Abstract
Here, we investigated the pre-steady-state deoxynucleoside triphosphate (dNTP) incorporation kinetics of primate foamy virus (PFV) reverse transcriptase (RT) in comparison with those of HIV-1 and MuLV RTs. PFV RT displayed a drastic reduction in primer extension at low dNTP concentrations where HIV-1 RT remains highly active, indicating a low dNTP binding affinity in the case of PFV RT. Indeed, kinetic analysis showed that, as observed with MuLV RT, PFV RT exhibits ∼10 to 80 times lower dNTP binding affinity than HIV-1 RT. These three RTs, however, show similar catalytic activities. In conclusion, PFV RT displays mechanistic distinctions in comparison to HIV-1 RT and shares close similarity to MuLV RT.
Unlike other retroviruses, foamy viruses (FVs), which have been isolated from various vertebrate animals, are considered nonpathogenic (8, 9, 11). FV naturally targets dividing cells, and FV vector systems have also been shown to efficiently transduce only cells with a mitotic index (13). In contrast, lentiviruses, including human immunodeficiency virus type 1 (HIV-1), uniquely replicate in nondividing cells, such as macrophages and microglia, which are not observed in other groups of retroviruses, including FVs and oncoretroviruses.
We recently demonstrated that the unique high affinity of binding of HIV-1 reverse transcriptase (RT) to deoxynucleoside triphosphate (dNTP) substrate contributes to viral infectivity in macrophages that contain very low dNTP pools (∼20 to 50 nM), compared to levels in the other natural target cell type, activated CD4+ T cells containing ∼2 to 5 μM dNTP (2). Our follow-up, pre-steady-state kinetic study revealed an unexpected kinetic difference between the RTs of HIV-1 and murine leukemia virus (MuLV). Indeed, HIV-1 RT has ∼7 to 123 times higher affinity of binding to dNTPs than MuLV RT (12). This and several other studies on reduced-dNTP-binding mutants of HIV-1 RT mimicking MuLV RT (2, 4, 12) suggested that the lower dNTP binding affinity of MuLV RT is still sufficient to support the replication of MuLV. This is because MuLV replicates only in dividing cells containing high cellular dNTP concentrations. An important implication of these studies is that reduction of the dNTP binding affinity of HIV-1 RT can limit viral infectivity to only those cell types containing high dNTP concentrations (2, 3). In this study, we investigated the dNTP incorporation mechanism of primate FV (PFV) RT, using pre-steady-state kinetic analysis, in comparison to those of HIV-1 and MuLV RTs. We hope that this biochemical analysis with PFV RT provides additional supporting evidence that the dNTP binding profile of RT is mechanistically linked with the cell tropism of retroviruses.
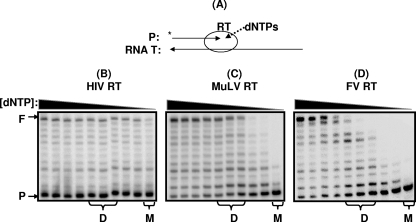
It was previously demonstrated that RTs with lower dNTP binding affinity, such as MuLV RT, exhibit reduced polymerase/primer extension activity at low dNTP concentrations. In environments with low dNTP concentrations, the RTs with higher dNTP binding affinity, such as HIV-1 RT, still remain active. However, in the case of MuLV RT, dNTP binding becomes a rate-limiting step at these low dNTP concentrations, leading to a decrease in polymerase activity (12, 14, 15). Therefore, we first compared the dNTP concentration-dependent DNA polymerase activity of PFV RT with those of MuLV and HIV-1 RTs. We performed primer extension assays (Fig. 1A), using a 5′-end, 32P-labeled, 23-mer T primer annealed to a 38-mer RNA template as previously described (12). Primer extension by the RT proteins was assayed in the presence of all four dNTPs at 10 different concentrations, ranging from 250 μM to 0.05 μM. First, we determined the amount of protein that yielded full extension of approximately 25 to 75% of the primer (Fig. 1, row F) with 250 μM dNTPs (for each dNTP) at 37°C for 5 min. Under these reaction conditions, the amount of primer extension is linear to the quantity of RT used. We intentionally employed higher activities for MuLV and PFV RTs showing 60 and 75% primer extension, respectively, for the reason described below. The primer extension reactions were repeated at decreasing dNTP concentrations, 125 to 0.05 μM, with the same amount of RNA-dependent DNA polymerization activity of RT proteins used in the 250 μM dNTP reactions. As shown in Fig. 1B, HIV-1 RT efficiently synthesizes DNA even at the low dNTP concentrations found in macrophages (i.e., 0.05 μM) (Fig. 1, columns M), whereas even with higher RT activity showing ∼60% primer extension, MuLV RT (Fig. 1C) showed decreased polymerization activity in reaction mixtures containing low dNTP concentrations. As shown in Fig. 1D, with a high RT activity showing 75% primer extension, PFV RT also displayed drastically reduced RNA=dependent DNA polymerization activity at low dNTP concentrations (i.e., 0.05 μM), as observed with MuLV RT. All three RTs, however, displayed efficient primer extension at the high dNTP concentrations found in many types of dividing cells, including primary and established cell lines (Fig. 1, columns D) (∼1 to 10 μM). Our previous studies demonstrated that other lentiviral RTs, such as simian immunodeficiency virus and feline immunodeficiency virus RTs, display similar high dNTP incorporation efficiencies at low dNTP concentrations, whereas two other oncoretroviral RTs, avian myeloblastosis virus and feline leukemia virus RTs, failed to show efficient DNA synthesis at low dNTP concentrations (10). Therefore, the data presented in Fig. 1 support that PFV RT harbors a dNTP utilization profile very different from that of lentiviral RTs but similar to that of oncoretroviral RTs.
FIG. 1.
dNTP concentration-dependent reverse transcription activity of RT proteins. (A) Schematic illustration of primer extension reaction by RT. A 5′-end, 32P-labeled, 23-mer T primer (P, 5′-CCGAATTCCCGCTAGCAATATTC-3′) annealed the 38-mer RNA template (T, 5′-GCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG-3′; template/primer ratio, 2.5:1) was extended by RTs of HIV-1 (B), MuLV (C), and PFV (D), showing approximately 25, 60, and 75% of primer extension (F), respectively, with 250 μM dNTPs (first lane) at 37°C for 5 min as previously described (12), and the reactions were repeated with decreasing dNTP concentrations (125, 50, 25,10, 5, 1, 0.2, 0.1, and 0.05 μM). The dNTP concentrations found in dividing cells (D) (1∼5 μM) and macrophages (M) (0.05 μM) are marked at the bottom of the figure. All three RT proteins used here were fused to the N-terminal His tag and purified from a bacterial overexpression system as described previously (7). PFV RT was purified from pET28a, containing the PFV PR-RT gene provided by Stephen Hughes (1). F, 38-nucleotide-long, fully extended product; P, 23-mer, unextended primer.
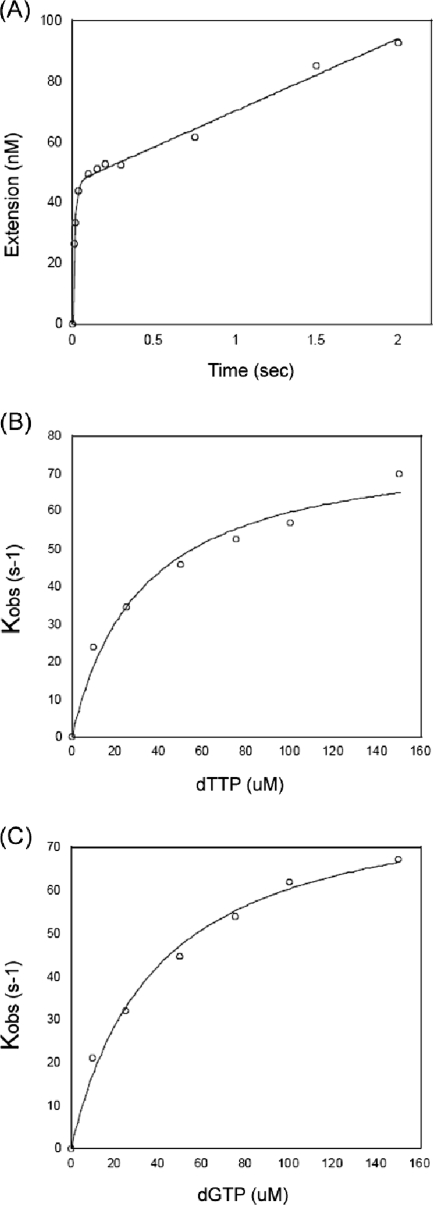
Next, we performed pre-steady-state kinetic analysis to determine Kd and kpol, which represent the dNTP binding affinity and conformational change/catalysis rate constants, respectively. This kinetic analysis requires a single round of nucleotide incorporation, which is initiated by mixing RT protein prebound to T/P with dNTPs (5, 6). Molar excess of the RT protein bound to T/P, which is defined as the active site of RT, prevents multiple rounds of primer extension. For this, we first determined the active site concentrations of PFV RT on a 32P-labeled, 23-mer T primer annealed to a 38-mer RNA template (T-T/P). The active-site concentration of RT is equal to the concentration of RT bound to T/P and capable of the first round of pre-steady-state dNTP incorporation, followed by a second round of steady-state dNTP incorporation. Under this condition, the amplitude of the burst in equation 1 (Fig. 2 legend) becomes the active-site RT concentration. We measured the product formation observed when the saturating 800 μM dTTP was mixed with RT (100 nM PFV RT protein concentration) prebound onto T/P (300 nM: excess T/P). By fitting these results to equation 1, we observed that ∼50% of the PFV RT protein (or 50 nM) was active on T/P. In our previous studies (12, 15), HIV-1 and MuLV RT proteins, purified using identical protocols, showed approximately 30 and 50% active concentrations, respectively. Additional data obtained from these burst experiments include measures for the rates of DNA polymerization during the pre-steady (kobs) and steady (kss) states. The PFV RT pre-steady-state rate of dTTP incorporation (kobs) was 71.5 s−1, and the rate during the steady state was 0.51 s−1. This implies that, similar to HIV-1 RT and other DNA polymerases, PFV RT incorporates dNTPs at a higher rate during the pre-steady state than during the steady state, and the steady-state reaction of PFV RT contains a major rate-limiting step(s) of the overall DNA polymerization reaction, such as product release.
FIG. 2.
Active-site determination and dNTP titration of PFV RT. (A) Pre-steady- and steady-state kinetics of PFV RT incorporating dTTP onto the 32P-labeled, 23-mer T primer annealed to the 38-mer template used in Fig. 1 were analyzed as previously described (12). Reactions were carried out at the indicated times by mixing together a solution of RT (100 nM protein concentration) prebound to T/P (300 nM) and a second solution with 800 μM dTTP under rapid quench conditions. The data were fit into the burst equation (equation 1), product concentration = A[1 − exp(−kobst) + ksst] (5, 6), which provides a measure of the active concentration of RT (Amp), the observed rate constant for the burst phase (kobs), and the rate constant for the linear phase (kss) for PFV RT. The pre-steady-state rates of dTTP incorporation onto T/P (kobs) for PFV RT were 71.5 ± 14 s−1, and their rates during the steady state were 0.51 s−1. (B) Pre-steady-state T and G titration by PFV RT. The 32P labeled, 23-mer dTTP and dGTP primers (12) annealed to the 38-mer template (50 nM) were extended with excess RT (200 nM active-site concentration) for single round of dTTP and dGTP incorporation at 10 μM, 25 μM, 50 μM, 75 μM, 100 μM, and 150 μM concentrations. These data were used for the determination of Kd and kpol values of PFV RT, using equation 2, kobsd = kpol(dNTP concentration)/(Kd + dNTP concentration) (5, 6), as summarized in Table 1.
Next, we performed single turnover experiments (100 nM active RT and 50 nM T/Ps) to obtain an actual measure for the dNTP incorporation rates at different concentrations of the individual dNTPs during the pre-steady state. By measuring the dependence-of-reaction rates (kobsd) at different dNTP concentrations (Fig. 2B and 2C for dTTP and dGTP, respectively), we were able to determine the kinetic parameters of Kd, which represents the affinity of binding of RT to the incoming dNTP substrate, and kpol, the maximum rate of dNTP incorporation. As summarized in Table 1, the affinities of binding (Kd) of PFV RT to dNTPs are 26 to 39 μM and the incorporation rates (kpol) are 75 to 83 s−1.
TABLE 1.
Pre-steady-state kinetic parameters of PFV, HIV-1, and MuLV RT proteinsa
| RT protein | dNTP | Kd (μM) | kpol (s−1) | kpol/Kd |
|---|---|---|---|---|
| HIV-1 | dATP | 0.610 ± 0.5 | 33.7 ± 0.6 | 55.3 |
| dCTP | 0.33 ± 0.4 | 37.5 ± 7.5 | 112.5 | |
| dGTP | 3.9 ± 0.04 | 39.4 ± 5.6 | 10.2 | |
| dTTP | 2.5 ± 1.4 | 36.6 ± 7.5 | 14.6 | |
| PFV | dATP | 26.3 ± 1.5 (43) | 77.0 ± 2.3 (2) | 2.9 |
| dCTP | 27.1 ± 10.6 (82) | 75.1 ± 3.6 (2) | 2.7 | |
| dGTP | 38.7 ± 7.5 (10) | 83.7 ± 9.1 (2) | 2.1 | |
| dTTP | 31.8 ± 0.4 (13) | 78.6 ± 1.3 (0.5) | 2.4 | |
| MuLV | dATP | 74.2 ± 1.2 (123) | 159.2 ± 4.7 (4.7) | 2.1 |
| dCTP | 18.1 ± 9.4 (55) | 47.2 ± 8.1(1.3) | 2.6 | |
| dGTP | 25.2 ± 8.3 (7) | 72.2 ± 3.8 (1.8) | 2.9 | |
| dTTP | 115.9 ± 9.3 (46) | 159.3 ± 9.3 (4.4) | 1.4 |
Next, we compared the Kd and kpol values of PFV RTs with those of MuLV and HIV-1 RTs, which have previously been determined under identical conditions (12). Interestingly, while the kpol values of all three RTs are similar, PFV RT is 10- to 80-fold less efficient at binding the incoming dNTP than HIV-1 RT but shows a dNTP affinity similar to that of MuLV RT (Table 1). These data suggest that PFV RT has mechanistic properties very different from those of HIV-1 RT but close similarity to MuLV RT in terms of dNTP incorporation reaction. This kinetic finding supports that the restricted DNA synthesis of PFV RT at low dNTP concentrations, which was observed in Fig. 1, results from the low dNTP binding affinity of PFV RT, making dNTP binding a rate-limiting step at low dNTP concentrations.
Our previous pre-steady-state kinetic analysis showed that the HIV-1 RT mutants with reduced dNTP binding affinity (Q151N and V148I) kinetically mimic MuLV RT (2, 3, 14). Interestingly, HIV-1 vectors containing these dNTP binding mutant RTs failed to transduce macrophages, though they retained the ability to transduce cells with elevated dNTP concentrations (2, 3). This observation supports the idea that HIV-1 might have evolved to harbor an RT with high dNTP binding affinity in order to efficiently replicate in macrophages. In contrast, since MuLV replicates exclusively in dividing cells containing high dNTP concentrations, this virus likely did not need to evolve to harbor a polymerase with high dNTP binding affinity. PFV also replicates only cells with a mitotic index, and therefore, the low dNTP binding affinity of PFV RT is still sufficient to support proviral DNA synthesis efficiently in environments with high cellular dNTP concentrations. In conclusion, the data presented in this report further support that RT is one of the mechanistic elements that can contribute to the target cell type specificity of retroviruses.
Acknowledgments
We thank Stephen Hughes (National Cancer Institute) for kindly sharing a construct overexpressing PFV RT and Pauline Chugh, Mark Skasko, Z. Kelley, and Edward Kennedy for critical reading of the manuscript.
This work was supported by National Institutes of Health (NIH) grant AI049781 to B.K. and NIH Education grant R25GM64133 to J.S.-V.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Boyer, P. L., C. R. Stenbak, P. K. Clark, M. L. Linial, and H. S. Hughes. 2004. Characterization of the polymerase and RNase H activities of human foamy virus reverse transcriptase. J. Virol. 786112-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond, T. L., M. Roshal, V. K. Jamburuthugoda, H. M. Reynolds, A. R. Merriam, K. Y. Lee, M. Balakrishnan, R. A. Bambara, V. Planelles, S. Dewhurst, and B. Kim. 2004. Macrophage tropism of HIV-1 depends upon efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 27951545-51553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamburuthugoda, V. K., P. Chugh, and B. Kim. 2006. Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J. Biol. Chem. 28113388-13395. [DOI] [PubMed] [Google Scholar]
- 4.Jamburuthugoda, V. K., D. Guo, J. E. Wedekind, and B. Kim. 2005. Kinetic evidence for interaction of human immunodeficiency virus type 1 reverse transcriptase with the 3′-OH of the incoming dTTP substrate. Biochemistry 4410635-10643. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, K. A. 1993. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 62685-713. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, K. A. 1995. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 24938-61. [DOI] [PubMed] [Google Scholar]
- 7.Kim, B. 1997. Genetic selection in Escherichia coli for active human immunodeficiency virus reverse transcriptase mutants. Methods 12318-324. [DOI] [PubMed] [Google Scholar]
- 8.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 731747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Löchelt, M., S. F. Yu, M. L. Linial, and R. M. Flügel. 1995. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology 206601-610. [DOI] [PubMed] [Google Scholar]
- 10.Operario, D. J., H. M. Reynolds, and B. Kim. 2005. Comparison of DNA polymerase activities between recombinant feline immunodeficiency and leukemia virus reverse transcriptases. Virology 335106-121. [DOI] [PubMed] [Google Scholar]
- 11.Saïb, A., and H. de Thé. 1996. Molecular biology of the human foamy virus. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13S254-260. [DOI] [PubMed] [Google Scholar]
- 12.Skasko, M., K. K. Weiss, H. M. Reynolds, V. Jamburuthugoda, K. Lee, and B. Kim. 2005. Mechanistic differences in RNA-dependent DNA polymerization and fidelity between murine leukemia virus and HIV-1 reverse transcriptases. J. Biol. Chem. 28012190-12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trobridge, G., and D. W. Russell. 2004. Cell cycle requirements for transduction by foamy virus vectors compared to those of oncovirus and lentivirus vectors. J. Virol. 782327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss, K. K., R. A. Bambara, and B. Kim. 2002. Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). Pre-steady state kinetic study of the Q151N HIV-1 RT mutant with increased fidelity. J. Biol. Chem. 27722662-22669. [DOI] [PubMed] [Google Scholar]
- 15.Weiss, K. K., R. Chen, M. Skasko, H. M. Reynolds, K. Lee, R. A. Bambara, L. M. Mansky, and B. Kim. 2004. A Role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry 434490-4500. [DOI] [PubMed] [Google Scholar]