Abstract
The prM protein of Japanese encephalitis virus (JEV) contains a single potential N-linked glycosylation site, N15-X16-T17, which is highly conserved among JEV strains and closely related flaviviruses. To investigate the role of this site in JEV replication and pathogenesis, we manipulated the RNA genome by using infectious JEV cDNA to generate three prM mutants (N15A, T17A, and N15A/T17A) with alanine substiting for N15 and/or T17 and one mutant with silent point mutations introduced into the nucleotide sequences corresponding to all three residues in the glycosylation site. An analysis of these mutants in the presence or absence of endoglycosidases confirmed the addition of oligosaccharides to this potential glycosylation site. The loss of prM N glycosylation, without significantly altering the intracellular levels of viral RNA and proteins, led to an ≈20-fold reduction in the production of extracellular virions, which had protein compositions and infectivities nearly identical to those of wild-type virions; this reduction occurred at the stage of virus release, rather than assembly. This release defect was correlated with small-plaque morphology and an N-glycosylation-dependent delay in viral growth. A more conservative mutation, N15Q, had the same effect as N15A. One of the four prM mutants, N15A/T17A, showed an additional defect in virus growth in mosquito C6/36 cells but not human neuroblastoma SH-SY5Y or hamster BHK-21 cells. This cell type dependence was attributed to abnormal N-glycosylation-independent biogenesis of prM. In mice, the elimination of prM N glycosylation resulted in a drastic decrease in virulence after peripheral inoculation. Overall, our findings indicate that this highly conserved N-glycosylation motif in prM is crucial for multiple stages of JEV biology: prM biogenesis, virus release, and pathogenesis.
Japanese encephalitis virus (JEV) is a member of the genus Flavivirus, which consists of ≈80 enveloped RNA viruses in the family Flaviviridae (5, 22, 36). The flaviviruses include many other clinically important human pathogens, such as dengue virus (DENV), yellow fever virus, West Nile virus (WNV), St. Louis encephalitis virus, Murray Valley encephalitis virus, and tick-borne encephalitis virus (TBEV). JEV is transmitted in an enzoonotic cycle between mosquito vectors and vertebrate hosts, with pigs and ardeid birds as the primary viremia-amplifying hosts and reservoirs, respectively, and with humans as incidental hosts (4). JEV is the most important cause of epidemic encephalitis in many Asian countries, leading to permanent neuropsychiatric sequelae and even death in children and young adults (13, 60, 64, 66). Over the past two decades, JEV has spread throughout the Indonesian archipelago (9, 75) to the Australian territories (41, 42, 65), attracting increasing attention in the arena of international public health.
JEV contains a single-stranded, positive-sense RNA genome of ≈11,000 nucleotides. The RNA genome has a cap structure at the 5′ end and lacks a poly(A) tail at the 3′ end (37). It consists of a single long open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs) that are important cis-acting elements for replication, transcription, and translation (43). The ORF encoding all of the viral proteins is translated into a large polyprotein, which is co- or posttranslationally processed by cellular and viral proteases into at least 10 mature proteins. The three structural proteins, the capsid (C; ≈11 kDa), premembrane (prM; ≈25 kDa), and envelope (E; ≈50 kDa) proteins, are encoded in the 5′-most third of the ORF. prM is a precursor of the membrane-anchored and virion-associated M protein (≈8 kDa). The seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, in the order in which the corresponding genes are arranged in the genome) are encoded in the remaining 3′ two-thirds (7, 77).
Like that of other flaviviruses, the genomic RNA of JEV is organized with multiple copies of the C protein, which forms a nucleocapsid surrounded by a host-derived lipid bilayer containing two viral surface proteins, prM/M and E (23, 45). During the synthesis of the nascent polyprotein, host signal cleavages generate the N and C termini of prM. The C terminus containing the adjacent transmembrane domains allows prM to anchor itself to the membrane and serves as the signal sequence for the translocation of E (7). While prM is able to form its native folded structure independently of the E protein, E requires the coexpression of prM to acquire its native conformation, suggesting a chaperone-like role for prM in the folding of E (35, 39). In addition, prM interacts with E to form prM-E heterodimers, which are important for the formation of immature virions (2, 11, 39, 71, 72). During flavivirus assembly and release, immature virions are generally formed by the budding of a viral nucleocapsid into the endoplasmic reticulum (ER), where prM-E heterodimers are acquired. In general, the mature virions are released into the extracellular compartment through the cellular secretory pathway (23, 40).
During its transport through the ER and Golgi apparatus to the cell surface, the prM of the prM-E heterodimers keeps E from undergoing the low-pH-induced conformational rearrangements required for membrane fusion and infectivity (17, 18, 21, 52, 61, 72, 78). Shortly before the flavivirus is released from the cell, the portion of prM that distinguishes it from M (the pr portion) is cleaved by furin or a related protease in the trans-Golgi apparatus (61). This cleavage reaction is inhibited by treatment with agents that raise the pHs of acidic intracellular compartments (17, 18, 21, 52). The cleavage of prM enables E to generate head-to-tail homodimers, which form a lattice-like structure layered on the surfaces of the mature virions (44). During flavivirus entry, E binds to an unidentified cellular receptor(s), followed by uptake into endosomes, where it undergoes low-pH-induced conformational changes, resulting in the fusion of the viral membranes with cellular membranes (10, 15, 32). Thus, the prM cleavage required for the generation of highly infectious viruses leads to two distinct forms of virions: intracellular immature virions containing uncleaved prM and E and extracellular mature virions containing exclusively cleaved M and E, with a trace amount of uncleaved prM (23, 45).
In the flaviviruses, prM contains one to three potential N-linked glycosylation sites (N-X-S/T) within the pr domain, and all of these sites have the sequence N-X-T (7). In the case of JEV, the prM protein contains only a single potential N-linked glycosylation site, N15-X16-T17, which is highly conserved among not only all JEV isolates but also other serologically related flaviviruses (WNV, Kunjin virus, St. Louis encephalitis virus, and Murray Valley encephalitis virus). To elucidate the functional importance of the two residues N15 and T17 and the role of the carbohydrate side chain in JEV replication and pathogenesis, we have now used the specific mutagenesis of infectious JEV cDNA to generate four sets of prM mutations in the context of the genomic RNA: three by substituting alanine for either N15 or T17 or both residues and one by introducing silent point mutations corresponding to all three residues in the glycosylation site. After transfecting BHK-21 (baby hamster kidney) cells with full-length RNA transcripts derived from the four prM mutant DNAs and the wild type (WT), we analyzed (i) the intracellular accumulation of the viral RNA and proteins, (ii) the average sizes of the plaques produced, and (iii) the infectivities and levels of production of extracellular virions released over time from the transfected cells. Subsequently, we examined the relative abundances of intracellular virions in the cytoplasm of BHK-21 cells infected with each of the four prM mutants and the WT. Also, we compared the four prM mutants to WT virus with respect to the capacities of the viruses for growth in three different cell types: BHK-21 cells, which are routinely used for JEV propagation in laboratories, and SH-SY5Y (human neuroblastoma) and C6/36 (mosquito) cells, which are biologically relevant to JEV pathogenesis and transmission. Finally, we assessed the pathogenicities of the four prM mutants in a murine infection model system. Our data demonstrate that the single N-linked glycosylation site within JEV prM plays an important role in multiple steps of the JEV life cycle: not only in viral replication, by maximizing virus release and optimizing prM protein biogenesis in a cell type-specific manner, but also in viral pathogenesis, by enhancing pathogenicity after inoculation via a peripheral route.
MATERIALS AND METHODS
Cell culture.
BHK-21 cells were cultivated in alpha minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, vitamins, and penicillin-streptomycin at 37°C in 5% CO2 (76). The human neuroblastoma SH-SY5Y cells were purchased from the American Type Culture Collection (ATCC) and maintained in a 1:1 mixture of MEM with Earle's salts and Ham's F-12 nutrient medium supplemented with 10% FBS, 0.1 mM nonessential amino acids, and penicillin-streptomycin at 37°C in 5% CO2. The mosquito Aedes albopictus C6/36 cells were obtained from the ATCC and propagated in a mixture of MEM with Earle's balanced salt solution, 10% FBS, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and penicillin-streptomycin at 28°C in 5% CO2.
Antibodies.
A mouse hyperimmune antiserum specific for JEV was obtained from the ATCC (76). Four polyclonal rabbit antisera specific for the JEV C protein (103 amino acids corresponding to nucleotides 96 to 404), the pr protein (92 amino acids corresponding to nucleotides 477 to 752), the M protein (44 amino acids corresponding to nucleotides 753 to 884), and the E protein (116 amino acids corresponding to nucleotides 978 to 1,325) were generated by expressing defined coding regions of the corresponding genes with pGEX-4T-1 (Amersham Biosciences) as glutathione S-transferase (GST) fusion proteins in Escherichia coli BL21 and then using the GST fusion proteins as immunogens. Two additional polyclonal rabbit antisera specific for the N-terminal and C-terminal eight residues of the JEV prM protein were also produced by immunization with the corresponding synthetic peptides conjugated at their N termini with a hapten carrier, keyhole limpet hemocyanin (KLH); these peptides were designated prM/N8 (KLH-MKLSNFQG) and prM/C8 (KLH-LLVAPAYS) (Invitrogen), respectively. All polyclonal antisera were generated in specific-pathogen-free New Zealand White rabbits over a 12-week period according to conventional procedures. Nucleotide positions refer to the complete nucleotide sequence of JEV CNU/LP2 (GenBank accession no. AY585243). The rabbit polyclonal anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) antiserum was purchased from LabFrontier Co., Ltd. (Seoul, South Korea). Alkaline phosphatase-conjugated goat anti-mouse and anti-rabbit immunoglobulin G antibodies were purchased from Jackson ImmunoResearch Laboratories Inc.
Site-directed mutagenesis of infectious JEV cDNA.
All plasmids were constructed according to standard molecular biology protocols (56). All four JEV prM mutants used in this study were generated from the infectious WT JEV cDNA molecular clone pBACSP6/JVFLx/XbaI (76), referred to herein as the WT. All mutations were introduced by PCR-based site-directed mutagenesis. All PCR-derived fragments were sequenced to confirm the desired mutations and exclude further off-site mutations. The oligonucleotides used for mutagenesis in this study are listed in Table 1. Details of the cloning procedures for the individual mutants and computer-readable sequence files are available upon request.
TABLE 1.
Oligonucleotides used for PCR-based site-directed mutagenesis in this study
| Oligonucleotide | Sequencea | Positionb | Polarity | Amino acid change(s)c |
|---|---|---|---|---|
| JEVpacIF | 5′-TAGTTAATTAACCTGCAGGGGGCTG-3′ | Vector | Sense | |
| JEVmluIR | 5′-ACCACGCGTTGACCATTGTTACTGC-3′ | 913-937 | Antisense | |
| N15AF | 5′-ATGACCATTGCCAACACGGA-3′ | 510-529 | Sense | N15A |
| N15AR | 5′-TCCGTGTTGGCAATGGTCAT-3′ | 510-529 | Antisense | |
| T17AF | 5′-TTAACAACGCGGACATTG-3′ | 517-534 | Sense | T17A |
| T17AR | 5′-CAATGTCCGCGTTGTTAA-3′ | 517-534 | Antisense | |
| DoubleF | 5′-ACCATTGCCAACGCGGACAT-3′ | 513-532 | Sense | N15A, T17A |
| DoubleR | 5′-ATGTCCGCGTTGGCAATGGT-3′ | 513-532 | Antisense | |
| SmF | 5′-CATTAATAATACCGACATTG-3′ | 515-534 | Sense | None (SM) |
| SmR | 5′-CAATGTCGGTATTATTAATG-3′ | 515-534 | Antisense |
Nucleotide changes that were introduced by site-directed mutagenesis are identified by boldface letters. Restriction enzyme recognition sites used for cDNA cloning are underlined.
Nucleotide positions refer to the complete nucleotide sequence of the JEV CNU/LP2 strain (GenBank accession number AY585243).
Mutagenesis resulted in the replacement of the Asn15 and/or Thr17 residue of JEV prM with Ala, as indicated. Numbering refers to the amino acid positions in the JEV prM protein.
For the N15A construct, two cDNA fragments were first isolated by PCR amplification of pBACSP6/JVFLx/XbaI (76) with two pairs of primers, JEVpacIF-N15AR and N15AF-JEVmluIR. These two cDNA fragments were then fused by a second round of PCR amplification with the primer pair JEVpacIF and JEVmluIR. The 1,064-bp PacI-MluI fragment of the resulting amplicons was ligated to the 4,647-bp MluI-BamHI and 12,854-bp BamHI-PacI fragments of pBACSP6/JVFLx/XbaI, generating the N15A construct. Similarly, the first two cDNA fragments for each of the other three constructs (T17A, N15A/T17A, and SM) were first isolated by PCR amplification of pBACSP6/JVFLx/XbaI with two pairs of primers, namely, JEVpacIF-T17AR and T17AF-JEVmluIR, JEVpacIF-DoubleR and DoubleF-JEVmluIR, and JEVpacIF-SmR and SmF-JEVmluIR, respectively. In all cases, these two fragments were then fused by a second round of PCR with primers JEVpacIF and JEVmluIR, and the 1,064-bp PacI-MluI fragment of the resulting amplicons was finally cloned into pBACSP6/JVFLx/XbaI as described above for the N15A construct.
In vitro synthesis of RNA transcripts.
The parental full-length infectious WT JEV cDNA and its derivatives were digested with XbaI and modified with mung bean nuclease for linearization. XbaI-linearized and mung bean nuclease-treated plasmids were used as DNA templates for in vitro runoff transcription using SP6 RNA polymerase, as described previously (76). In brief, ≈200 ng of the template DNA was added to a 25-μl reaction mixture consisting of the buffer supplied by the manufacturer (New England Biolabs) plus 0.6 mM cap analog [m7G(5′)ppp(5′)A; New England Biolabs]; 0.5 μM [3H]UTP (1.0 mCi/ml and 50 Ci/mmol; New England Nuclear Corp.); 10 mM dithiothreitol; 1 mM (each) UTP, GTP, CTP, and ATP; 40 U of RNaseOUT; and 15 U of SP6 RNA polymerase. The reaction mixtures were incubated at 37°C for 1 h. RNA transcripts were quantified on the basis of [3H]UTP incorporation, as measured by RNA adsorption to DE-81 filter paper (Whatman). The integrity of the RNA transcripts was verified by 1% agarose gel electrophoresis and ethidium bromide staining.
Transfection with RNA transcripts and infectious-center assays.
Electroporation was used to transfect BHK-21, SH-SY5Y, and C6/36 cells with in vitro-transcribed RNAs. Subconfluent cells were collected by trypsinization, washed three times with ice-cold RNase-free phosphate-buffered saline (PBS), and resuspended at 2 × 107 cells/ml in PBS. In vitro-transcribed RNA (2 μg) was mixed with a 400-μl aliquot of the cell suspension (8 × 106 cells) in a 2-mm-gap cuvette (BTX), and the suspension was immediately pulsed under conditions determined previously to be optimal (980 V, a 99-μs pulse length, and five pulses for BHK-21 cells; 780 V, a 99-μs pulse length, and five pulses for SH-SY5Y cells; and 800 V, a 99-μs pulse length, and five pulses for C6/36 cells) by using a model no. ECM 830 electroporator as recommended by the manufacturer (BTX). After a 10-min recovery at room temperature, the electroporated mixture was diluted with fresh complete medium to obtain a volume of 1 ml. A 100-μl aliquot was taken to determine the specific infectivity of the RNA as described below, and the remainder was plated onto two 150-mm dishes and incubated at 37°C for 48 to 72 h before viral harvest.
RNA infectivity was quantified by an infectious-center assay (76). A 1/10 portion of the 1-ml electroporation mixture was serially diluted 10-fold with fresh complete medium and plated onto monolayers of naïve BHK-21 cells (5 × 105 cells/well) in a six-well plate. The cells were allowed to recover and attach to the plate by incubation at 37°C with 5% CO2 for 6 h, after which the medium was removed and replaced with 0.5% SeaKem LE agarose overlay. Plates were incubated for 4 to 5 days at 37°C with 5% CO2, and the infectious centers of plaques were visualized by staining with 1% (wt/vol) crystal violet (in 50% ethanol).
Real-time quantitative RT-PCR.
Total RNA was isolated from duplicate wells with Trizol reagent (Invitrogen); to normalize total RNA levels, 50 ng of total cellular RNA was used for the reverse transcription (RT) reaction involving primers specific for the JEV NS3 region and for the BHK-21 β-actin RNA. JEV and BHK-21 β-actin cDNAs were synthesized by RT at 45°C for 30 min, followed by the inactivation of the reverse transcriptase at 95°C for 10 min. JEV-specific and BHK-21 β-actin-specific cDNAs were amplified with the iQ Supermix quantitative PCR system (Bio-Rad Laboratories) and detected with the iCycler iQ multicolor real-time PCR detection system (Bio-Rad Laboratories). One-tenth of the RT reaction mixture was used for PCR amplification, with 45 cycles of 95°C for 15 s and 60°C for 1 min. The JEV forward and reverse primers were 5′-ATCCAACTCAACCGCAAGTC-3′ and 5′-TCTAAGATGGTGGGTTTCACG-3′, respectively. The probe sequence (nucleotides 5837 to 5856) was 5′-6-carboxyfluorescein-CATCTCTGAAATGGGGGCTA-Black Hole Quencher 1-3′ (Integrated DNA Technologies). The forward and reverse primers for BHK-21 β-actin were 5′-ACTGGCATTGTGATGGACTC-3′ and 5′-CATGAGGTAGTCTGTCAGGTC-3′, respectively. The probe sequence was hexachlorofluorescein-CCAGCCAGGTCCAGACGCAGG-Black Hole Quencher 2-3′ (Integrated DNA Technologies). The 2−ΔΔCT method was used to analyze relative changes in JEV RNA levels from real-time quantitative RT-PCR experiments (59, 74).
Preparation of cell- and virion-associated viral proteins.
To isolate cell-associated JEV proteins, cells were directly lysed in 0.2 ml of 1× sample loading buffer (80 mM Tri-HCl [pH 6.8], 2.0% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue) at 6, 12, 18, and 24 h after RNA transfection. Cell lysates were sheared by repeated passage through a 26-gauge needle. For virion-associated JEV proteins, cell culture supernatants were harvested at 24 h posttransfection and cellular debris was removed by centrifugation at 3,000 rpm for 30 min at 4°C in a Hitachi CF7D2 centrifuge. Virus particle-containing supernatants were concentrated by ultracentrifugation through a 20% sucrose cushion in a Beckman Coulter Optima LE-80K SW28 rotor at 20,000 rpm (72,128 × g) for 2 h at 4°C. Viral pellets were resuspended in the 1× sample loading buffer. Samples were boiled for 10 min prior to separation by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Immunoblotting.
Protein samples of equal amounts were resolved by either the glycine-SDS-PAGE (56) or the tricine-SDS-PAGE (58) system for low-molecular-mass forms of the JEV C and M proteins. Proteins were transferred onto methanol-activated polyvinylidene difluoride membranes by using a Trans-Blot SD electrophoretic transfer cell machine (Bio-Rad Laboratories). Membranes were blocked with 5% nonfat dried milk in washing solution (0.2% Tween 20 in PBS) at room temperature for 1 h. They were then washed three times with the washing solution and incubated at room temperature for 2 h with one of the following primary antibodies: a mouse JEV-specific hyperimmune antiserum (1:1,000 dilution), rabbit anti-C antiserum C3 (1:1,000 dilution), rabbit anti-pr antiserum PR2 (1:4,000 dilution), rabbit anti-M antiserum M5 (1:2,000 dilution), rabbit anti-E antiserum EN1 (1:500 dilution), rabbit anti-prM/N8 antiserum (1:100 dilution), rabbit anti-prM/C8 antiserum (1:100 dilution), or rabbit anti-GAPDH antiserum (1:5,000 dilution). The primary antibody-decorated membranes were washed three times with the washing solution and incubated at room temperature for 2 h with an alkaline phosphatase-conjugated goat anti-mouse or anti-rabbit immunoglobulin G antibody (1:5,000 dilution), as appropriate. The membranes were washed three times with the washing solution and once with PBS. JEV or GAPDH protein bands were visualized by incubation with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium as substrates.
Immunoprecipitation and endoglycosidase digestion.
For the immunoprecipitation of JEV proteins, BHK-21 cells were used for RNA transfection as described above. At 10 h posttransfection, cells (2 × 106) were washed once and incubated in methionine-free MEM for 30 min and then labeled with the same medium supplemented with 2% FBS and 200-μCi/ml l-[35S]methionine (PerkinElmer Life and Analytical Sciences) for 8 h. Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (0.15 M NaCl, 0.05 M Tris-HCl [pH 7.2], 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitors). Cellular debris was removed by centrifugation. The supernatants were immunoprecipitated for 10 h at 4°C with mouse JEV-specific hyperimmune antiserum that had been preabsorbed with protein A-Sepharose beads. Immunoprecipitated pellets were washed five times in RIPA lysis buffer, solubilized in 1× sample loading buffer, boiled for 10 min, separated using the 15% tricine-SDS-PAGE system, and analyzed by autoradiography.
For endoglycosidase treatment of JEV proteins, immune complexes were eluted from protein A-Sepharose pellets after the final washing step by being boiled in 60 μl of 1× glycoprotein-denaturing buffer (0.5% SDS and 1% β-mercaptoethanol) for 10 min. The supernatant was then split into three equal portions. For endoglycosidase H (endo H) digestion, 20 μl of one aliquot was added to a 40-μl reaction mixture consisting of 1× G5 buffer (50 mM sodium citrate [pH 5.5]) and 250 U of endo H (New England Biolabs). For N-glycosidase F (PNGase F) digestion, another 20 μl was added to a 40-μl reaction mixture consisting of 1× G7 buffer (50 mM sodium phosphate [pH 7.5]), 1% NP-40, and 250 U of PNGase F (New England Biolabs). The mixtures containing these two aliquots and the corresponding mock-treated controls lacking the enzymes were incubated for 6 h at 37°C. After incubation, the samples were boiled for 10 min in 1× sample loading buffer and analyzed by 15% tricine-SDS-PAGE and autoradiography.
Growth curves.
The growth properties of the JEV prM mutant viruses in BHK-21, SH-SY5Y, and C6/36 cells were analyzed. For this purpose, cells were preseeded into 35-mm dishes at 3 × 105 per dish for 12 to 18 h and then infected with viruses at a multiplicity of infection (MOI) of 0.01 for 1 h at 37°C with frequent agitation. After incubation, the inoculum was removed and the monolayers were washed once and incubated in fresh complete medium. Supernatants were collected at the indicated time points (6, 12, 18, 24, 36, 48, 72, and 96 h postinfection for BHK-21 and SH-SY5Y cells and the additional time points of 120 and 144 h postinfection for C6/36 cells), and aliquots were stored at −80°C. The titers of supernatants were determined by plaque assays on BHK-21 cell monolayers (76).
Electron microscopy (EM).
BHK-21 cells were infected at an MOI of 10 with the WT or one of the prM mutant viruses. At 18 h after infection, cells were gently rinsed with PBS and fixed in a 2.5% paraformaldehyde-glutaraldehyde mixture buffered with 0.1 M phosphate (pH 7.2) for 1 h at 4°C. Fixed cells were scraped from the plates, collected by low-speed centrifugation (200 × g for 10 min at 4°C), postfixed in 1% osmium tetroxide in the same buffer for 1 h, dehydrated in graded ethanol and propylene oxide, and embedded in Epon-812. Ultrathin sections cut with an ULTRACUTE ultramicrotome (Leica, Austria) were stained with uranyl acetate and lead citrate and examined under a CM 20 electron microscope (Philips, The Netherlands).
Mouse experiments.
Three-week-old female ICR mice (Charles River Laboratories) were used for pathogenesis studies. Animals were handled according to the regulations of the Chungbuk National University Medical School's animal experimentation facility. Mice were anesthetized with methoxylfluorane and inoculated intracerebrally (i.c.) or intraperitoneally (i.p.) with doses of virus stock serially diluted 10-fold in MEM (n = 10 mice per dose) in a volume of 30 or 50 μl, respectively. Control groups were inoculated with an equivalent volume of supernatant from an uninfected cell culture at a comparable dilution via the same route. Mice were observed every 12 h until death or were maintained over a 5-week period, during which morbidity and mortality rates were recorded. The 50% lethal doses (LD50s) were determined by the method of Reed and Muench (53). Whenever possible, sera were collected and stored at −80°C until being used for plaque reduction neutralizing tests. All mice that died after i.p. inoculation showed typical signs of clinical encephalitis, and the replication of JEV genomic RNA in brain tissue was confirmed by RT-PCR; all mice that survived showed variable levels of antibody response, and all the mock-infected mice survived with no signs of disease. All the mice that died after i.c. inoculation displayed typical signs of clinical encephalitis, and JEV amplification in their brain tissues was confirmed by plaque assays after the experiment; the control group of mock-infected mice all survived the 5-week period with no signs of disease. For statistical analysis, survival curves were created by the Kaplan-Meier method and analyzed by the log-rank test in order to identify any significant differences between the WT and each of the four prM mutants. A P value of <0.05 was considered statistically significant. All procedures were performed using the SPSS software package, version 10.0 (SPSS, Inc.).
RESULTS
Mutations substituting alanine for either or both of the N15 and T17 residues in the single potential N-linked glycosylation site N15-X16-T17 within JEV prM result in significant reductions in virus titer and plaque size, with no detectable changes in viral RNA replication or protein expression.
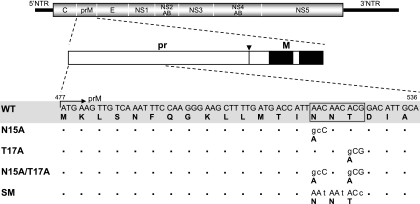
The pr region of JEV prM protein contains a single potential N-linked glycosylation site that is highly conserved in 444 of the 447 JEV strains whose sequences are currently available from the GenBank database (data not shown). Although the canonical consensus sequence is N-X-T/S, all of these sites found within JEV prM invariably display the sequence N-X-T at amino acid positions 15 to 17. To investigate the functional significance of the evolutionarily conserved N15 and T17, we introduced mutations replacing either or both of these two residues with alanine in full-length infectious JEV cDNA (the WT) to produce three mutants, N15A, T17A, and N15A/T17A (Fig. 1). In parallel, we also engineered three silent point mutations corresponding to all three amino acid positions (15 to 17) to generate an additional mutant, SM, to allow for the possibility that cis-acting RNA sequences required for replication are located in this region of the viral genome (Fig. 1).
FIG. 1.
Schematic representation of JEV prM N-glycosylation mutants constructed in the context of a full-length infectious JEV cDNA molecular clone. The RNA genome structure of JEV is schematically outlined at the top. Coding regions corresponding to viral proteins are indicated, with thick solid lines at both termini representing the 5′ and 3′ UTRs (NTR) of the viral genome. Below the diagram of the viral genome, the major regions of the JEV prM protein are outlined. The soluble pr portion of JEV prM is cleaved from the membrane-anchored virion-associated M protein by furin or a related cellular protease (61). The cleavage site is indicated by an arrowhead, and a pair of transmembrane domains located at the C terminus of the prM protein are indicated by solid boxes. Four JEV prM N-glycosylation mutants (N15A, T17A, N15A/T17A, and SM) were constructed from full-length infectious JEV cDNA (WT), as described in Materials and Methods. Nucleotide and amino acid sequence alignments corresponding to amino acid positions 1 to 20 of the WT and the four prM mutants are shown. The location of the single potential N-linked glycosylation consensus sequence (N15-X16-T17) is indicated by an open box. Nucleotide changes introduced into this region are indicated by lowercase letters, and the resulting amino acid alterations are indicated by bold uppercase letters. An arrow indicates the N terminus of JEV prM protein. Bullets represent identical residues.
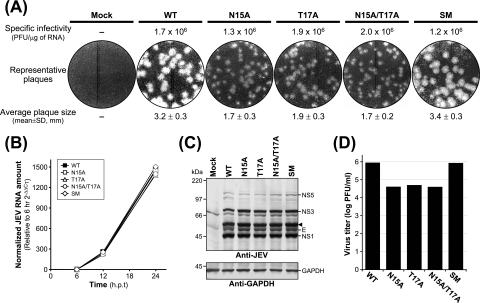
We first examined the effects of these mutations on the specific infectivity and viability of JEV genomic RNA. For this purpose, we transfected naïve BHK-21 cells by electroporation with RNA transcripts derived from the WT and each of the four prM mutant cDNA templates (N15A, T17A, N15A/T17A, and SM) in order to analyze the numbers and sizes of the plaques formed in RNA-transfected cells. At 4 to 5 days after transfection, the RNA transcripts produced from all four mutant cDNA constructs had specific infectivities ranging from 1.2 × 106 to 2.0 × 106 PFU/μg, similar to that of the WT (1.7 × 106 PFU/μg) (Fig. 2A). Thus, none of the mutations introduced at this potential N-linked glycosylation site altered the infectivity or viability of the JEV genomic RNA. However, the sizes of the plaques formed in mutant RNA-transfected cells were different from those of the plaques formed in WT RNA-transfected cells (Fig. 2A). BHK-21 cells transfected with mutant RNAs derived from N15A, T17A, and N15A/T17A displayed a homogeneous population of plaques, ≈2-fold smaller than those formed in the cells transfected with WT-derived RNA. In contrast, BHK-21 cells transfected with the SM-derived RNA exhibited large homogeneous plaques, phenotypically indistinguishable from those formed in the cells transfected with WT RNA.
FIG. 2.
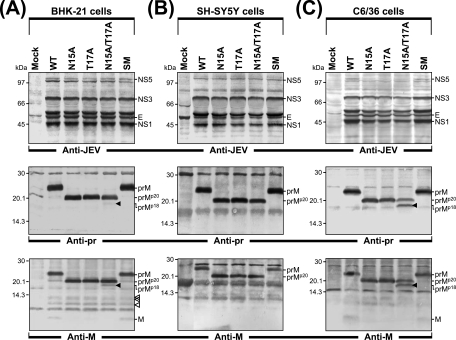
Characterization of JEV prM N-glycosylation mutants: specific infectivities, plaque morphologies, viral RNA and protein expression profiles, and virus yields. Naïve BHK-21 cells were mock transfected (mock) or transfected with the synthetic RNA transcripts derived from either the WT or one of four JEV prM mutant cDNA templates (N15A, T17A, N15A/T17A, or SM) by electroporation. (A) Specific infectivities of the RNA transcripts and representative plaque morphologies. The specific infectivities of the RNA transcripts were estimated by infectious-center assays as described in Materials and Methods (results are expressed as numbers of PFU per microgram of RNA). To evaluate representative plaque morphologies, the cells were overlaid with agarose and stained with crystal violet 4 to 5 days after RNA transfection. The average plaque sizes (mean diameters ± standard deviations) were estimated by counting 20 representative plaques formed by each RNA transcript. (B) Levels of production of JEV RNA. Total cellular RNA was isolated from the transfected cells at 6, 12, and 24 h posttransfection (h.p.t.). Real-time quantitative RT-PCR was utilized for the quantitation of JEV-specific RNA and β-actin RNA for the normalization of total RNA levels. The 2−ΔΔCT method was used to analyze the changes in JEV RNA levels relative to those measured at 6 h posttransfection in real-time quantitative PCR experiments. (C) Accumulation of the JEV proteins. Equal amounts of protein extracts collected at 24 h posttransfection were separated by 12% glycine-SDS-PAGE. Viral proteins were visualized by immunoblotting with a polyclonal JEV-specific mouse hyperimmune antiserum (anti-JEV). In parallel, GAPDH protein, used as a loading and transfer control, was detected with a polyclonal anti-GAPDH rabbit antiserum (anti-GAPDH). The positions of the JEV-specific proteins, a viral protein-related cleavage intermediate (arrowhead), and GAPDH are indicated to the right. Molecular mass markers, in kilodaltons, are indicated to the left. (D) Levels of production of infectious virions. Virus titers in the culture supernatants harvested from monolayers of the transfected cells at 24 h posttransfection were determined by plaque assays on naïve BHK-21 cells. Data from one of three independent experiments, which yielded similar results, are shown.
We then assessed the levels of production of the viral RNA and proteins in BHK-21 cells transfected with each of the RNAs. Real-time quantitative RT-PCR using a JEV-specific probe complementary to the sequence of nucleotides 5837 to 5856, corresponding to JEV NS3, showed that in all cases, the accumulation levels of the RNAs relative to those at 6 h posttransfection were invariably increased by ≈250-fold at 12 h posttransfection and subsequently increased by ≈1,400- to 1,500-fold at 24 h posttransfection (Fig. 2B). Thus, all the viral genomic RNAs were produced at similar levels in RNA-transfected cells over a period of 24 h after transfection. Consistent with this RNA production, immunoblotting using a JEV-specific hyperimmune antiserum revealed equivalent amounts and identical patterns of JEV-specific proteins (such as E, NS1, NS3, and NS5) in WT RNA-transfected cells and cells transfected with each of the four prM mutant RNAs (Fig. 2C). Using real-time quantitative RT-PCR to examine the levels of production of the viral RNA has the potential for overestimating the amounts of full-length genomic RNAs, because in our assay this method detected only a 158-nucleotide region (nucleotides 5757 to 5914) within the genome. However, since comparable amounts of JEV-specific proteins were detected in the cells transfected with the WT and each of the four prM mutant RNAs, it is unlikely that there was undetected preferential degradation of the WT or prM mutant RNAs.
Next, we determined the virus titers, measuring the infectivities of the viruses produced from BHK-21 cells transfected with the WT and each of the four prM mutants. Following 24 h of incubation after transfection, equal volumes of culture supernatants were used to infect naïve BHK-21 cells, and the virus titers were estimated by plaque assays (Fig. 2D). Three prM mutants, N15A (4.2 × 104 PFU/ml), T17A (5.1 × 104 PFU/ml), and N15A/T17A (4.0 × 104 PFU/ml), all showed ≈20-fold reductions (17- to 22-fold) in titers compared to the WT titer (8.8 × 105 PFU/ml). The SM mutant produced a titer (8.3 × 105 PFU/ml) similar to the WT titer. Taken together, these results indicated that a step(s) other than viral RNA synthesis and protein expression was primarily affected by mutations introduced into the single potential N-linked glycosylation site in JEV prM.
A single potential N-linked glycosylation site, N15-X16-T17, in JEV prM is utilized for the addition of oligosaccharides.
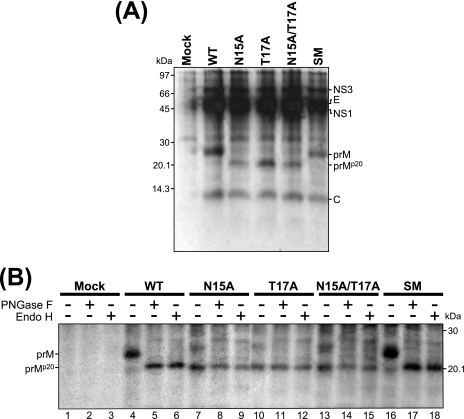
To characterize the JEV-specific proteins produced by the four JEV prM mutants, we transfected or mock-transfected BHK-21 cells with the RNAs derived from the WT and each of four prM mutant cDNAs and then metabolically labeled the cells with [35S]methionine for 8 h and immunoprecipitated the cell lysates with a polyclonal JEV-specific mouse hyperimmune antiserum (Fig. 3A). The WT JEV prM protein migrated as a species of ≈25 kDa. In the case of the three prM mutants lacking the single potential N-linked glycosylation site (N15A, T17A, and N15A/T17A), a smaller form of ≈20 kDa, designated prMp20, was instead observed. As expected, the SM prM protein comigrated with the WT prM protein, demonstrating that the three silent point mutations introduced into the potential N-linked glycosylation site did not alter the apparent molecular size. These results were consistent with those of mobility shift assays in which equal amounts of cell lysates were detected with a polyclonal anti-pr rabbit antiserum (data not shown).
FIG. 3.
Biochemical and endoglycosidase analyses of JEV prM N-glycosylation mutants. Naïve BHK-21 cells were mock transfected (mock) or transfected with the synthetic RNA transcripts derived from either the WT or one of four JEV prM mutant cDNA templates (N15A, T17A, N15A/T17A, or SM). At 10 h posttransfection, cells were metabolically labeled with [35S]methionine for 8 h and lysed with 1× RIPA lysis buffer. (A) Immunoprecipitation of metabolically labeled JEV proteins. Cell lysates were immunoprecipitated with a polyclonal JEV-specific mouse hyperimmune antiserum. Samples were analyzed by 15% tricine-SDS-PAGE and visualized by autoradiography. The positions of JEV-specific proteins are indicated to the right, and molecular mass markers, in kilodaltons, are indicated to the left. (B) Endoglycosidase digestion of cell-associated JEV prM proteins. Cell lysates were immunoprecipitated with a polyclonal JEV-specific mouse hyperimmune antiserum. Denatured immunoprecipitates were digested with PNGase F (lanes 2, 5, 8, 11, 14, and 17) or endo H (lanes 3, 6, 9, 12, 15, and 18) or incubated without either endoglycosidase (lanes 1, 4, 7, 10, 13, and 16) and analyzed by 15% tricine-SDS-PAGE followed by autoradiography. Glycosylated (prM) and unglycosylated (prMp20) forms of the WT and four mutant prM proteins are indicated to the left. +, present; −, absent.
To determine whether the mobility shift observed for the JEV prM mutant proteins actually reflected the loss of glycans at the single potential N-linked glycosylation site, we treated or mock-treated immunoprecipitated lysates with either endo H, which removes high-mannose-content N-linked glycans, or PNGase F, which cleaves all types of N-linked glycans (Fig. 3B). The digestion of the ≈25-kDa WT prM with either endoglycosidase yielded a product of ≈20 kDa, whose migration pattern was invariably identical to those of all three mutant prMp20 proteins lacking the potential N-linked glycosylation site (those of the N15A, T17A, and N15A/T17A mutants) that had been mock treated. In addition, the mobilities of these three mutant prMp20 proteins were identical before and after digestion with either endoglycosidase. As expected, the sensitivity of the SM mutant prM protein to digestion with either endoglycosidase was the same as that of WT prM. Therefore, we concluded that the cell-associated prM protein is glycosylated primarily with high-mannose oligosaccharides at its single potential N-linked glycosylation site, N15-X16-T17.
Reductions in the plaque size and virus titer observed with JEV prM N-glycosylation-defective mutants are correlated with a decrease in the production of extracellular virus particles.
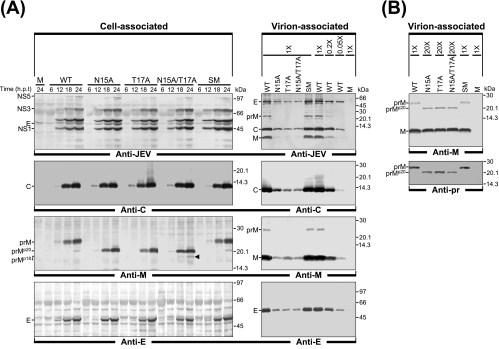
One possible explanation for the effects of alanine substitution that we observed is that JEV prM N-glycosylation-defective mutants are defective in virus assembly or release. Alternatively, the mutations may affect viral infectivity, altering the ratio of virus particles to PFU. To test these two possibilities, we used immunoblotting to analyze the relative production levels and profiles of three JEV structural proteins (C, prM/M, and E) that were released into culture supernatants in the first 24 h after the transfection of BHK-21 cells with the relevant RNAs.
BHK-21 cells were transfected with the RNAs derived from the WT and each of four prM mutant cDNAs under our optimized electroporation conditions (yielding >90% transfection efficiency). Cell lysates were collected at 6, 12, 18, and 24 h after transfection, and the JEV particles released into the culture supernatants were harvested at 24 h by ultracentrifugation through a 20% sucrose cushion. The three JEV structural proteins in the cell lysates and released viral particles were analyzed by immunoblotting with a JEV-specific mouse hyperimmune antiserum and four rabbit antisera specific for the C, pr, M, and E proteins of JEV. In the lysates of all four prM mutants, the expression levels of all three structural proteins were nearly identical to WT levels (Fig. 4A). In contrast, when JEV particles purified from equal volumes of the culture supernatants were analyzed, the accumulation levels of all three JEV structural proteins of the three prM N-glycosylation-defective mutants (N15A, T17A, and N15A/T17A) were invariably ≈20-fold lower than those of the WT proteins (≈5% of the WT levels) (Fig. 4A). This reduction directly paralleled the average ≈20-fold decrease in virus particle infectivity in these samples (Fig. 2). As expected, the SM mutant revealed no obvious mutant phenotype, yielding WT levels of virion-associated JEV structural proteins (Fig. 4A). Also, immunoblotting with an anti-M rabbit antiserum revealed a trace amount of unglycosylated prMp20, together with a large amount of glycosylated prM, in the cells transfected with the WT or control mutant SM RNA (Fig. 4A). Unexpectedly, we also noted a detectable amount of an M-reactive protein of ≈18 kDa (smaller than the unglycosylated prMp20), designated prMp18, only in the N15A/T17A RNA-transfected cells (Fig. 4A; also see Fig. 7 and 8 for a detailed description).
FIG. 4.
Analysis of virus particle release by JEV prM N-glycosylation mutants. Naïve BHK-21 cells were mock transfected (M) or transfected with the synthetic RNA transcripts derived from either the WT or one of four prM mutant cDNAs (N15A, T17A, N15A/T17A, or SM). (A) Comparison of cell-associated JEV proteins with virion-associated JEV proteins. For cell-associated JEV proteins, total cell lysates were prepared by directly lysing monolayers of transfected cells at 6, 12, 18, and 24 h posttransfection (h.p.t.). For virion-associated JEV proteins, equal volumes of culture supernatants were collected from RNA-transfected cells at 24 h posttransfection and extracellular viral particles were pelleted by ultracentrifugation through a 20% sucrose cushion. Equivalent portions of total cell lysates and pelleted viral particles were subjected to either 12% glycine-SDS-PAGE or 15% tricine-SDS-PAGE for the separation of the JEV C and prM/M proteins with small molecular sizes. For the relative quantitation of virus particle release, concentrated WT viral particles were diluted 1:1 (1×), 1:5 (0.2×), and 1:20 (0.05×) and used as a reference. Cell-associated and virion-associated JEV proteins were visualized by immunoblotting using a polyclonal JEV-specific mouse hyperimmune antiserum (anti-JEV) and a panel of three polyclonal rabbit antisera specific for the JEV C (anti-C), M (anti-M), and E (anti-E) proteins. Indicated to the left of each panel are the positions of the JEV-specific proteins, including two forms of JEV prM, glycosylated (prM) and unglycosylated (prMp20). The position of an M-reactive protein of ≈18 kDa (prMp18), found only in the N15A/T17A RNA-transfected cells, is highlighted with an arrowhead; molecular mass markers, in kilodaltons, are indicated to the right of each panel. (B) Detection and comparison of glycosylated prM or unglycosylated prMp20 proteins associated with released viral particles. Viral particles pelleted from clarified supernatants 24 h after transfection were subjected to immunoblot analysis. For three prM mutants (N15A, T17A, and N15A/T17A), protein samples were adjusted to contain equivalent amounts of viral particles prior to 15% tricine-SDS-PAGE by overloading the gel with 20-fold-larger amounts of N15A, T17A, and N15A/T17A viral particle-containing pelleted materials than of WT and SM mutant particle-containing materials. Samples were transferred onto two membranes: one was blotted with a polyclonal anti-M rabbit antiserum (anti-M), and the other was blotted with a polyclonal anti-pr rabbit antiserum (anti-pr). Indicated to the left of each panel are the positions of JEV M and/or two forms of JEV prM, glycosylated (prM) and unglycosylated (prMp20). Numbers to the right are molecular masses, in kilodaltons.
FIG. 7.
Abnormal prM protein biogenesis by JEV prM N-glycosylation mutants in three different cell types. BHK-21 (A), SH-SY5Y (B), and C6/36 (C) cells were mock transfected (mock) or transfected with the synthetic RNA transcripts derived from the WT or one of four prM mutant cDNAs (N15A, T17A, N15A/T17A, or SM). After a 24-h incubation, monolayers of the transfected cells were directly lysed with 1× sample loading buffer. Equal amounts of cell lysates were separated by 12% glycine-SDS-PAGE (for staining with anti-JEV) or 15% tricine-SDS-PAGE (for staining with anti-pr and anti-M) and transferred onto membranes. Membranes were probed with a polyclonal JEV-specific mouse hyperimmune antiserum (anti-JEV) or two polyclonal rabbit antisera, one specific for the JEV pr (anti-pr) and the other specific for the M protein (anti-M). Molecular mass markers, in kilodaltons, are indicated to the left of each panel, and immunoreactive JEV proteins are shown to the right. Also highlighted are a pr- and M-reactive protein band of ≈18 kDa (prMp18, indicated by a black arrowhead) seen in the BHK-21 and C6/36 cells transfected with N15A/T17A-derived RNA and three additional M-reactive protein bands (indicated by three white arrowheads) detected in the BHK-21 cells transfected with the RNAs derived from the WT or any of the four prM mutants.
FIG. 8.
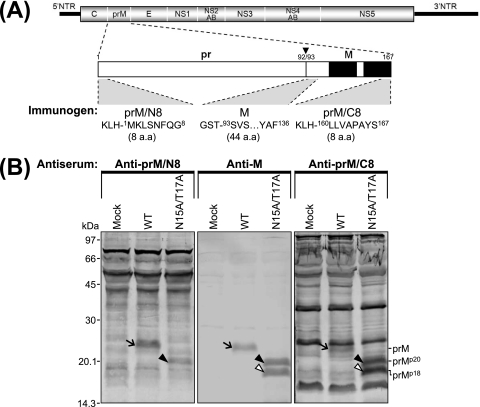
The prMp18 protein lacks the N terminus of unglycosylated prMp20. (A) Diagram indicating the three immunogens used to generate the corresponding polyclonal rabbit antisera against JEV prM. Shown at the top is the organization of the JEV genomic RNA; the regions corresponding to the viral proteins are indicated, with thick solid lines at both termini corresponding to the 5′ and 3′ UTRs (NTR) of the genome. Below is an expanded outline of the 167-amino-acid prM protein. An arrowhead indicates the site at which prM is cleaved to yield the pr and M proteins, occurring between amino acid positions 92 and 93; two solid boxes indicate a pair of transmembrane domains located at the C terminus of the prM protein. Of the three immunogens used, two were the 8-amino-acid (a.a.) synthetic peptides corresponding to the N terminus (prM/N8) and the C terminus (prM/C8) of the uncleaved prM, with KLH conjugated to the N termini of these peptides; the third immunogen was the E. coli-produced recombinant protein encoding the 44-amino-acid N-terminal region of the cleaved M, with GST fused to its N terminus. (B) Detection of the unglycosylated prMp20 and prMp18 proteins by immunoblotting using three polyclonal rabbit antisera specifically recognizing various regions of JEV prM protein. C6/36 cells were mock transfected (mock) or transfected with the synthetic RNA transcripts derived from the WT or N15A/T17A mutant cDNA. After a 24-h incubation, monolayers of the transfected cells were directly lysed with 1× sample loading buffer. Equal amounts of cell lysates were separated by 15% tricine-SDS-PAGE and transferred onto membranes, which were probed with one of three polyclonal rabbit antisera: anti-prM/N8, anti-M, or anti-prM/C8 antiserum. Molecular mass markers are given in kilodaltons; shown within the panels are the positions of the prM (arrows), prMp20 (black arrowheads), and prMp18 (white arrowheads) proteins.
When we analyzed purified JEV particles by immunoblotting with two polyclonal rabbit antisera detecting the JEV pr (data not shown) and M (Fig. 4A) proteins, we observed trace amounts of uncleaved prM protein in the WT and control mutant SM samples. This finding is consistent with previous results for other flaviviruses (19, 47, 71), although the biological significance of this release of uncleaved prM from the infected cells is unclear. We were unable to detect any such uncleaved prMp20 protein in the case of the three prM N-glycosylation-defective mutants, when JEV particles purified from equal volumes of culture supernatant were analyzed (Fig. 4A). However, when we increased the amounts of JEV particles used for immunoblotting by loading the gels with 20 times the amount of WT particles used, we found that the three prM N-glycosylation-defective mutants displayed uncleaved prMp20 and that the ratios of uncleaved prMp20 to cleaved M proteins were similar to those of uncleaved prM to cleaved M proteins in the WT and control mutant SM samples (Fig. 4B). Thus, these results indicated that although JEV prM N-glycosylation-defective mutants had a severe defect in virus particle assembly and release, the virus particles they released had a protein composition analogous to that of the WT particles.
In principle, the decrease in the levels of virion-associated structural proteins in the culture supernatants may reflect either inefficient virus assembly and release or the instability of the released virus particles. To distinguish between these two possibilities, we used trichloroacetic acid to precipitate the total protein from the culture supernatants and then analyzed the levels of the three JEV structural proteins in the precipitates by immunoblotting. Again, the levels of each of the three proteins were reduced by ≈20-fold compared to the WT level in the case of all three prM N-glycosylation-defective mutants (N15A, T17A, and N15A/T17A), indicating that the reduction was due to a defect in virus assembly and release rather than a decrease in the stability of the released particles (data not shown). Taken together, our results demonstrated that the inactivation of N glycosylation at the N15-X16-T17 site in JEV prM resulted in the defective assembly and release of JEV particles. This finding was further confirmed by the results obtained for an additional mutation replacing N15 with Q. This mutation is chemically more conservative than replacement with Ala, in that both Asn and Gln contain an amide group in their side chains; the N15Q mutant had a defect in virus assembly and release, as did the N15A mutant (see Fig. S1 in the supplemental material).
The N glycosylation of JEV prM promotes the release of intracellular virions accumulated in the lumina of reorganized membrane-bound vesicles.
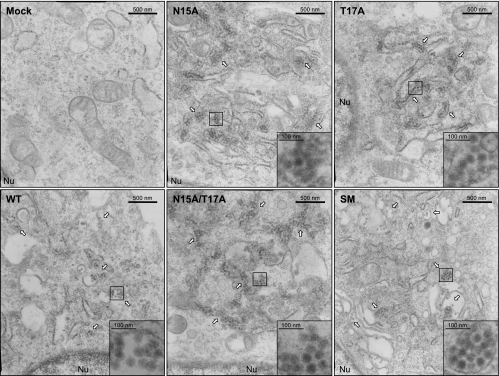
To determine at the morphological level whether the reduction in the production of extracellular virus particles by JEV prM N-glycosylation-defective mutants occurs at the stage of assembly or release, we performed thin-section EM analysis of BHK-21 cells infected at an MOI of 10 with the WT or one of four prM mutant viruses and then fixed 18 h after infection (Fig. 5). In all cases, large numbers of intact intracellular virions (≈50 nm), as evidenced by the presence of an electron-dense core of ≈30 nm surrounded by a lipid envelope (45, 46, 70, 73), were observed in the lumina of reorganized membrane-bound vesicles induced by viral replication. In the case of the three prM N-glycosylation-defective mutants (N15A, T17A, and N15A/T17A), there was no obvious difference between the appearances of their intact intracellular virions, which showed typical flavivirus morphology, and those of the WT and control mutant SM virions. However, we noted an increase in the numbers of these structures within the lumina of reorganized membrane-bound vesicles in the cells infected with any of the three prM N-glycosylation-defective mutant viruses compared to the numbers seen in the cells infected with the WT or control mutant SM virus. Of particular note was our observation that thin sections of cells infected with each of the three prM N-glycosylation-defective mutant viruses featured large bundles of intracellular virions tightly condensed within the lumina of elongated vesicles, whereas thin sections of cells infected with the WT or control mutant SM virus revealed mainly small aggregates of intracellular virions loosely packed or dispersed within the lumina of vesicles with diverse morphologies, suggesting that the release of the prM N-glycosylation-defective mutant virions assembled in the lumina of intracellular membrane compartments was impaired.
FIG. 5.
Thin-section EM analysis of JEV prM N-glycosylation mutants. Electron micrographs show representative thin sections of BHK-21 cells mock infected (mock) or infected with the WT or one of four JEV prM N-glycosylation mutant viruses (N15A, T17A, N15A/T17A, or SM) at an MOI of 10; cells were collected at 18 h after infection for examination by EM. White arrows point to aggregates of assembled intracellular virions, as identified by an electron-dense core surrounded by a lipid envelope, accumulating within the lumina of JEV-induced reorganized membrane-bound vesicles. Also indicated is the edge of the nucleus (Nu), to assist in orienting each of the electron micrographs. Original magnifications, ×27,500 for each panel and ×150,000 for each inset (boxed region) inside the corresponding panels.
To obtain a more quantitative assessment of the release defect of the prM N-glycosylation-defective mutants, we estimated the relative abundances of the intracellular virions for the WT and each of the four prM mutants. To quantify the intracellular virions in thin sections of cells infected with each of the corresponding viruses, the intracellular virions in 15 to 20 cells randomly selected from each sample were counted; the number of intracellular virions per cell was then determined. This EM analysis was independently repeated three times. As summarized in Table 2, the relative abundances of the intracellular virions of the three prM N-glycosylation-defective mutants were found to be significantly higher (by ≈2-fold; range, 1.60- to 2.02-fold) than that of the WT; in contrast, no such difference between the WT and the control mutant SM was seen. As in the case of the biochemical experiments described above, the N15Q mutant was indistinguishable from the N15A mutant with respect to the appearance and number of intracellular virions when assessed at the morphological level (data not shown). Taken together, these results indicate that JEV prM N-glycosylation-defective mutants are defective at the stage of virus release, rather than assembly.
TABLE 2.
Relative abundances of JEV WT and prM mutant intracellular virionsa
| Virus | First expt
|
Second expt
|
Third expt
|
Total no. of intracellular virions counted | Total no. of cells examined | No. of intracellular virions/cell | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of intracellular virions counted | No. of cells examined | No. of intracellular virions/cell | No. of intracellular virions counted | No. of cells examined | No. of intracellular virions/cell | No. of intracellular virions counted | No. of cells examined | No. of intracellular virions/cell | ||||
| WT | 3,101 | 18 | 172 | 4,809 | 18 | 267 | 3,955 | 15 | 264 | 11,865 | 51 | 233 |
| N15A | 5,721 | 15 | 381 | 8,282 | 17 | 487 | 8,695 | 17 | 511 | 22,698 | 49 | 463 |
| T17A | 6,932 | 20 | 347 | 6,376 | 18 | 354 | 6,398 | 15 | 427 | 19,706 | 53 | 372 |
| N15A/T17A | 8,852 | 20 | 443 | 8,384 | 17 | 493 | 9,629 | 20 | 481 | 26,865 | 57 | 471 |
| SM | 3,924 | 16 | 245 | 3,044 | 15 | 203 | 5,471 | 20 | 274 | 12,439 | 51 | 244 |
JEV intracellular virions were identified by EM using morphological criteria. BHK-21 cells were infected at an MOI of 10 with the WT or one of four prM N-glycosylation mutant viruses (N15A, T17A, N15A/T17A, or SM). At 18 h postinfection, thin sections of the infected cells were examined by EM to quantify the intracellular JEV virions, which were identified by an electron-dense core of ≈30 nm surrounded by a lipid envelope (46, 70, 73). The number of intracellular virions per cell was then determined.
The JEV prM N-glycosylation defect has differential effects on virus growth in three different cell types.
The four JEV prM mutants described above were examined for their abilities to grow and establish a productive infection in three cell types: mammalian BHK-21 cells, which are commonly used for virus propagation in laboratories, and human neuroblastoma SH-SY5Y and mosquito C6/36 cells, which are both biologically relevant for JEV pathogenesis and transmission. Monolayers of each of the three cell lines were infected at an MOI of 0.01 with the WT and each of four prM mutant viruses in parallel, and the levels of the virus particles released were monitored over time by plaque assays. This viral growth experiment was independently repeated twice, yielding similar results.
In the case of BHK-21 (Fig. 6A) and SH-SY5Y (Fig. 6B) cells, the yields of all four JEV prM mutants peaked at 48 to 72 h postinfection, as did that of the WT. However, the three prM N-glycosylation-defective mutants (N15A, T17A, and N15A/T17A) produced virus yields ≈20-fold lower than that of the WT during this time period. As expected, the SM mutant grew with WT kinetics in both cell types. In the case of C6/36 cells (Fig. 6C), however, the growth kinetics of the parental WT and all four prM mutants exhibited a significant delay compared to the kinetics in BHK-21 and SH-SY5Y cells, peaking at 120 to 144 h postinfection. It is important that during this period of time, N15A/T17A produced a drastically lower yield than the other two N-glycosylation-defective mutants, N15A and T17A (≈500-fold lower than the WT yield in the case of N15A/T17A, versus ≈50-fold lower in the cases of N15A and T17A). As expected, the yield for the SM mutant was similar to that for the WT, indicating no impairment in virus growth. Again, the growth properties of the N15Q mutant were nearly identical to those of the N15A mutant (see Fig. S2 in the supplemental material). Overall, this analysis showed that a lack of N-linked glycosylation on JEV prM was associated with a delay in virus growth and a reduction in virus yield in all three cell types tested, with the delay and reduction being considerably more pronounced in C6/36 cells than in BHK-21 or SH-SY5Y cells. Of note is the fact that the yield of N15A/T17A virus was ≈10-fold lower than that of N15A and T17A in C6/36 cells but not BHK-21 or SH-SY5Y cells. Thus, the N15 residue, along with T17, apparently plays an additional important role in JEV biology, affecting virus growth in a cell type-specific manner.
FIG. 6.
Comparison of the virus growth properties of JEV prM N-glycosylation mutants in three different cell types. BHK-21 (A), SH-SY5Y (B), and C6/36 (C) cells were infected at an MOI of 0.01 with the WT or one of four JEV prM N-glycosylation mutant viruses (N15A, T17A, N15A/T17A, or SM). Culture supernatants were collected at the indicated hour postinfection (h.p.i.), and virus titers were determined by plaque assays on naïve BHK-21 cells. The data shown are from one of two independent experiments yielding similar results.
The double mutation N15A/T17A, but not N15A or T17A, causes a subtle difference in JEV prM protein biogenesis in a cell type-specific fashion, leading to a significant defect in virus growth.
To determine whether the unexpected cell type-specific effect of the N15A/T17A mutation on virus growth was associated with improper proteolytic processing of JEV prM, we transfected BHK-21, SH-SY5Y, and C6/36 cells with the RNAs transcribed from the WT and four prM mutant cDNAs under our optimized electroporation conditions. After a 24-h incubation, we compared the profiles and expression levels of intracellular JEV proteins by immunoblotting using the polyclonal anti-JEV antiserum and two rabbit antisera recognizing the JEV pr and M proteins.
In all three cell types, we detected small but nonsignificant differences in the intracellular accumulation levels of JEV proteins (such as E, NS1, NS3, and NS5) among the cells transfected with each of the four prM mutant RNAs and the WT RNA-transfected cells (Fig. 7). Of the three prM N-glycosylation-defective mutants, however, only N15A/T17A produced a band corresponding to an additional pr- and M-reactive protein of ≈18 kDa, designated prMp18 (Fig. 7). The N15A/T17A mutant produced the highest-level accumulation of prMp18 in C6/36 cells (Fig. 7C), with a 1:1 ratio of prMp20 to prMp18; the same mutant produced no detectable prMp18 in SH-SY5Y cells (Fig. 7B) and only barely detectable levels in BHK-21 cells (Fig. 7A). Since this prMp18 protein was recognized by two antisera, one specific for the pr protein that we mutated and the other specific for the M protein, which we did not mutate, the difference in the amount of the prMp18 protein detected was not caused by the loss of reactive epitopes in the prM, prMp20, or prMp18 protein. Another difference between BHK-21 cells and the two biologically relevant cell types was also noted. In BHK-21 cells, trace amounts of unglycosylated prMp20 protein, as well as large amounts of glycosylated prM, were recognized in the cases of the WT and the control mutant SM; detectable amounts of three additional M-reactive protein bands were seen in the WT and all four prM mutant samples (Fig. 7A). These protein bands, however, were not detectable in SH-SY5Y or C6/36 cells (Fig. 7B and C). From these data, it appears that a subtle difference in JEV prM protein biogenesis, particularly that observed for N15A/T17A in three different cell types, can lead to a significant defect in virus growth that is cell type dependent and prM glycosylation independent.
The prMp18 protein lacks the N-terminal region of unglycosylated prMp20.
Because the two polyclonal rabbit antisera reactive with prMp18 were raised against the region corresponding to the N-terminal 44 residues of cleaved M (for anti-M antiserum) and the region corresponding to the N-terminal 92 residues of the pr portion of uncleaved prM (for anti-pr antiserum), with GST fused to their N termini (Fig. 7), it was not known which terminus was missing from the prMp18 protein compared with the unglycosylated prMp20 protein. To answer this question, we generated an additional pair of polyclonal rabbit antisera specific for the N terminus (anti-prM/N8) and the C terminus (anti-prM/C8) of uncleaved prM by immunization with one of the two corresponding 8-amino-acid synthetic peptides conjugated to a hapten carrier, KLH, at their N termini (Fig. 8A). As seen with the anti-M antiserum, immunoblotting using the anti-prM/C8 antiserum detected prMp18, together with unglycosylated prMp20, in C6/36 cells transfected with N15A/T17A RNA (Fig. 8B). However, the prMp18 protein band was not detected by immunoblotting using the anti-prM/N8 antiserum in the extract of N15A/T17A RNA-transfected C6/36 cells expressing unglycosylated prMp20 (Fig. 8B). As expected, both the anti-prM/N8 and anti-prM/C8 antisera recognized glycosylated prM in WT RNA-transfected C6/36 cells (Fig. 8B). These results demonstrated that the prMp18 protein lacked the N terminus of unglycosylated prMp20, mostly likely as a result of aberrant processing(s) of unglycosylated prMp20. Based on the estimated molecular mass of the prMp18 protein, the aberrant cleavage site(s) appeared to be located within the N-terminal 50-amino-acid region of prM. This region, however, lacks the minimal cleavage site sequence, R-X-X-R (34, 48), for the Golgi apparatus resident protease furin, which is involved in the cleavage of prM into pr and M (61). Given our current understanding that uncleaved prM has a chaperone-like role, preventing E from undergoing an acid-catalyzed conformational change during the transport of nascent virions through the secretory pathway to the cell surface, our data suggest that the improper processing of unglycosylated prMp20 by an unknown cellular protease(s), as observed for the N15A/T17A mutant in C6/36 cells, is able to reduce virus growth in a prM glycosylation-independent manner.
The N glycosylation of JEV prM is crucial for viral pathogenicity in a murine infection model system.
To compare the lethalities of the WT and four prM mutant viruses, we first performed a pilot experiment by i.p. inoculating groups of 3-week-old ICR mice (n = 20 per group) with 5 × 102 PFU of each virus. The results of this pilot study indicated that the SM mutant produced a mortality rate similar to that produced by the WT virus, whereas all three prM N-glycosylation-defective mutants (N15A, T17A, and N15A/T17A) gave mortality rates lower than that produced by the WT virus (data not shown), suggesting that the N glycosylation of JEV prM might influence viral pathogenicity in mice. For a more quantitative comparison, we then determined the LD50s for i.p. inoculation of the viruses into 3-week-old ICR mice (Table 3). As expected, the WT virus was highly virulent, yielding an LD50 of 1.4 PFU; the control mutant SM virus retained the virulent phenotype of the WT virus, with an LD50 of 1.1 PFU. However, the LD50s of the three prM N-glycosylation-defective mutants were significantly higher than that of the WT virus: N15A, 29.4 PFU; T17A, 50.0 PFU; and N15A/T17A, 497.6 PFU. Thus, N15A/T17A exhibited an LD50 that was ≈10- to 15-fold higher than that of either N15A or T17A. Although no significant differences in clinical symptoms were noted among the mice inoculated i.p. with 5 × 102 PFU of the WT or any of the four prM mutant viruses, the three prM N-glycosylation-defective mutants corresponded to mean survival times (shown with standard deviations) longer than those associated with the WT or control mutant SM virus: WT-infected mice, 6.70 ± 0.37 days; N15A-infected mice, 10.55 ± 0.40 days; T17A-infected mice, 13.45 ± 0.70 days; N15A/T17A-infected mice, 12.00 ± 0.41 days; and SM-infected mice, 8.75 ± 1.73 days (Table 3). The mean survival times of WT- and SM mutant-infected mice were not significantly different in pairwise comparisons; however, highly significant differences in survival times between mice infected with the WT and those infected with the N15A, T17A, and N15A/T17A mutants were observed (in all cases, P < 0.0001).
TABLE 3.
Pathogenicity of JEV WT and prM mutants in 3-week-old ICR mice
| Virus | i.p. inoculation
|
i.c. inoculation
|
||||
|---|---|---|---|---|---|---|
| LD50 (PFU) | Mean survival time (days ± SD) | P valuea | LD50 (PFU) | Mean survival time (days ± SD) | P valuea | |
| WT | 1.4 | 6.70 ± 0.37 | <1.5 | 5.35 ± 0.24 | ||
| N15A | 29.4 | 10.55 ± 0.40 | <0.0001 | <1.5 | 6.55 ± 0.17 | 0.0003 |
| T17A | 50.0 | 13.45 ± 0.70 | <0.0001 | <1.5 | 6.75 ± 0.19 | <0.0001 |
| N15A/T17A | 497.6 | 12.00 ± 0.41 | <0.0001 | <1.5 | 6.25 ± 0.40 | 0.0342 |
| SM | 1.1 | 8.75 ± 1.73 | 0.2338 | <1.5 | 5.00 ± 0.22 | 0.2465 |
P values refer to pairwise comparisons to the value for the WT.
Having demonstrated an impairment in the pathogenicity of the prM N-glycosylation-defective mutants following i.p. inoculation, we proceeded to analyze the lethality of the viruses when administered to 3-week-old ICR mice by i.c. inoculation. Both the WT and the four prM mutant viruses were highly lethal in mice when administered by the i.c. route, as evidenced by the fact that the mortality rates at doses of 1.5 PFU and higher were invariably 100%, yielding LD50s below 1.5 PFU (Table 3). However, the mean survival times for mice inoculated with each of the three prM N-glycosylation-defective mutants at a dose of 1.5 PFU were significantly longer than those for mice inoculated with the WT virus (Table 3): WT-infected mice, 5.35 ± 0.24 days; N15A-infected mice, 6.55 ± 0.17 days (P = 0.0003 versus the result for WT-infected mice); T17A-infected mice, 6.75 ± 0.19 days (P < 0.0001); and N15A/T17A-infected mice, 6.25 ± 0.40 days (P = 0.0342). As expected, no significant difference in the mean survival time between mice infected with the WT and those infected with the control mutant SM virus (5.00 ± 0.22 days) was seen (P = 0.2465). Thus, all three prM N-glycosylation-defective mutant viruses displayed strong lethality that was nearly indistinguishable from that of WT virus, but the mean survival times were longer for mice receiving these mutant viruses than for mice receiving the WT by an i.c. route at the low dose of 1.5 PFU.
Overall, these results indicated that the inhibition of JEV prM N glycosylation resulted in neuroattenuation in mice, with a more profound effect being seen after inoculation via a peripheral route than after inoculation via a direct route. The difference in the virulence that we observed was found to be due primarily to the lack of prM N glycosylation, as confirmed by the results obtained for the N15Q mutant, which displayed a phenotype similar to that of the N15A mutant (LD50, 27.5 PFU, and mean survival time of infected mice, 14.20 ± 0.88 days after i.p. inoculation; LD50, <1.5 PFU, and mean survival time of infected mice, 6.70 ± 0.23 days after i.c. inoculation [data not shown]). Of particular note was the fact that the neuroattenuation observed in the case of N15A/T17A was more severe than that seen for either N15A or T17A after inoculation via the peripheral route. Paralleling the defect in virus growth that was observed in the case of N15A/T17A in mosquito cells (C6/36) but not in the two mammalian cell types (SH-SY5Y and BHK-21), this result also suggests that the N15 residue, along with T17, plays an additional role in JEV biology, affecting viral pathogenicity in mice.
DISCUSSION
In the present study, we have examined the role of the highly conserved N-glycosylation site in JEV prM, N15-X16-T17, in virus replication and pathogenesis. We constructed four prM N-glycosylation mutants by replacing either or both of the N15 and T17 residues with alanine or by introducing silent point mutations corresponding to all three residues and then analyzed the four prM mutants in three different cell types that are relevant to JEV biology and in a murine infection model system. Our results indicated that (i) the N15 and T17 residues were equally essential for the addition of N-linked glycans to JEV prM; (ii) the loss of JEV prM N glycosylation had a significant effect on the release of infectious virions; (iii) unexpectedly, the replacement of both the N15 and T17 residues simultaneously, but of neither individual residue alone, caused abnormal prM protein biogenesis that was cell type dependent and glycosylation status independent, with a subtle difference in prM biogenesis having a profound impact on the efficiency of virus growth; and (iv) the elimination of JEV prM N glycosylation resulted in a marked reduction in pathogenicity compared to that of the WT when the virus was administered to mice via a peripheral route. These findings indicate that the highly conserved N-glycosylation motif of JEV prM and its N-glycan substituents play a range of critical roles in viral replication by facilitating prM biogenesis and the release of infectious virions in infected cells, as well as contributing to viral pathogenesis in infected mice.
We found that although the intracellular levels of accumulated viral RNA and structural proteins remained unchanged, the elimination of JEV prM N glycosylation resulted in an ≈20-fold reduction in the production of extracellular virus particles, primarily as the result of the inefficient release of intracellular virions that had been preformed in intracellular membrane compartments in infected cells. This release defect was evidenced by the ≈2-fold increase in the relative abundance of intracellular virions at a given time point after infection in a given thin section of cells infected with one of three prM N-glycosylation-defective mutants; the magnitude of this increase would be even greater if the three-dimensional volume of the infected cells were taken into consideration, together with the cumulative incubation time after infection. Our findings are consistent with those described in recent reports of a defect in the secretion of virus-like particles for TBEV (16) and WNV (19) when glycosylation-deficient prM is ectopically expressed along with WT E in the absence of other viral proteins. However, the magnitudes of the reductions that were observed in these previous studies varied significantly, ranging from ≈2-fold for TBEV to ≈10- to 30-fold for WNV, depending on the lineage of the virus strain being used. Also, the removal of the N-glycosylation site in WNV prM led to an ≈10 to 40% decrease in the number of viral replicon particles produced by the transcomplementation of a subgenomic replicon, with all the structural proteins provided in trans (19). Our data, together with the data in these previous reports, indicate that the N glycosylation of the flavivirus prM protein plays a critical role in modulating the efficiency of virus release but has little effect on the protein compositions and infectivities of the released virus particles, at least in the three cell types analyzed in our study. The assembly and release of infectious virions or virus-like particles have also been shown previously to be affected by the N glycosylation of the other surface protein, E, in other flaviviruses (16, 19, 38, 39), indicating that this posttranslational modification of the two viral surface proteins is important for maximizing the efficiency of virus assembly and release in flavivirus-infected cells.
The highly conserved N-glycosylation site in JEV prM is in the pr region, which is cleaved from the protein as it moves through the ER and the Golgi apparatus to the plasma membrane (23, 61). Given that the removal of the pr region is required for the generation of highly infectious virus (23, 45), the primary roles of such oligosaccharide modifications are thought to be associated with a late, rather than early, stage of viral infection, facilitating protein folding, oligomerization, quality control, and transport in the cells. It should be noted that the first step in N-linked glycosylation involves the addition of mannose-rich carbohydrate to newly synthesized polypeptides in the ER, and these moieties are occasionally further modified as a result of catalytic activity by a series of glycosidases and carbohydrate transferases in the ER and Golgi apparatus (25). These glycosidases and transferases work in coordination with chaperones such as calreticulin, calnexin, and ERp57 to assemble a network, facilitating the correct folding and sorting of misfolded glycoproteins within the cell (20, 24). In the cases of other flaviviruses, prM is thought to play a chaperone-like role in facilitating the proper folding, maturation, and transport of E (2, 35, 39), thereby preventing the premature conformational changes that can be induced by the low-pH environment in the trans-Golgi network until the protein is released from the infected cells (1, 18, 21, 62). However, whether or how this chaperone-like role of prM is directly related to the defect in virus release is still unknown. Further investigation of this issue may provide new insights into the role of N glycosylation for viral and cellular glycoproteins.
The observation that in the three cell types (BHK-21, SH-SY5Y, and C6/36) used in our study, infectious virions derived from JEV prM N-glycosylation-defective mutants were nearly as infectious as the WT suggests that the primary role of the N glycans added to JEV prM is to facilitate protein folding and virus release; however, it is also possible that this modification is involved in an early stage of flavivirus infection, such as the attachment or binding of the virus particles to the cell surface. Recently, the presence of a single N-glycosylation site in the WNV prM or E protein has been shown to be involved in DC-SIGNR-mediated infection, potentially altering viral tropism (12). In this previous study, it was noted that WNV infection was more efficiently promoted by DC-SIGNR than by DC-SIGN when the virus was cultivated in human cell types. Similarly, DC-SIGN and DC-SIGNR have been implicated in the enhancement of DENV infection (49, 55, 63). The precise role of prM N glycosylation in the early stage of flavivirus infection promoted by DC-SIGN and/or DC-SIGNR, however, requires further investigation.
In the light of the finding that the removal of the N-glycosylation site from JEV prM had a significant effect on virus release, we examined the growth of the prM N-glycosylation mutants in three different cell types: not only the BHK-21 cells that are commonly used for virus propagation in laboratories but also two cell types that are biologically relevant to JEV transmission and pathogenesis. It is interesting that the viral growth defect in the mosquito C6/36 cells was more severe than that in the BHK-21 or SH-SY5Y cells. A similar differential growth defect has also been described previously for WNV prM N-glycosylation-defective mutants grown on C6/36 cells, as opposed to BHK-21 and Vero cells (19). We also noted that of the three JEV prM N-glycosylation-defective mutants, only N15A/T17A showed an additional defect in viral growth when grown on C6/36 cells but not on BHK-21 or SH-SY5Y cells. This cell type-specific, glycosylation status-independent defect was subsequently attributed to the abnormal biogenesis of the prM protein that produced an unexpected prMp18 form lacking the N terminus of the unglycosylated prMp20 protein. In this context, it is interesting that enhancing prM cleavability adversely affects virus production while having only a minimal effect on infectivity (30). These data suggest that the N-glycosylation motif of prM, in addition to the N glycan added to this protein, is involved in the late stage of viral morphogenesis, which includes the processing of the prM protein, an event that may be influenced by a cell type-specific factor(s). The importance of this glycosylation for virus production may explain the high level of conservation of the JEV prM N-glycosylation consensus sequence in all JEV strains and other closely related mosquito-borne flaviviruses.
While a large number of genetic determinants of viral pathogenesis in the gene encoding the E protein have been found previously, few genetic determinants for the prM protein have been identified (for a recent review, see references 8 and 27). The examination of the prM protein has been limited primarily to sequence comparisons of proteins from different viral strains (6, 68, 69) and analyses of the prM furin cleavage and prM-E signalase cleavage sites (26, 50, 51). In the present study, we have shown in a murine infection model system that infectious virions generated from all three of our prM N-glycosylation-defective mutants had low levels of pathogenicity when administered via a peripheral route. Therefore, it is conceivable that the N glycans attached to JEV prM may play a critical role in viral entry into the central nervous system (CNS) from the periphery by an unknown mechanism(s). Alternatively, it is also possible that the low levels of pathogenicity observed for all three of the prM N-glycosylation-defective mutants were largely a result of their failure to replicate to a level similar to that of the WT. Thus, the question of whether the JEV prM N glycosylation per se is a determinant of viral pathogenesis requires further investigation. Although the mechanisms by which flaviviruses enter into the CNS are as yet unclear, this process is thought to involve one or more of three possible scenarios, which may occur individually or in combination: (i) the migration of leukocytes capable of supporting productive virus infection through the blood-brain barrier into the CNS; (ii) the direct translocation of the viruses across the blood-brain barrier into the CNS via the endothelium, involving the productive infection of the endothelium and/or the internalization of the virions or increased vascular permeability occurring as a result of virus-induced endothelial cell damage; and/or (iii) the retrograde axonal transport of the virions via the peripheral nervous system (for a recent review, see references 33 and 57).
Although the pr domain bearing the carbohydrate moiety is largely absent from mature virions, we found that in the case of JEV, detectable amounts of glycosylated prM in the WT and unglycosylated prMp20 in the three prM N-glycosylation-defective mutants were associated with released virus particles. This incomplete cleavage of prM has also been described previously for other flaviviruses, including DENV (47, 52, 54, 71), WNV (19), Kunjin virus (31), and Langat virus (17, 28). The DENV prM/M protein has been shown previously to actively induce protective immunity; the passive administration of anti-prM antibodies protects mice against a lethal challenge (3, 14, 29, 67). Some evidence has suggested that monoclonal antibodies directed against prM provide protective immunity, perhaps because of their cross-reactivity with the E protein (14, 29) or because they are able to neutralize uncleaved prM protein present on the surfaces of the extracellular virions (29). In the case of Langat virus, evidence for protection by a nonneutralizing antibody recognizing the prM/M protein has been reported previously (28). The precise roles of the prM/M and/or prMp20/M proteins in the early stage of viral infection and in virus transmission into the CNS await further investigation.
In summary, we have demonstrated that the N glycosylation of JEV prM is required for productive virus propagation in cell culture. The process plays an important role in the release of infectious virions, probably by facilitating the correct folding of the prM protein and possibly the E protein and by promoting the transport of immature virions from the ER to the plasma membrane. We have further shown that the highly conserved N-glycosylation motif of JEV prM has a differential effect on virus growth that is cell type dependent and reflects the abnormal biogenesis of the prM protein. This elucidation of a critical role of prM N glycosylation and/or of the N-glycosylation consensus sequence in the release of infectious virions and in the pathogenesis of the virus may contribute to the design of vaccines against JEV and other flaviviruses.
Supplementary Material
Acknowledgments
This work was supported by a Korea Research Foundation grant (KRF-2003-015-E00104) funded by the Korean government (MOEHRD) and a Korea Science and Engineering Foundation grant (R01-2007-000-10346-0) funded by the Korean government (MOST).
We especially thank Yong-Dae Kim and Chul-Ho Lee for statistical analysis and Deborah McClellan for editorial assistance during manuscript preparation.
Footnotes
Published ahead of print on 4 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 695816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray, M., and C. J. Lai. 1991. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology 185505-508. [DOI] [PubMed] [Google Scholar]
- 4.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, PA.
- 5.Calisher, C. H., and E. A. Gould. 2003. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 591-19. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, M. S., and A. G. Pletnev. 2000. Infectious cDNA clones of Langat tick-borne flavivirus that differ from their parent in peripheral neurovirulence. Virology 269225-237. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44649-688. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, T. J., and M. S. Diamond. 2003. Pathogenesis of flavivirus encephalitis. Adv. Virus Res. 60273-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W. R., R. Rico-Hesse, and R. B. Tesh. 1992. A new genotype of Japanese encephalitis virus from Indonesia. Am. J. Trop. Med. Hyg. 4761-69. [DOI] [PubMed] [Google Scholar]
- 10.Chu, J. J., and M. L. Ng. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 7810543-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courageot, M. P., M. P. Frenkiel, C. D. Dos Santos, V. Deubel, and P. Despres. 2000. α-Glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, C. W., H. Y. Nguyen, S. L. Hanna, M. D. Sanchez, R. W. Doms, and T. C. Pierson. 2006. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 801290-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endy, T. P., and A. Nisalak. 2002. Japanese encephalitis virus: ecology and epidemiology. Curr. Top. Microbiol. Immunol. 26711-48. [DOI] [PubMed] [Google Scholar]
- 14.Falconar, A. K. 1999. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch. Virol. 1442313-2330. [DOI] [PubMed] [Google Scholar]
- 15.Gollins, S. W., and J. S. Porterfield. 1986. pH-dependent fusion between the flavivirus West Nile and liposomal model membranes. J. Gen. Virol. 67157-166. [DOI] [PubMed] [Google Scholar]
- 16.Goto, A., K. Yoshii, M. Obara, T. Ueki, T. Mizutani, H. Kariwa, and I. Takashima. 2005. Role of the N-linked glycans of the prM and E envelope proteins in tick-borne encephalitis virus particle secretion. Vaccine 233043-3052. [DOI] [PubMed] [Google Scholar]
- 17.Guirakhoo, F., F. X. Heinz, C. W. Mandl, H. Holzmann, and C. Kunz. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 721323-1329. [DOI] [PubMed] [Google Scholar]
- 18.Guirakhoo, F., R. A. Bolin, and J. T. Roehrig. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna, S. L., T. C. Pierson, M. D. Sanchez, A. A. Ahmed, M. M. Murtadha, and R. W. Doms. 2005. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J. Virol. 7913262-13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert, D. N., S. C. Garman, and M. Molinari. 2005. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 15364-370. [DOI] [PubMed] [Google Scholar]
- 21.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198109-117. [DOI] [PubMed] [Google Scholar]
- 22.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moorman, C. M. Rice, and H.-J. Thiel. 2000. Family Flaviviridae, p. 859-878. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, CA.
- 23.Heinz, F. X., and S. L. Allison. 2003. Flavivirus structure and membrane fusion. Adv. Virus Res. 5963-97. [DOI] [PubMed] [Google Scholar]
- 24.Helenius, A., E. S. Trombetta, D. N. Hebert, and J. F. Simons. 1997. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 7193-200. [DOI] [PubMed] [Google Scholar]
- 25.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 2912364-2369. [DOI] [PubMed] [Google Scholar]
- 26.Holbrook, M. R., H. Ni, R. E. Shope, and A. D. Barrett. 2001. Amino acid substitution(s) in the stem-anchor region of Langat virus envelope protein attenuates mouse neurovirulence. Virology 28654-61. [DOI] [PubMed] [Google Scholar]
- 27.Hurrelbrink, R. J., and P. C. McMinn. 2003. Molecular determinants of virulence: the structural and functional basis for flavivirus attenuation. Adv. Virus Res. 601-42. [DOI] [PubMed] [Google Scholar]
- 28.Iacono-Connors, L. C., J. F. Smith, T. G. Ksiazek, C. L. Kelley, and C. S. Schmaljohn. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43125-136. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman, B. M., P. L. Summers, D. R. Dubois, W. H. Cohen, M. K. Gentry, R. L. Timchak, D. S. Burke, and K. H. Eckels. 1989. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 41576-580. [DOI] [PubMed] [Google Scholar]
- 30.Keelapang, P., R. Sriburi, S. Supasa, N. Panyadee, A. Songjaeng, A. Jairungsri, C. Puttikhunt, W. Kasinrerk, P. Malasit, and N. Sittisombut. 2004. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J. Virol. 782367-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 725967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura, T., S. W. Gollins, and J. S. Porterfield. 1986. The effect of pH on the early interaction of West Nile virus with P388D1 cells. J. Gen. Virol. 672423-2433. [DOI] [PubMed] [Google Scholar]
- 33.King, N. J., D. R. Getts, M. T. Getts, S. Rana, B. Shrestha, and A. M. Kesson. 2007. Immunopathology of flavivirus infections. Immunol. Cell Biol. 8533-42. [DOI] [PubMed] [Google Scholar]
- 34.Klenk, H. D., and W. Garten. 1994. Activation cleavage of viral spike proteins by host proteases, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Konishi, E., and P. W. Mason. 1993. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 671672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuno, G., G. J. Chang, K. R. Tsuchiya, N. Karabatsos, and C. B. Cropp. 1998. Phylogeny of the genus Flavivirus. J. Virol. 7273-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, PA.
- 38.Lorenz, I. C., J. Kartenbeck, A. Mezzacasa, S. L. Allison, F. X. Heinz, and A. Helenius. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J. Virol. 774370-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenz, I. C., S. L. Allison, F. X. Heinz, and A. Helenius. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 765480-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 7510787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackenzie, J. S. 2005. Emerging zoonotic encephalitis viruses: lessons from Southeast Asia and Oceania. J. Neurovirol. 11434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackenzie, J. S., C. A. Johansen, S. A. Ritchie, A. F. van den Hurk, and R. A. Hall. 2002. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr. Top. Microbiol. Immunol. 26749-73. [DOI] [PubMed] [Google Scholar]
- 43.Markoff, L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302248. [DOI] [PubMed] [Google Scholar]
- 45.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 313-22. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, F. A. 1980. Togavirus morphology and morphogenesis, p. 241-316. In R. W. Schlesinger (ed.), The togaviruses: biology, structure, replication. Academic Press, New York, NY.
- 47.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74175-182. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro-Sanchez, E., R. Altmeyer, A. Amara, O. Schwartz, F. Fieschi, J. L. Virelizier, F. Arenzana-Seisdedos, and P. Despres. 2003. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pletnev, A. G., M. Bray, and C. J. Lai. 1993. Chimeric tick-borne encephalitis and dengue type 4 viruses: effects of mutations on neurovirulence in mice. J. Virol. 674956-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pryor, M. J., R. C. Gualano, B. Lin, A. D. Davidson, and P. J. Wright. 1998. Growth restriction of dengue virus type 2 by site-specific mutagenesis of virus-encoded glycoproteins. J. Gen. Virol. 792631-2639. [DOI] [PubMed] [Google Scholar]
- 52.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174450-458. [DOI] [PubMed] [Google Scholar]
- 53.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 54.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246317-328. [DOI] [PubMed] [Google Scholar]
- 55.Sakuntabhai, A., C. Turbpaiboon, I. Casademont, A. Chuansumrit, T. Lowhnoo, A. Kajaste-Rudnitski, S. M. Kalayanarooj, K. Tangnararatchakit, N. Tangthawornchaikul, S. Vasanawathana, W. Chaiyaratana, P. T. Yenchitsomanus, P. Suriyaphol, P. Avirutnan, K. Chokephaibulkit, F. Matsuda, S. Yoksan, Y. Jacob, G. M. Lathrop, P. Malasit, P. Despres, and C. Julier. 2005. A variant in the CD209 promoter is associated with severity of dengue disease. Nat. Genet. 37507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 57.Samuel, M. A., and M. S. Diamond. 2006. Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J. Virol. 809349-9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 59.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285194-204. [DOI] [PubMed] [Google Scholar]
- 60.Solomon, T. 1997. Viral encephalitis in Southeast Asia. Neurol. Infect. Epidemiol. 2191-199. [Google Scholar]
- 61.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 718475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 708142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tassaneetrithep, B., T. H. Burgess, A. Granelli-Piperno, C. Trumpfheller, J. Finke, W. Sun, M. A. Eller, K. Pattanapanyasat, S. Sarasombath, D. L. Birx, R. M. Steinman, S. Schlesinger, and M. A. Marovich. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai, T. F. 2000. New initiatives for the control of Japanese encephalitis by vaccination: minutes of a W.H.O. CVI meeting, Bangkok, Thailand, 13-15 October 1998. Vaccine 181-25. [DOI] [PubMed] [Google Scholar]
- 65.Van Den Hurk, A. F., B. L. Montgomery, J. A. Northill, I. L. Smith, P. Zborowski, S. A. Ritchie, J. S. Mackenzie, and G. A. Smith. 2006. Short report: the first isolation of Japanese encephalitis virus from mosquitoes collected from mainland Australia. Am. J. Trop. Med. Hyg. 7521-25. [PubMed] [Google Scholar]
- 66.Vaughn, D. W., and C. H. Hoke, Jr. 1992. The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol. Rev. 14197-221. [DOI] [PubMed] [Google Scholar]
- 67.Vázquez, S., M. G. Guzmán, G. Guillen, G. Chinea, A. B. Pérez, M. Pupo, R. Rodriguez, O. Reyes, H. E. Garay, I. Delgado, G. García, and M. Alvarez. 2002. Immune response to synthetic peptides of dengue prM protein. Vaccine 201823-1830. [DOI] [PubMed] [Google Scholar]
- 68.Wallner, G., C. W. Mandl, M. Ecker, H. Holzmann, K. Stiasny, C. Kunz, and F. X. Heinz. 1996. Characterization and complete genome sequences of high- and low-virulence variants of tick-borne encephalitis virus. J. Gen. Virol. 771035-1042. [DOI] [PubMed] [Google Scholar]
- 69.Wang, E., K. D. Ryman, A. D. Jennings, D. J. Wood, F. Taffs, P. D. Minor, P. G. Sanders, and A. D. Barrett. 1995. Comparison of the genomes of the wild-type French viscerotropic strain of yellow fever virus with its vaccine derivative French neurotropic vaccine. J. Gen. Virol. 762749-2755. [DOI] [PubMed] [Google Scholar]
- 70.Wang, J. J., C. L. Liao, Y. W. Chiou, C. T. Chiou, Y. L. Huang, and L. K. Chen. 1997. Ultrastructure and localization of E proteins in cultured neuron cells infected with Japanese encephalitis virus. Virology 23830-39. [DOI] [PubMed] [Google Scholar]
- 71.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 732547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 632521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitfield, S. G., F. A. Murphy, and W. D. Sudia. 1973. St. Louis encephalitis virus: an ultrastructural study of infection in a mosquito vector. Virology 5670-87. [DOI] [PubMed] [Google Scholar]
- 74.Winer, J., C. K. S. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 27041-49. [DOI] [PubMed] [Google Scholar]
- 75.Wuryadi, S., and T. Suroso. 1989. Japanese encephalitis in Indonesia. Southeast Asian J. Trop. Med. Public Health 20575-580. [PubMed] [Google Scholar]
- 76.Yun, S.-I., S.-Y. Kim, C. M. Rice, and Y.-M. Lee. 2003. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 776450-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yun, S.-I., and Y.-M. Lee. 2006. Japanese encephalitis virus: molecular biology and vaccine development, p. 225-271. In M. Kalitzky and P. Borowski (ed.), Molecular biology of the flavivirus. Horizon Scientific Press, Norwich, United Kingdom.
- 78.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 222604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.