Sapovirus (SaV) belongs to the family Caliciviridae and is a causative agent of gastroenteritis in children and adults (6, 2). On the basis of complete capsid gene sequences, SaV can be divided into five genogroups, among which GI, GII, GIV, and GV infect humans, whereas SaV GIII infects porcine species (1). Porcine SaV was initially isolated from a nursing pig with diarrhea, and the isolate induced diarrhea, viremia, and seroconversion in experimentally infected gnotobiotic pigs (3). Porcine SaV is currently under consideration as a potential zoonotic agent, due to the identification of a porcine SaV isolate that is similar to human SaV in its genomic sequence and to the emergence of recombinant porcine SaVs (8). Although a few outbreaks of SaV in humans have been reported (6, 5, 7), a case of its outbreak has never been found in swine populations. To the best of our knowledge, few reports so far indicate that SaV strains infect humans or swine in China.
We report the first outbreak of gastroenteritis caused by porcine SaV in piglets in China. This outbreak occurred in February 2008 on a small commercial pig farm that lies in a Shanghai suburb. Seven stool specimens were collected from seven piglets which showed symptoms of diarrhea and vomiting in two neighboring farrows on the farm. These piglets from the two farrows were 21 and 25 days old, respectively. We also collected 12 stool specimens from the rest of the 12 piglets in the two farrows that showed no evident illness. In order to avoid sample contamination, specimens were obtained directly from the swine anus and disposable materials were used during sampling. We first examined the seven specimens from the ill piglets for porcine circovirus, porcine rotavirus, porcine transmissible gastroenteritis virus, and porcine epidemic diarrhea virus using reverse transcription-PCR (RT-PCR), and all specimens showed negative results (data not shown). At the same time, 20 and 60 stool specimens were collected from 20 piglets in four other farrows from the same farm and 60 piglets from three other pig farms nearby, respectively. All specimens were then examined for SaV by using RT-PCR as described previously (1).
Our results showed that all seven piglets that showed clinical symptoms were positive for SaV RNA, suggesting this outbreak of gastroenteritis may have been due to porcine SaV infection. Although the rest of the 12 piglets in the two farrows indicated no evident clinic symptoms (these piglets were observed in an isolation room throughout this outbreak), one of them tested positive for SaV RNA. This confirms that pigs could be subclinically infected with porcine SaV (9). None of the other 80 specimens tested positive for SaV RNA. Broader research should be carried out to elucidate the infection status of porcine SaV in general swine populations of China.
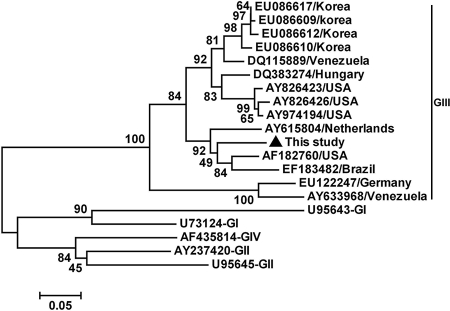
The eight expected DNA bands specific for SaV were excised from the gel, purified with the AxyPrep DNA gel extraction kit (Axygen, CA), cloned into pMD T vector (TaKaRa, Japan), and sequenced (Dalian Baoshengwu, China). Sequence analysis showed that the eight 331-nucleotide (nt) SaV sequences shared 100% nucleotide identity and likely represented the same SaV strain (Ch-sw-sav1; GenBank accession no. EU599212). Phylogenetic analysis based on this sequence and 19 other referenced representative strains from different regions (GenBank no. and source of regions are shown in Fig. 1) indicated that the strain we isolated in the present study belonged to GIII SaV and closely clustered with another three strains (Fig. 1), forming a subgroup in the GIII cluster. The identities between the sequence determined in the present study and the other GIII SaV isolates we referenced ranged from 76% to 91%. This Chinese SaV strain, Ch-sw-sav1, shared the highest nucleotide sequence homology (91%) with a porcine U.S. SaV strain (AF182760), which was isolated from a 27-day-old diarrheic nursing pig (4), suggesting they may come from a common source of infection.
FIG. 1.
Phylogenetic tree constructed by alignment of the 331-nt sequence of open reading frame 1 of the porcine SaV isolate in this study and the 19 references from swine and human SaV isolates, using the neighbor-joining method, and evaluated using the interior branch test method with Mega 4 software. Percent bootstrap support is indicated at each node. The scale bar represents nucleotide substitutions per base. The GenBank accession no. and country of origin are indicated. The isolates identified in this study are marked with a triangle.
In order to elucidate whether the fecal samples contained infectious SaV, the eight SaV-positive fecal specimens were converted to 20% (wt/vol) suspensions in phosphate-buffered saline (PBS) (0.01 M, pH 7.2 to 7.4) and clarified by centrifugation at 10,000 × g for 10 min. The supernatants were then subjected to purification by passage through microfilters with a pore size of 0.22 μm (Millex-GV; Millipore) to remove possible bacteria and parasites. A 1-ml aliquot of each suspension was used to infect eight 10-day-old piglets through oral inoculation. Another three piglets were inoculated with PBS as a control. The piglets were investigated every 2 hours during the first day, and fecal samples were collected from each piglet every day. Our results showed that all eight piglets in the experimental group indicated evident symptoms of diarrhea and vomiting after 14 to 20 h. These clinical symptoms persisted for 2 to 5 days, and no piglet died. The three piglets in the control group showed no clinical symptoms. SaV RNA was detected in the feces of all the eight piglets in the experimental group after 4 to 5 days of infection but not in the control pigs.
In conclusion, this is the first report that SaV infects piglets in China and leads to an outbreak of gastroenteritis.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 1491309-1323. [DOI] [PubMed] [Google Scholar]
- 2.Farkas, T., X. Deng, G. Ruiz-Palacios, A. Morrow, and X. Jiang. 2006. Development of an enzyme immunoassay for detection of sapovirus-specific antibodies and its application in a study of seroprevalence in children. J. Clin. Microbiol. 443674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo, M., J. Hayes, K. O. Cho, A. V. Parwani, L. M. Lucas, and L. J. Saif. 2001. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J. Virol. 759239-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, M., K. O. Chang, M. E. Hardy, Q. Zhang, A. V. Parwani, and L. J. Saif. 1999. Molecular characterization of a porcine enteric calicivirus genetically related to Sapporo-like human caliciviruses. J. Virol. 739625-9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansman, G. S., H. Saito, C. Shibata, S. Ishizuka, M. Oseto, T. Oka, and N. Takeda. 2007. Outbreak of gastroenteritis due to sapovirus. J. Clin. Microbiol. 451347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson, P. J., K. Bergentoft, P. A. Larsson, G. Magnusson, A. Widell, M. Thorhagen, and K. O. Hedlund. 2005. A nosocomial sapovirus-associated outbreak of gastroenteritis in adults. Scand. J. Infect. Dis. 37200-204. [DOI] [PubMed] [Google Scholar]
- 7.Phan, T. G., Q. D. Trinh, F. Yagyu, K. Sugita, S. Okitsu, W. E. Müller, and H. Ushijima. 2006. Outbreak of sapovirus infection among infants and children with acute gastroenteritis in Osaka City, Japan during 2004-2005. J. Med. Virol. 78839-846. [DOI] [PubMed] [Google Scholar]
- 8.Wang, Q. H., M. G. Han, J. A. Funk, G. Bowman, D. A. Janies, and L. J. Saif. 2005. Genetic diversity and recombination of porcine sapoviruses. J. Clin. Microbiol. 435963-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu, J. N., M. Y. Kim, D. G. Kim, S. E. Kim, J. B. Lee, S. Y. Park, C. S. Song, H. C. Shin, K. H. Seo, and I. S. Choi. 2008. Prevalence of hepatitis E virus and sapovirus in post-weaning pigs and identification of their genetic diversity. Arch. Virol. 153739-742. [DOI] [PubMed] [Google Scholar]