b strain. A similar pattern with transient viremia or absence of viremia was observed following three subsequent challenges with polyclonal genotype 2a virus (strain HC-J6) (35) (Fig. 1B). Cloning of the E1-E2 region of virus recovered from the chimpanzee at weeks 4 (18 clones) and 10 (17 clones), respectively, of the first 2a challenge with 100 CID and at weeks 4 (12 clones) and 8 (12 clones) of the subsequent 2a challenge with 1,000 CID (Fig. 1B) confirmed the infection with strain HC-J6 and showed a similar homogeneous virus population after each challenge (data not shown). Peripheral and intrahepatic CD4+ responses were detected throughout the genotype 1b and 2a challenges (Fig. 1B), even though we used only genotype 1a antigens for these assays.
FIG. 3.
HVR1 sequences of viruses recovered from CH1494 at weeks 1 and 4, respectively, after the first rechallenge with 100 CID of the HC-J4 (genotype 1b) pool. The sequence of multiple clones was compared with the different virus sequences previously identified in the HC-J4 challenge virus (36).
Persistent infection in CH1494 following a final challenge with the monoclonal H77C virus.
After the animal had cleared HCV following the last challenge with the 2a virus, it was challenged with 320 CID50 of the virus used for the initial rechallenges, the monoclonal H77C virus. Interestingly, the animal became persistently infected (Fig. 1C). The animal was viremic during weeks 1 to 3 (peak titer, ∼103 GE/ml), but the virus was not detected in the serum during weeks 4 to 9. Viremia reappeared at week 10, and the animal remained infected during 85 weeks of follow-up. Liver enzyme levels remained normal. We did not detect significant levels of neutralizing antibodies in the H77C pseudoparticle assay throughout follow-up. It is noteworthy that the peripheral and intrahepatic CD4+ responses were weak or not detected, even during periods with nondetectable viremia or low-titer viremia. We analyzed the ORF sequences of viruses recovered at weeks 2, 25, 35, 43, and 50 following the final challenge (Table 3). The polyprotein sequence of viruses recovered at week 2 had several changes compared with the challenge virus (Table 4) (Fig. 1C). These changes were detected also in viruses recovered at later time points. In addition, the virus recovered at each subsequent week analyzed had new amino acid changes. Thus, persistence after multiple rechallenges was associated with early emergence of virus variants that might represent T-cell escape mutants.
TABLE 3.
Nucleotide changes of consensus ORF sequences of HCV variants recovered from chimpanzee 1494 after homologous monoclonal rechallengea
| Sample | Nucleotide at indicated position of HCV genome region
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Core | E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
||||||||||||||||||
| 725 | 1515 | 1591 | 1732 | 2470 | 2546 | 2640 | 2718 | 3018 | 3389 | 3399 | 3539 | 3818 | 4385 | 5618 | 5859 | 6246 | 6977 | 7390 | 7707 | 7788 | 7973 | 8766 | 8770 | 8901 | |
| H77C challenge | C | G | A | A | C | G | T | A | C | C | T | C | C | T | C | G | A | G | A | C | C | A | A | C | A |
| Wks CH1494 post- challenge/totalb | |||||||||||||||||||||||||
| 2/216 | • | A | G/a | A/G | T | • | C | A/g | • | • | C | • | • | • | • | A | G | G/a | • | • | T | G | • | • | • |
| 25/239 | C/T | A | G | A/G | T | • | C | G/A | • | T | C | T | T | • | • | A | G/a | G/A | • | • | T | G | • | • | • |
| 35/249 | T | A | G | G | T | • | C | G | C/t | • | C | T | T | • | C/t | A | G | A | G | T | T | G | • | T | T |
| 43/257 | T | A | G | G | T | G/T | C | G | T | • | C | T | T | T/c | T | A | G | A | G | T | T | G | • | T | T |
| 50/264 | T | A | G | G | T | T | C | G | T | • | C | T | T | C | T | A | G | A | G | T | T | G | A/G | C/T | T |
The sequence of the H77 challenge virus is shown on the top. Nucleotides identical with challenge virus are indicated with dots. Dominant changes are indicated by capital letters. Minor sequences are indicated with lowercase letters.
Weeks after final challenge followed by total weeks after initial challenge.
TABLE 4.
Amino acid changes of HCV variants recovered from chimpanzee 1494a
| Sample | Amino acid encoded by HCV genome region at indicated polyprotein position
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2
|
p7
|
NS2
|
NS4B
|
NS5A
|
NS5B
|
||||||||||
| 392 | 417 | 464 | 710 | 735 | 767 | 793 | 1020 | 1840 | 1969 | 2350 | 2483 | 2809 | 2810 | 2854 | |
| H77C | A | N | D | A | L | S | M | S | G | T | K | H | T | T | I |
| HCV from 1494 at wks postchallenge/totalb | |||||||||||||||
| 2/216 | T | S/n | D/G | V | • | P | M/v | P | S | A | • | Y | • | • | • |
| 25/239 | T | S | D/G | V | • | P | M/V | P | S | A/t | • | Y | • | • | • |
| 35/249 | T | S | G | V | • | P | V | P | S | A | R | Y | • | I | L |
| 43/257 | T | S | G | V | L/F | P | V | P | S | A | R | Y | • | I | L |
| 50/264 | T | S | G | V | F | P | V | P | S | A | R | Y | T/A | T/I | L |
The sequence of the H77 challenge virus is shown on the top. Amino acids identical with the challenge virus are indicated with dots. Dominant changes are indicated by capital letters. Minor sequences are indicated with lowercase letters.
Weeks after final challenge followed by total weeks after initial challenge.
Lack of protective immunity in CH96A008 following homologous rechallenge.
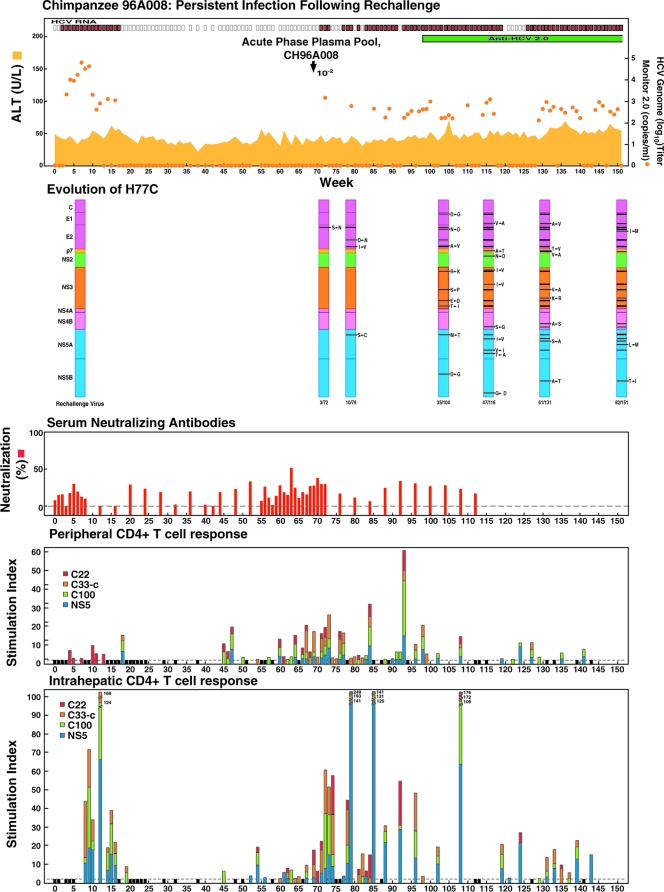
In the second chimpanzee studied with resolved HCV infection, homologous monoclonal rechallenge resulted in infection that persisted. CH96A008 had originally been infected with an HCV mutant derived from our infectious H77C clone that lacked the E2 HVR1 (16). During the acute infection, this animal had peak serum HCV titers of 104 to 105 GE/ml (Fig. 4). It did not develop antibodies against HCV as measured by the second-generation ELISA. More importantly, we did not detect neutralizing antibodies in the in vitro neutralization test using pseudoparticles bearing the H77C envelope proteins (Fig. 4). The chimpanzee had evidence of mild acute viral hepatitis with necro-inflammatory changes in liver biopsy specimens. The infection was resolved at week 18. In this animal we were able to study both the peripheral and intrahepatic CD4+ T-cell responses. CD4+ T cells isolated from PBMC or CD4+ T cells polyclonally expanded from liver biopsy specimens were tested for HCV-specific proliferative responses with a panel of recombinant genotype 1a HCV proteins (from strain HCV-1), including core, NS3, NS4, and NS5 (31), and the clearance of HCV in CH96A008 was associated with a peripheral and intrahepatic proliferative CD4+ T-cell response (Fig. 4). As reported previously, this animal also had peripheral and intrahepatic multispecific CD8+ T-cell responses during the acute infection (31).
FIG. 4.
Homologous rechallenge experiments in CH96A008 with resolved monoclonal HCV infection following inoculation with H77C virus (genotype 1a) lacking the HVR1 region. About 12 months after viral clearance the chimpanzee was rechallenged with homologous HCV collected during the acute phase of infection. The animal became persistently infected. To evaluate the course of infection, serum samples collected weekly were tested for HCV RNA by in-house RT-nested PCR with 5′-untranslated region primers and/or by the Roche Monitor Test 2.0. Red rectangle, positive by RT-nested PCR and/or by Monitor; empty rectangle, negative by RT-nested PCR; orange dots, HCV Monitor titers, in GE/ml; (samples with no registered value are shown as not detected); green horizontal bar, seroconversion in the second-generation ELISA; yellow-orange shaded area, serum ALT. For evolution of H77C-delta HVR1, the H77C genome is shown as a vertical bar with core at the top and NS5B at the bottom. Solid black lines with capital letters indicate new amino acid changes that were identified when a sequence was compared with the sequence obtained at the previous time point. Solid lines without capital letters represent amino acid changes that persisted. The week analyzed is indicated at the bottom of each genome. For neutralizing antibodies, the percent neutralization of retroviral pseudoparticles bearing the H77C envelope proteins is shown (>50% considered significant). For peripheral and intrahepatic CD4+ T-cell responses, the CD4+ T-cell responses to core (red), NS3 (orange), NS3-NS4 (green), and NS5 (blue) HCV-1 (genotype 1a) antigens are shown as the specific SI. A specific SI of >2 was considered significant. At weeks tested in which the SI was ≤2 against all four antigens, the negative result is indicated by a black bar (with a value of 2).
About 12 months after the original infection was cleared, as evidenced by the lack of detectable HCV RNA in serum and liver biopsy samples, CH96A008 was experimentally challenged with 1 ml of a 10−2 dilution (containing <103 genome copies) of the homologous virus collected at peak titer during the primary infection. The animal was viremic during weeks 2 to 4 (peak titer, ∼103 GE/ml), but the virus was not detected in the serum during weeks 5 to 7. However, viremia reappeared at week 8 and the animal remained infected for more than 2 years of follow-up (Fig. 4). Throughout follow-up there were short periods in which HCV RNA could not be detected, and HCV RNA titers remained at low levels. At the 1:25 dilution tested, we did not detect significant neutralizing antibodies in the in vitro neutralization tests using H77C pseudoparticles (Fig. 4).
In this animal, the virus persisted despite anamnestic multispecific intrahepatic proliferative CD4+ T-cell responses (Fig. 4). Peripheral CD4+ responses were also detected, but similar responses were observed prior to rechallenge. Finally, peripheral and intrahepatic CD8+ T-cell responses were detected, but only a few samples were tested. A peripheral CD8+ T-cell response (percent gamma interferon-producing CD8+ cells in response to peptide stimulation) was detected at the six time points tested during the first 17 weeks after reinoculation against four epitopes in core, NS4, and NS5 that were also targeted during the original acute resolving infection (31): week 1 (NS5, 2,290; 3.2%), week 2 (NS4, 1,667; 13.3%), week 3 (NS5, 2,290; 3.2%), week 7 (core, 169; 4.4%), week 16 (NS5, 2,575; 3.7%), and week 17 (core 169; 33.3%). In addition, intrahepatic CD8+ T cells were tested for HCV-specific reactivity at weeks 1, 12, 16, 23, 24, 25, 38, 39, and 43 after reinoculation. Of note, HCV-specific gamma interferon-producing CD8+ T-cell responses were detectable at weeks 16, 24, 25, 39, and 43 after stimulation with autologous Epstein-Barr virus-transformed immortalized B-cell lines that were infected with recombinant HCV vaccinia viruses vHCV 1-1488 (0.3%, 0.2%, 0.4%, 1.61%, and 0.15%, respectively) or vHCV 827-3011 (0.42%, 1.0%, 0.73%, 0.35%, and 0.16%, respectively). Thus, we observed low-level viral persistence following a true homologous virus challenge in the presence of primed helper and cytotoxic T-cell responses.
The entire polyprotein sequence of viruses recovered at weeks 3, 10, 35, 47, 61, and 82 postchallenge, when viral titers spiked (but did not exceed 104 GE/ml), was determined and compared with the sequence of the virus recovered during the initial infection (Table 5). A single change was identified at week 3, whereas this change and multiple additional changes were identified at each subsequent week analyzed (Fig. 4; Table 5), suggesting that HCV might have persisted through an immune escape mechanism.
TABLE 5.
Amino acid changes in HCV variants recovered from chimpanzee 96A008 after homologous monoclonal rechallengea
| Sample | Amino acid encoded by HCV genome region at indicated H77C nucleotide and amino acid position
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1
|
E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
|||||||||||||||||||||||||||||
| A994G 218 | A1097T 252 | C1420T 360 | T1435C 365 | G1597A 419 | A1683G 448 | C1757G 472 | G1881A 514 | G2169A 610 | C2470T 710 | A2481G 714 | G2697A/ C2698T 786 | T2701C 787 | A2934G 865 | G3604A 1088 | A3633G 1098 | G3769A 1143 | A4194G 1285 | T4269C 1310 | T4435C 1365 | T4443C 1368 | A4825G 1495 | G4943T 1534 | C5191T 1617 | G5994T 1885 | A6189G 1950 | T6505C 2055 | A6510T 2057 | A6687G 2116 | T6876G 2179 | C6957A 2206 | G7200A 2287 | A7410G 2357 | A8296G 2652 | G8613A/ C8614T 2758 | G9229A 2963 | |
| H77C | D | K | A | V | S | N | I | V | D | A | I | A | V | N | R | I | R | I | Y | V | S | K | E | T | A | S | M | S | I | S | L | V | T | D | A | G |
| HCV from 96A008, wks postchallenge/totalb,c | ||||||||||||||||||||||||||||||||||||
| 4/4 | • | N | • | • | • | • | • | M | • | • | • | • | • | • | • | • | • | • | H | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| 9/9 | • | N | • | • | • | • | • | M | • | • | • | • | • | • | • | • | H/r | • | H | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Infection after rechallenge | ||||||||||||||||||||||||||||||||||||
| 3/72 | • | N | • | • | N | • | • | M | • | • | • | • | • | • | • | • | • | • | H | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| 10/79 | • | N | • | • | N | • | • | M | N | • | V | • | • | • | • | • | • | • | H | • | • | • | • | • | • | • | • | C/s | • | • | • | • | • | • | • | • |
| 35/104 | G | N | • | • | N | D | • | M | N | V | V | • | • | • | K | • | • | • | H | • | P | • | D | I | • | • | T | C | • | • | • | • | • | G | • | • |
| 47/116 | G | N | • | A | N | D | • | M | N | • | V | T | • | D | K | V | • | V | H | • | P | • | D | I | • | G | T | C | V | • | • | I | A | • | • | D/g |
| 61/131 | G | N | V | • | N | D | • | M | N | • | V | V/m | A | • | K | • | • | V | H | A | P | R | D | I | S | G | T | C | V | A | • | I | • | • | T | • |
| 82/151 | G | N | V | • | N | D | M/i | M | N | • | V | A/v | A | • | K | • | • | V | H | A | P | R | D | I | S | G | T | C | V | A | M | I | • | • | I | • |
The rechallenge virus was an acute-phase sample collected from CH96A008 at week 7. The sequence of H77C is shown on the top. Positions refer to the H77C polyprotein. All recovered viruses lacked the HVR1. Amino acids identical to the challenge virus are indicated by a dot. Dominant changes are indicated by capital letters. Minor sequences are indicated with lowercase letters. Associated nucleotide change is indicated.
Weeks after final challenge followed by total weeks after initial challenge.
Samples collected during primary infection (weeks 4/4 and 9/9) and after homologous rechallenge (weeks 3/72, 10/79, 35/104, 47/116, 61/131, and 82/151) were analyzed.
DISCUSSION
This study demonstrated absence of significant levels of neutralizing antibodies in acute resolving HCV infections in two experimentally infected chimpanzees. Furthermore, such neutralizing antibodies were not detected after homologous rechallenges that produced no detectable infection, a transient viremia, or viral persistence. This apparent absence of neutralizing antibodies was not due to genetic heterogeneity of the virus, since the envelope sequence of the challenge virus and of the HCV pseudoparticles used to detect neutralizing antibodies were both strain H77C. The absence of neutralizing antibodies in one chimpanzee with apparent sterilizing immunity was confirmed in an in vitro neutralization experiment of the H77C virus. We cannot rule out that a low titer (reciprocal titer, <25) of neutralizing antibodies developed, but the lack of correlation of neutralizing antibodies with viral clearance is consistent with a recent study of patients with acute HCV (19). In contrast, proliferative T-cell responses correlated with viral control following homologous rechallenges, but detected responses did not necessarily prevent the subsequent development of persistence following homologous rechallenge. We cannot, however, based on our data, rule out the possibility that effector function (such as production of various cytokines) might have been compromised, since proliferation may be the last function to be lost as anergy is established. Viral persistence was associated with an evolving HCV polyprotein sequence.
The only HCV vaccine for which a significant decrease in chronicity rate in vaccinated chimpanzees has been demonstrated is based on the envelope proteins (14, 18), and this vaccine led to the induction of high-titer neutralizing antibodies as determined in a pseudoparticle assay using ppH77 (J. C. Meunier, unpublished data). We found that viral clearance occurred multiple times in CH1494 following homologous challenges in the absence of detectable neutralizing antibodies. Therefore, most likely effective vaccine-induced cellular immune responses that mimic those seen after a resolved HCV infection might be expected to control HCV and lower the chronicity rate, as was suggested also in a recent study of a T-cell-based vaccine candidate (13).
Chimpanzee 1494 was originally infected with RNA transcripts from a molecular clone (H77C) with the consensus sequence of the acute-phase H77 virus collected from patient H with acute posttransfusion hepatitis. Rechallenge of CH1494 with monoclonal homologous virus that was collected from a chimpanzee infected from the H77C clone resulted in transient viremia in two of three cases. In both instances we could demonstrate that these short-lived infections most likely were caused by neutralization failures, since the polyprotein sequences of recovered viruses had no amino acid changes compared with the challenge virus. However, after repeated homologous rechallenges with the H77C monoclonal virus, the animal had what appeared to be sterilizing immunity against challenge with the polyclonal H77 virus. Following the challenge with 640 CID50, we could not detect HCV RNA at days 2, 4, and 7 postchallenge. However, given the apparent lack of neutralizing antibodies, it is likely that a nondetectable infection occurred, since the anamnestic cellular immune responses, most likely responsible for the observed immunity, would require processing and presentation of viral antigens. It is noteworthy that viral clearance following rechallenge occurred without evidence of hepatitis, suggesting that noncytolytic mechanisms might play an important role in HCV clearance or that the number of infected cells was very low.
The sterilizing immunity that initially developed in CH1494 appeared to be strain or subtype specific, since subsequent heterologous rechallenge with viruses of another subtype of genotype 1 (genotype 1b) resulted in 12 weeks of viremia, and analysis of recovered sequences suggested neutralization failure, since the various virus species previously identified in the 1b challenge pool could be recovered. Furthermore subsequent rechallenge with the 1b virus resulted in what appeared to be sterilizing immunity, and the following rechallenge with virus of another genotype (genotype 2a) again resulted in viremia. Thus, the induced T-cell immunity appeared to be sequence specific, which is in agreement with a recent study that analyzed protective immunity following heterologous challenges with various genotypes (27). We did not analyze T-cell responses against 1b and 2a antigens during the 1b and 2a challenges. It has been reported that strain-specific T-cell suppression existed after reexposure to HCV with the potential of promoting persistence (30), but in CH1494 we observed clearance following the 1b and 2a challenges that followed resolved 1a infections. It is noteworthy that even though CH1494 had developed such effective immunity, the detected intrahepatic T-cell responses were not as vigorous as those detected in CH96A008 that developed persistent infection after the first rechallenge. The reason for this difference remains unknown, but this might reflect the different genetic backgrounds of these animals or other currently unknown virological and immunological factors.
Since we used monoclonal viruses for our homologous rechallenges that eventually resulted in persistence in both chimpanzees, it was possible to determine whether the recovered virus had a changed polyprotein sequence. Experimental infections with polyclonal viruses usually result in recovery of different variant HCV sequences. In contrast, we have previously demonstrated that the recovered viruses in five animals experimentally infected with the monoclonal H77C virus had the same polyprotein consensus sequence (15). Thus, the changes observed in the virus initially recovered from CH1494 and CH96A008 in infections that persisted might have resulted from immune selection of minor variants present in this challenge pool or were selected de novo. We have not been able to study the mechanism for this evolution, but they might represent immune escape variants as previously described in primary experimental infections (reviewed in reference 4) or in CD4+-depleted recovered animals that were rechallenged (17).
It was previously found that depletion of CD4+ cells, but not of CD8+ cells, resulted in persistence in animals that had previously demonstrated protective immunity (17, 29). It is noteworthy that we observed the same phenomenon in an animal (CH96A008) with anamnestic peripheral and intrahepatic CD4+ T-cell responses. However, in CH1494 viral persistence after multiple rechallenges developed in the apparent absence of vigorous CD4+ responses, suggesting potential T-cell suppression. In both cases, however, the virus was initially controlled, as evidenced by 3 to 6 weeks of serum HCV RNA negativity. Thus, the host responses associated with protective immunity against HCV and the lack thereof require more detailed studies, such as a comprehensive microarray analysis of the mRNA profile in the liver.
In previously published homologous rechallenge studies in chimpanzees it was reported that induced immunity prevented viral persistence (2, 21, 27). Our study demonstrated that this is not necessarily the case. However, following rechallenge the virus persists at low levels with periods in which the virus cannot be detected. Such low-level persistence following homologous rechallenge might actually have developed in some previously reported animals. For example, low-level viremia was apparently present in two of three animals rechallenged with homologous monoclonal virus after more than 6 months of follow-up (25). As in natural infections that persist, the virus titer is controlled at lower levels than those seen during the acute infection. In persistent infection in humans this control might be partly induced by neutralizing antibodies, since those are present at high levels and the virus develops neutralization escape mutants (32). This was apparently not the case in our persistently infected chimpanzees, since they did not develop detectable neutralizing antibodies during the chronic infection. Thus, the underlying mechanism for this control requires further studies; although we cannot rule out that the mutant nature of the virus infecting CH96A008 affected the outcome, it is possible that the robust T-cell responses detected in this animal played an important role in the observed viral control.
Overall, our data in CH1494 confirm previous observations of protective immunity, since clearance was observed multiple times, including clearance of homologous and heterologous viruses. Our demonstration of chronicity after a first rechallenge (CH96A008) and after multiple rechallenges (CH1494) is an important confirmation of the observations in repeatedly exposed individuals. Persistence can occur even when the challenge virus is identical to the virus that was originally inoculated into the animals, and even in an animal that has demonstrated protective immunity against this virus and viruses of two other genotypes. These results indicate that the failure to prevent chronicity is not due only to the heterologous nature of the reinfecting virus. Studies in CH96A008 demonstrated low-level viral persistence following a true homologous virus challenge in the presence of primed T-cell responses.
Supplementary Material
Acknowledgments
We thank Doris C. Wong and Ronald E. Engle (Hepatitis Viruses Section, LID, NIAID, NIH) for technical assistance.
Jens Bukh is the recipient of a professorship at the University of Copenhagen with external funding from the Lundbeck Foundation. This research was supported by NIH, NIAID contract number NO1-AO-062713, and by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 11 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 10014199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassett, S. E., B. Guerra, K. Brasky, E. Miskovsky, M. Houghton, G. R. Klimpel, and R. E. Lanford. 2001. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology 331479-1487. [DOI] [PubMed] [Google Scholar]
- 3.Bowen, D. G., and C. M. Walker. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436946-952. [DOI] [PubMed] [Google Scholar]
- 4.Bowen, D. G., and C. M. Walker. 2005. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J. Exp. Med. 2011709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh, J. 2004. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology 391469-1475. [DOI] [PubMed] [Google Scholar]
- 6.Bukh, J., C. L. Apgar, R. Engle, S. Govindarajan, P. A. Hegerich, R. Tellier, D. C. Wong, R. Elkins, and M. C. Kew. 1998. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: genetic analysis of the virus and generation of a standardized challenge pool. J. Infect. Dis. 1781193-1197. [DOI] [PubMed] [Google Scholar]
- 7.Bukh, J., X. Forns, S. U. Emerson, and R. H. Purcell. 2001. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology 44132-142. [DOI] [PubMed] [Google Scholar]
- 8.Bukh, J., R. Thimme, S. Govindarajan, X Forns, W Satterfield, G Eder, K.-M. Chang, M Yanagi, S. U. Emerson, F. V. Chisari, and R. H. Purcell. 2002. Monoclonal hepatitis C virus infection in chimpanzees, p. 336-340. In H. S. Margolis, M. J. Alter, T. Liang, and J. Dienstag (ed.), Viral hepatitis and liver disease. International Medical Press, London, United Kingdom.
- 9.Farci, P., H. J. Alter, S. Govindarajan, D. C. Wong, R. Engle, R. R. Lesniewski, I. K. Mushahwar, S. M. Desai, R. H. Miller, N. Ogata, et al. 1992. Lack of protective immunity against reinfection with hepatitis C virus. Science 258135-140. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., H. J. Alter, D. C. Wong, R. H. Miller, S. Govindarajan, R. Engle, M. Shapiro, and R. H. Purcell. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. USA 917792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farci, P., A. Shimoda, D. Wong, T. Cabezon, G. D. De, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 9315394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinstone, S. M., H. J. Alter, H. P. Dienes, Y. Shimizu, H. Popper, D. Blackmore, D. Sly, W. T. London, and R. H. Purcell. 1981. Non-A, non-B hepatitis in chimpanzees and marmosets. J. Infect. Dis. 144588-598. [DOI] [PubMed] [Google Scholar]
- 13.Folgori, A., S. Capone, L. Ruggeri, A. Meola, E. Sporeno, B. B. Ercole, M. Pezzanera, R. Tafi, M. Arcuri, E. Fattori, A. Lahm, A. Luzzago, A. Vitelli, S. Colloca, R. Cortese, and A. Nicosia. 2006. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat. Med. 12190-197. [DOI] [PubMed] [Google Scholar]
- 14.Forns, X., J. Bukh, and R. H. Purcell. 2002. The challenge of developing a vaccine against hepatitis C virus. J. Hepatol. 37684-695. [DOI] [PubMed] [Google Scholar]
- 15.Forns, X., P. J. Payette, X. Ma, W. Satterfield, G. Eder, I. K. Mushahwar, S. Govindarajan, H. L. Davis, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Vaccination of chimpanzees with plasmid DNA encoding the hepatitis C virus (HCV) envelope E2 protein modified the infection after challenge with homologous monoclonal HCV. Hepatology 32618-625. [DOI] [PubMed] [Google Scholar]
- 16.Forns, X., R. Thimme, S. Govindarajan, S. U. Emerson, R. H. Purcell, F. V. Chisari, and J. Bukh. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. USA 9713318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui, A., N. H. Shoukry, D. J. Woollard, J. H. Han, H. L. Hanson, J. Ghrayeb, K. K. Murthy, C. M. Rice, and C. M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302659-662. [DOI] [PubMed] [Google Scholar]
- 18.Houghton, M., and S. Abrignani. 2005. Prospects for a vaccine against the hepatitis C virus. Nature 436961-966. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, D. E., K. Sugimoto, K. Newton, M. E. Valiga, F. Ikeda, A. Aytaman, F. A. Nunes, M. R. Lucey, B. A. Vance, R. H. Vonderheide, K. R. Reddy, J. A. McKeating, and K. M. Chang. 2007. Discordant role of CD4 T-cell response relative to neutralizing antibody and CD8 T-cell responses in acute hepatitis C. Gastroenterology 132654-666. [DOI] [PubMed] [Google Scholar]
- 20.Lanford, R. E., B. Guerra, D. Chavez, C. Bigger, K. M. Brasky, X. H. Wang, S. C. Ray, and D. L. Thomas. 2004. Cross-genotype immunity to hepatitis C virus. J. Virol. 781575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Major, M. E., K. Mihalik, M. Puig, B. Rehermann, M. Nascimbeni, C. M. Rice, and S. M. Feinstone. 2002. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J. Virol. 766586-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta, S. H., A. Cox, D. R. Hoover, X. H. Wang, Q. Mao, S. Ray, S. A. Strathdee, D. Vlahov, and D. L. Thomas. 2002. Protection against persistence of hepatitis C. Lancet 3591478-1483. [DOI] [PubMed] [Google Scholar]
- 23.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 1024560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen, M., and J. Bukh. 2007. Current status of a hepatitis C vaccine: encouraging results but significant challenges ahead. Curr. Infect. Dis. Rep. 994-101. [DOI] [PubMed] [Google Scholar]
- 25.Nascimbeni, M., E. Mizukoshi, M. Bosmann, M. E. Major, K. Mihalik, C. M. Rice, S. M. Feinstone, and B. Rehermann. 2003. Kinetics of CD4+ and CD8+ memory T-cell responses during hepatitis C virus rechallenge of previously recovered chimpanzees. J. Virol. 774781-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prince, A. M., B. Brotman, T. Huima, D. Pascual, M. Jaffery, and G. Inchauspe. 1992. Immunity in hepatitis C infection. J. Infect. Dis. 165438-443. [DOI] [PubMed] [Google Scholar]
- 27.Prince, A. M., B. Brotman, D. H. Lee, W. Pfahler, N. Tricoche, L. Andrus, and M. T. Shata. 2005. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J. Infect. Dis. 1921701-1709. [DOI] [PubMed] [Google Scholar]
- 28.Sakai, A., S. Takikawa, R. Thimme, J. C. Meunier, H. C. Spangenberg, S. Govindarajan, P. Farci, S. U. Emerson, F. V. Chisari, R. H. Purcell, and J. Bukh. 2007. In vivo study of the HC-TN strain of hepatitis C virus recovered from a patient with fulminant hepatitis: RNA transcripts of a molecular clone (pHC-TN) are infectious in chimpanzees but not in Huh7.5 cells. J. Virol. 817208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 1971645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimoto, K., D. E. Kaplan, F. Ikeda, J. Ding, J. Schwartz, F. A. Nunes, H. J. Alter, and K. M. Chang. 2005. Strain-specific T-cell suppression and protective immunity in patients with chronic hepatitis C virus infection. J. Virol. 796976-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 9915661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Hahn, T., J. C. Yoon, H. Alter, C. M. Rice, B. Rehermann, P. Balfe, and J. A. McKeating. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132667-678. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, A. J., X. Paliard, M. J. Selby, A. Medina-Selby, D. Coit, S. Nguyen, J. Kansopon, C. L. Arian, P. Ng, J. Tucker, C. T. Lee, N. K. Polakos, J. Han, S. Wong, H. H. Lu, S. Rosenberg, K. M. Brasky, D. Chien, G. Kuo, and M. Houghton. 2001. Intrahepatic genetic inoculation of hepatitis C virus RNA confers cross-protective immunity. J. Virol. 757142-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 948738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262250-263. [DOI] [PubMed] [Google Scholar]
- 36.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244161-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.