Abstract
Triggering uncontrolled cellular proliferation, chronic inflammation, and/or disruption of p53 activity is critical for tumorigenesis initiated by latent viral oncogenes. The adenovirus type 5 (Ad5) early genes E1A and E1B can maintain lifelong latency in the lungs of patients with chronic pulmonary diseases. To determine the in vivo effects of the latent Ad5 E1A and E1B oncogenes, we have examined the influence of Ad5 E1A and E1B gene products on mouse lung carcinogenesis and inflammation by generation and characterization of lung-specific transgenic mouse models. Here, we show that either the Ad5 E1A 243-amino-acid (aa) protein or the E1B 58-kDa protein was dominantly expressed in the transgenic lung. Preferential expression of Ad5 E1A 243-aa protein alone was not sufficient to induce lung carcinogenesis but resulted in low-grade cellular proliferation and high-grade lymphoproliferative inflammation in the lung. The presence of Ad5 E1B dramatically enhanced the expression of the E1A 243-aa protein, in addition to impairing p53 and apoptosis response, resulting in uncontrolled cellular proliferation, lymphoproliferative inflammation, and metastatic carcinomas in the lung after a period of latency. Our studies may provide clues to understanding the potential in vivo biological effects of Ad5 E1A and E1B latent in the lung and a new scope for assessing in vivo functions of viral genes latent in the infection target tissue.
Many human viruses such as hepatitis B virus, Epstein-Barr virus, human papillomavirus, Kaposi's sarcoma-associated herpesvirus, and human immunodeficiency virus play a role in oncogenesis by triggering uncontrolled cell proliferation and chronic inflammation and/or disrupting p53 activity in the tissues where the virus causes latent or chronic infection. Human adenovirus type 5 (Ad5) can latently infect human pulmonary tissues. Ad5 early region 1A and 1B (E1A and E1B) genomic DNA can be integrated into the human cell chromosome (12). Either the DNA or the gene products of Ad5 E1A can be frequently detected in the lungs of patients with chronic pulmonary diseases or lung carcinomas (1, 7, 11, 14). However, the role of latent action by the Ad5 E1A and E1B genes in lung carcinogenesis and lymphoproliferative inflammation is unclear.
Ad5 E1A and E1B DNA is located within the left-hand 11% of the adenovirus genome. The Ad5 E1A gene encodes two major proteins, the 243-amino-acid (aa) protein and the 289-aa protein, by alternative splicing; likewise, the E1B gene encodes a 58-kDa protein and other, smaller proteins in cell culture (13, 17, 18, 23-25). Ad5 E1A is thought to be responsible for the induction of cellular DNA synthesis, inactivation of tumor suppressor pRb, stimulation of cellular transformation, increase in the level of cellular tumor suppressor p53, and cellular apoptosis by means of a series of protein-protein interactions with cellular transcriptional factors, while Ad5 E1B is responsible for the inactivation of tumor suppressor p53 and the repression of cellular apoptosis, leading to promotion of cell transformation in cell culture (13, 17, 18, 20, 23-25). However, Ad5 E1A and E1B have not been reported to induce a tumorigenic phenotype in transformants resulting from infection of cells in cell culture (in vitro) or in animals (in vivo), except in the immunoincompetent animal. Nevertheless, we lack knowledge of the in vivo biological function of Ad5 E1A and E1B.
In this study, we mimicked Ad5 E1A and E1B latency in the lung by lung-specific expression of the transgene Ad5 E1A or E1A+E1B and then assessed the in vivo effects of Ad5 E1A and E1B. The Ad5 E1A 243-aa and E1B 58-kDa proteins were dominantly expressed in the mouse lung. Transgenic expression of Ad5 E1A 243-aa protein alone resulted in low-grade cellular mitosis and chronic lymphoproliferative inflammation but was not sufficient to induce lung carcinomas. Transgenic expression of Ad5 E1B and Ad5 E1A 243-aa proteins led to lung carcinogenesis, high-grade lymphoproliferative inflammation, and impaired p53 response. This study offers a clue to understanding the potential role of latent Ad5 E1A and E1B in promoting lung carcinogenesis and lymphoproliferative inflammation upon reactivation.
MATERIALS AND METHODS
Generation of transgenic mice.
SPCE1A or SPCE1A+E1B was constructed by cloning an entire Ad5 E1A or E1A+E1B genomic fragment into the 3.7SPC/simian virus 40 vector (24), respectively. A 5.6-kb transgene E1A or a 9.2-kb transgene E1A+E1B DNA fragment was microinjected into fertilized B6/SJL mouse eggs, respectively, to generate transgenic mice. Transgenic founder mice were identified by Southern blot analysis of mouse tail DNA with a probe of 32P-labeled Ad5 E1A genomic fragment. Founders were bred to C57BL/6 mice, and transgene-heterozygous mice were used for our studies. A minimum of six serial lung sections from at least three mice of each line were stained in E1A immunostaining, bromodeoxyuridine (BrdU) incorporation, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL), and leukocyte marker assays.
mRNA analysis.
Total RNA was isolated from mouse lung. Twenty micrograms of each total RNA was loaded on an agarose gel, transferred to a Hybond N+ membrane, and hybridized with an Ad5 E1A+E1B genomic DNA probe. The mouse endogenous 18S RNA was used as total RNA loading control. Further, the expression of E1A and E1B was determined by isolation of E1A and E1B mRNAs using an mRNA purification kit [Poly(A) Purist kit; Ambion, Austin, TX]. The mRNAs purified from 50 μg of total RNA and (as a control) 12.5 μg of total RNA were used and hybridized with Ad5 E1A and E1B genomic DNA probes. The level of E1A or E1B mRNA in each lane was quantified by densitometry (ImageQuant; Molecular Dynamics, Sunnyvale, CA).
RT-PCR analysis.
Total RNA was extracted as described above and treated with RNase-free DNase (Roche Applied Science, Indianapolis, IN). Three micrograms of the total RNA was used to generate first-strand cDNA with oligo(dT) and Superscript reverse transcriptase (Invitrogen, San Diego, CA). PCR amplification was performed for 35 cycles using a 1/10 volume of first-strand cDNA and Taq DNA polymerase (Invitrogen, San Diego, CA). Primers used to amplify Ad5 E1B 19-kDa cDNA were as follows: forward primer, 5′-GAGCTCTAACAGTACCTCTTGG-3′, and reverse primer, 5′-GGGTTTCTTCGCTCCATTTATCC-3′ (the size of the amplified product was 266 bp). The primers used to amplify Ad5 E1b 58-kDa cDNA were as follows: forward primer, 5′-GATGCTGACCTGCTCGGACG-3′, and reverse primer, 5′-CCAGCATCACAGGCTGGTTCC-3′ (the size of the amplified product was 337 bp). The primers used to amplify actin cDNA were as follows: forward primer, 5′-TACCACAGGCATTGTGATGG-3′, and reverse primer, 5′-ATCGTACTCCTGCTTGCTGA-3′ (the amplified fragment was 649 bp). Reverse transcription-PCR (RT-PCR) products were analyzed on a 1.5% agarose gel.
Immunoblot analysis.
Fresh mouse lung lysate was immunoblotted with anti-Ad2/Ad5 E1A antibody (Santa Cruz Biotechnology, Santa Cruz, CA; BD Pharmingen, San Diego, CA) or anti-p53 polyclonal antibodies; the loading control used the mouse housekeeping gene product Actin to normalize the expression of E1A as previously described (24).
Immunohistochemistry.
The serial frozen lung sections (5 μm) were stained with anti-Ad2/Ad5 E1A antibody (Santa Cruz; BD Pharmingen), and serial paraffin-embedded lung sections (4 μm) were stained with anti-mouse p53 antibody (CM5; Novocastra, Newcastle, United Kingdom) as previously described (24). The stained nuclei from more than 2,000 lung cells (at least three mice) were scored under a light microscope (Zeiss, Thornwood, NY) with a 40× objective.
BrdU incorporation.
Mice received intraperitoneal injections of 100 μg/g of body weight of BrdU (Sigma, St. Louis, MO) and 10 μg/g of 5-fluoro-2′-deoxyuridine (Sigma). After 2 h, lung tissues were fixed in 10% neutral-buffered formalin. Serial paraffin-embedded lung sections (4 μm) were detected as previously described (24). BrdU-positive cells from more than 2,000 lung cells (at least three mice) were scored under a light microscope (Zeiss) with a 40× objective.
TUNEL assay.
TUNEL-positive nuclei on the fresh frozen lung sections were analyzed with TdT enzyme (Roche Applied Science, Indianapolis, IN) and aminoethylcarbazole (Genzyme, Cambridge, MA), as suggested by the manufacturers. The apoptotic cells were quantified based on the TUNEL assay and morphological analysis. The apoptotic cells from more than 2,000 lung cells (at least three mice) were scored under a light microscope (Zeiss) with a 40× objective.
Lung histopathologic analyses.
All mice were housed in a barrier environment in the animal facility of the Hospital for Sick Children. Animal work was carried out in accordance with institutional guidelines. Nontransgenic mice were derived from the same litters as controls. The lung and other tissues were examined during autopsy. The representative lungs and other tissues with abnormal lung morphology were sectioned and stained with hematoxylin and eosin and further analyzed by microscopy.
Statistical analyses.
Statistical differences between the mouse lines were analyzed using Student's t test (SigmaStat; Systat Software Inc., San Jose, CA). Statistical significance for this study was set at two sides, P < 0.05.
RESULTS
Expression of Ad5 E1A and E1B leads to metastatic lung carcinogenesis.
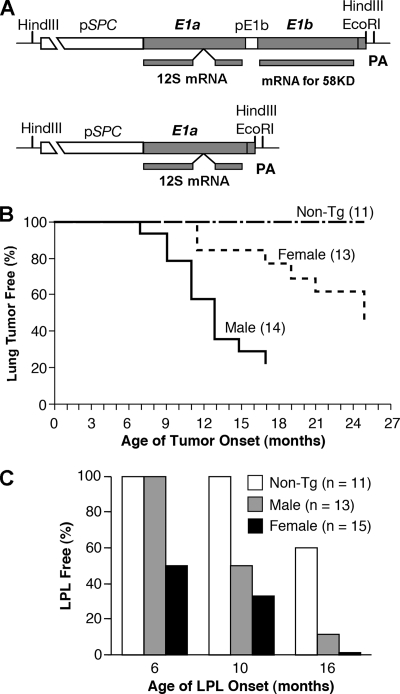
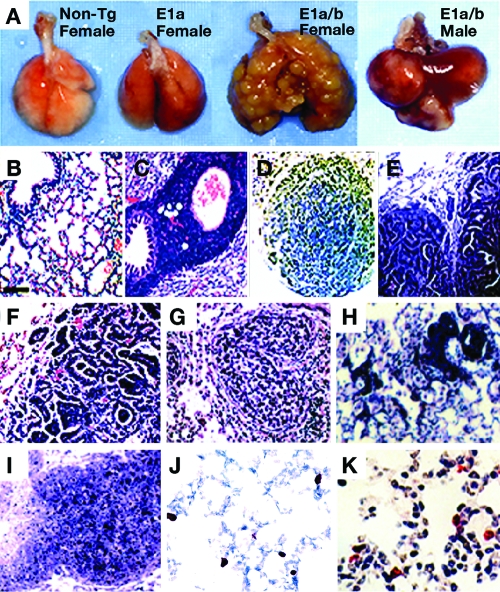
To best mimic the latency of the Ad5 E1A and E1B genes in the human respiratory system, we generated Ad5 E1A+E1B or E1A transgenic mice using Ad5 E1A and E1B genomic DNA controlled by a human pulmonary epithelial cell-specific promoter of surfactant protein C regulatory element (pSPC) (Fig. 1A). Expression of the transgene was detected by RT-PCR in the lungs of the transgenic mice with two to four copies of the transgene and one or two integration sites but not in the heart, liver, kidney, and spleen as previously reported (24). The E1A+E1B transgenic animal founders (AB2, AB5, and AB10) suffered from multiple lung adenocarcinomas or carcinomas (Fig. 1B and 2A). The lung carcinogenesis initiated as focal neoplasias or focal adenomas and then progressed to malignant adenomas or adenocarcinomas (Fig. 2B, D, and E). The aged mice died from malignant lung adenocarcinomas or carcinomas after a long latency. The malignant lung carcinomas were highly peripherally metastatic (Fig. 2F and G), along with the simultaneous appearance of adenomas or adenocarcinomas in the spleen and liver (unpublished data). The lung carcinogenesis differed in tumor morphology, latency, and incidence rate depending on the sex of the mice (Fig. 1B and 2). The lung adenocarcinomas started in the transgenic male mice before the age of 9 months, while the morphological diversity of lung carcinomas appeared in the transgenic female mice after the age of 12 months. In general, about half of the transgenic mice in all three Ad5 E1A+E1B transgenic lines were bearing lung tumors by the age of 25 months. For instance, the incidence rate of lung tumors in the male AB2 mice was 78.6% by 17 months and that in female AB2 mice was 53.8% by 25 months (P < 0.01). Moreover, the lymphoproliferative lesions (LPL) appeared in E1A+E1B transgenic mouse lungs after the age of 4 months (data not shown).
FIG. 1.
Transgenes induce lung carcinogenesis and lymphoproliferative lesions. (A) Schematic of transgenes E1A+E1B and E1A used for generation of the transgenic mice and predominantly spliced E1A 12S and E1B large mRNAs. pE1b, E1B promoter region; PA, simian virus 40 small t intron and polyadenylation sequence. (B) Kinetics of lung tumor formation in the E1A+E1B transgenic male and female mice versus nontransgenic mice (Non-Tg) (P = 0.002, log rank test). (C) Lung LPL onset in the E1A transgenic male and female mice versus nontransgenic mice (Non-Tg) (P = 0.002, log rank test). Age of onset is the time at which the mouse first appeared sick from visible lung tumors (B) and possessed a lung LPL (C). n, number of mice examined.
FIG. 2.
Histopathologic analyses of lung tumors and LPL. (A) Nontumorous lungs in aged nontransgenic mouse (non-Tg) and aged E1A transgenic mouse and tumorous lungs in 14-month-old female and 8-month-old male E1A+E1B transgenic mice. (B) Normal histopathology of non-Tg mouse lung. (C) LPL in aged E1A transgenic mouse lung; proliferating mononuclear lymphocytes aggregate and surround alveolar bronchioles and alveolar blood vessels. Tumors in E1A+E1B transgenic male lungs. (D) Separate adenoma with a solid nodular or papillary growth pattern. (E) Primary adenocarcinoma with a tubular papillary growth pattern. (F) Malignant carcinoma; note that highly metastatic, poorly differentiated tumor cells form separate cystic papillary-tubular and solid nodular islands. (G) Glandular carcinoma in E1A+E1B transgenic female lung; note that discrete glandular sheets display moderate differentiation and diffuse into the blood vessel or bronchial lumen. (H) Mass LPL in the terminal alveoli of E1A transgenic mouse lung. (I) Lymphoma-like lesions developed in the lung of 20-month-old A97 founder. (J and K) Cellular proliferation measured by BrdU incorporation (red nuclei) in young E1A+E1B transgenic mouse lung (J) and in aged E1A transgenic mouse lung (K). (B to K) Hematoxylin-eosin staining (original magnification, ×40).
The E1A transgenic mice from the founders (A2, A20, and A97) suffered from abnormal lung LPL and constant alveolar epithelial cell hyperproliferation, but no tumor was found in the lungs (Fig. 1C and 2A) as previously described (24). Up to the age of 16 months, compared with 27.3% of nontransgenic mice, 89.3% of E1A transgenic mice, starting at 10 months, developed LPL in the lungs (P < 0.01) (Fig. 1C and 2C and H), detected by histochemical staining of the lymphocyte marker CD3 and BrdU incorporation (previously described in detail [24]). The aged E1A transgenic mice, such as founder A97, died of lung perivascular and peribranchiolar lymphoma-like illness at the age of 20 months (Fig. 2I). In addition, the LPL were also present in the spleens and livers much later than those that appeared in the lung.
The presence of Ad5 E1B leads to a high level of Ad5 E1A 243-aa protein.
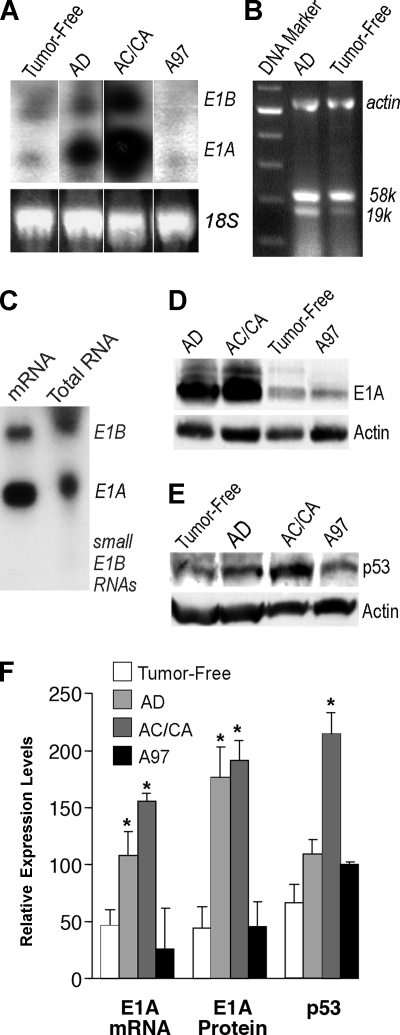
Since Ad5 E1A has the capacity to transform rodent cells in cell culture (8, 19), we evaluated the correlation between carcinogenesis and the levels of Ad5 E1A expression. Interestingly, E1A 243-aa protein was preferentially expressed in the lungs of all transgenic lines (Fig. 3A, C, and D) (also as described in reference 24). Likewise, it seems that E1B 58-kDa protein was dominantly expressed in the lungs of E1A+E1B transgenic mice because E1B 58-kDa protein could be detected in substantial amounts by Northern blotting (Fig. 3A and C), immunoblotting, or RT-PCR, while other, small E1B products such as E1B 19-kDa protein apparently could be detected neither by Northern blotting nor by immunoblotting with an appropriate antibody, except by RT-PCR, in which the large transcript of E1B 58-kDa protein was abundant (Fig. 3B). In addition, both transgenic products were present within the same tumor cells in the lung adenomas, detected by immunohistochemistry with anti-E1A and anti-E1B antibodies (unpublished data). The expression levels of E1A 12S mRNA or E1A 243-aa protein were variable and correlated with the tumorigenesis phenotype in the transgenic lung of AB2 mice, in which the lungs with tumors bore remarkably high levels of E1A 12S mRNA, 2.5 to 4 times higher than those in the lungs without tumors (P < 0.01) or four to six times higher than those in A97 transgenic mouse lung (P < 0.01) (Fig. 3A and F). Likewise, the levels of E1B mRNA in the lungs with tumors were 0.6 to 1.8 times higher than those in the lungs without tumors in AB2 mice (P < 0.01) (Fig. 3A). Moderate levels of E1A mRNA were detected only in the lung with higher proliferation in young A97 mice and low levels from adult A97 mice. Consistent with the mRNA levels, the levels of E1A 243-aa protein in the lungs with tumors in AB2 transgenic mice were four to five times higher than those in the lungs without tumors or those in young A97 mice (P < 0.01), in which E1A 243-aa protein was preferentially expressed at moderate or low levels (Fig. 3D and F). In addition, the number of cells highly expressing E1A 243-aa protein was assessed dynamically by immunostaining of lung sections with anti-E1A antibody (Fig. 4A and B). Increased expression of E1A 243-aa protein was localized in focal adenomas, adenomas and adenocarcinomas, or peripheral areas of malignant carcinomas (86%, 96%, and 65% or 63% of the excessively stained E1A protein-positive cells, respectively) but in few excessively stained E1A protein-positive cells in the central areas of malignant carcinomas (8%) and nontumor alveolar areas (5.4%) in the transgenic lungs, though most cells were faintly or less moderately stained with anti-E1A antibody. The results suggest that the presence of E1B leads to accumulation of E1A 243-aa protein in the cells. Overall, the findings of the quantitation of E1A expression resulting from three individual assays were concordant and all suggested that the presence of E1B leads to a significant increase in E1A expression in the lungs of Ad5 E1A+E1B transgenic mice.
FIG. 3.
Analysis of aberrant expression of E1A, E1B, and p53 in the lungs with tumors. Higher levels of E1A transcript in the lung with tumors (A and C) as analyzed by Northern blotting. Note the tumor-free lung; AD, adenomatous lung; AC/CA, adenocarcinomatous lung or lung with carcinoma lung mixed with adenoma in Ad5 E1A+E1B transgenic mice; A97, Ad5 E1A transgenic mouse lung. The amount of total RNA loaded in each lane was quantified and controlled by mouse 18S rRNA in panel A. (B) Both the large transcript of E1B 58-kDa protein and the small transcript of E1B 19-kDa protein were analyzed for dominant expression by RT-PCR using RNase-free DNase-treated total RNA from lungs with adenomas or tumor-free lungs of Ad5 E1A+E1B transgenic mice. (C) mRNA was purified from the total RNA, which was prepared from four Ad5 E1A+E1B transgenic mouse lungs with adenoma or adenocarcinoma tumors. (D) Higher levels of E1A 243-aa protein in the lungs with tumors detected by Western blotting. (E) Higher levels of p53 in the lung with malignant tumors detected by Western blotting. The same lungs were used in panels B to E. (F) Comparative quantitation of transgene E1A transcript, E1A 243-aa protein, and p53 protein in the E1A+E1B transgenic tumor-free lungs, adenomatous lungs, lungs with adenocarcinoma or carcinoma mixed with adenoma, and A97 E1A transgenic mouse lungs, respectively. Columns, means (n = 2 or 3); error bars, standard deviations; *, P < 0.05 versus E1A+E1B transgenic tumor-free lungs.
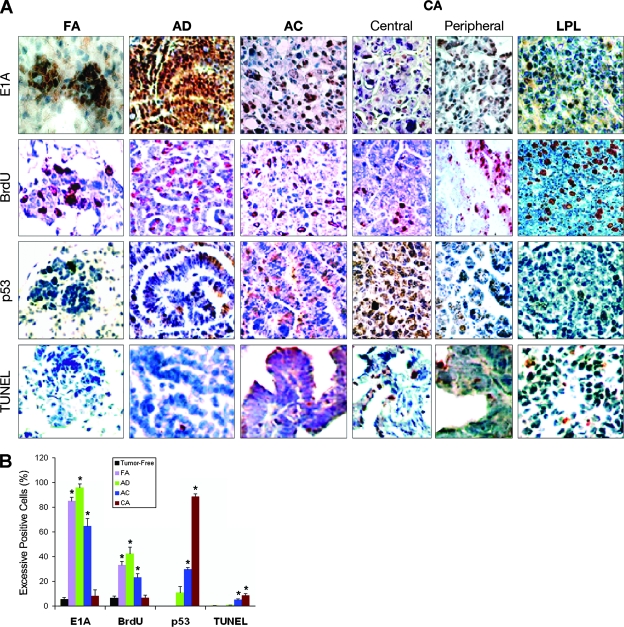
FIG. 4.
High levels of E1A 243-aa protein lead to high levels of proliferation rather than high levels of p53 and apoptosis. (A) Representative lung sections of focal adenomas (FA), adenomas (AD), adenocarcinomas (AC), and central and peripheral areas of carcinomas (CA) from E1A+E1B transgenic mice and LPL from A97 E1A transgenic mice stained for E1A 243-aa protein, BrdU-incorporating proliferation, p53, and TUNEL-positive apoptosis. Excessively stained E1A protein-positive nuclei (brown) or BrdU-positive nuclei (red) presented in tumor cells. BrdU-positive nuclei also existed in the proliferating lymphocytes of LPL. Excessively p53-positive nuclei (brown) mainly appeared in the tumor cells of AC or the CA central area. Few tumor cells in AC or CA showed TUNEL-positive nuclei (red). Original magnifications: ×100 (FA) and ×40 (AD, AC, CA, and LPL). The percentages of excessively stained E1A and p53-, BrdU-, and TUNEL-positive cells in tumor-free lung and FA, AD, AC, and CA central area of E1A+E1B transgenic mouse lungs are dynamically shown in panel B. Columns, means (n = 3); error bars, standard deviations; *, P < 0.025 versus tumor-free lungs.
High levels of E1A 243-aa protein integrated with E1B responsible for the uncontrolled cellular proliferation response.
To assess whether Ad5 E1A 243-aa protein directly correlated with cellular proliferation rather than cell death (13, 24), we directly analyzed BrdU incorporation and apoptosis using tumorous lung sections from transgenic mice of various ages. In 10-month-old AB2 mice, the percentages of BrdU-positive cells were 33% in focal adenomas, 42% in adenomas, 23% in adenocarcinomas, 32% in peripheral areas of malignant carcinomas, 6.5% in central areas of malignant carcinomas, and 6% in nontumorous alveolar areas (Fig. 2J and Fig. 4), compared with about 1% in the lungs of nontransgenic mice as previously reported (24). In A97 transgenic mice, BrdU-positive cells were detected at a higher rate in the alveoli of 3-month-old mice (12%) (24) than in 10-month-old mice (3 to 4%) (Fig. 2K). The results suggest that the higher levels of E1A 243-aa protein correlated with the higher levels of BrdU incorporation in the lung tumors of transgenic mice. Interestingly, the in situ cell death detection (TUNEL) results indicated that a relatively high number of apoptotic cells (9.6%) appeared in the central areas of malignant carcinomas but not in the focal adenomas (0%) or adenomas (0.9%), a result similar to the 0.7% apoptotic alveolar cells in the nontumorous lung (data not shown) (Fig. 4A and B), suggesting that a high level of E1A 243-aa protein does not lead to a high level of apoptosis. In addition, the higher number of BrdU-positive cells (30 to 60%) in the LPL but the lower number of apoptotic cells (2%) suggests an inflammation with lymphocyte proliferation (Fig. 4A, lane LPL).
E1B integrated with E1A leads to impaired p53 and apoptosis responses.
Whether a high level of E1A 243-aa protein directly induces high levels of p53 was investigated. Unlike the expression of the E1A 243-aa protein alone, high levels of p53 were detected only in the lung lysates with adenocarcinomas or carcinomas but not in the lung lysates with adenomas (Fig. 3E and F). The results of immunostaining serial tumorous lung sections revealed that the percentage of excessively p53 protein-positive cells was 89% in the central area of malignant carcinomas, whereas only 8% of the cells were excessively E1A 243-aa protein positive. In contrast, excessively p53-positive cells were not detected in the focal adenomas and were fewer (11%) in the adenomas or the peripheral areas of malignant carcinomas, although the numbers of excessively E1A 243-aa protein-positive cells were high (86%, 96%, and 63%, respectively) (Fig. 4A and B). The results suggest that a high level of p53 in carcinoma was not directly correlated with a high level of E1A 243-aa protein but was rather a response to uncontrolled cellular proliferation. We previously reported that apoptosis relevant to expression of E1A 243-aa protein was p53 dependent (24). The observation of low numbers of apoptotic cells and high numbers of excessively p53-positive cells in the malignant carcinomas suggested that p53 was inactivated in the E1A+E1B transgenic mouse lung.
DISCUSSION
Ad5 E1A 243-aa protein was preferentially expressed in the lungs of all transgenic lines, and E1B 58-kDa protein seemed to be dominantly expressed in the lungs of Ad5 E1A+E1B transgenic mouse lines. In the Ad5 E1A transgenic mice, Ad5 E1A 243-aa protein alone induced only low or moderate levels of cellular proliferation and obstructed lymphoproliferative inflammation. Furthermore, in the absence of the E1B gene, ongoing cellular proliferation was restricted by Ad5 E1A-induced p53-dependent growth arrest and apoptosis (13, 24). Therefore, expression of Ad5 E1A alone in the lung epithelial cells was not sufficient to initiate lung carcinogenesis. In Ad5 E1A+E1B transgenic mice, the presence of Ad5 E1B remarkably enhanced the expression of Ad5 E1A 243-aa protein and cellular proliferation but resulted in an impaired p53 response. The malignant lung carcinomas lost high levels of Ad5 E1A 243-aa protein but gained high levels of p53 which did not increase in tumor cell killing by apoptosis (Fig. 4). Thus, one possibility is that after lung carcinogenesis initiated by transient high expression of E1A 243-aa protein, tumors that relapsed despite the loss of E1A expression acquired a malignant property to maintain ongoing carcinogenesis (15). Due to the inactivation of p53-dependent apoptosis and growth arrest and the repression of cellular apoptosis by the presence of E1B (integrated synergic effects of E1B 58-kDa protein and E1B 19-kDa protein) (3, 8, 17, 20, 25), Ad5 E1A and E1B cooperation leads to uncontrolled cell growth and impaired p53 and apoptosis response; furthermore, the adenomas formed and progressed into adenocarcinomas or carcinomas. The effect of high levels of Ad5 E1A 243-aa protein and E1B products argues a direct correlation with uncontrolled cellular proliferation and initiation of lung carcinogenesis, rather than high levels of p53 and apoptosis. In general, Ad5 E1A and E1B are defined as nontumorigenic. The primary rodent cells can be transformed partially by Ad5 E1A alone. Also Ad5 E1A cooperation with Ad5 E1B dramatically increases in the transformation of either rodent or human cells in cell culture, in addition to cells becoming completely transformed. However, Ad5 E1A and E1B have not been reported to induce a tumorigenic phenotype in transformants resulting from infection of cells in animals or in cell culture (8, 22). Thus, our findings may imply that a distinctive biological effect of Ad5 E1A and E1B occurred in vivo which is different from that in vitro.
Our primary study with generation of these models aimed to investigate in vivo biological effects of Ad5 E1A and E1B on lung carcinogenesis and inflammation. The extensive investigations are necessary to understand the mechanism by which the presence of E1B enhances the expression of E1A 243-aa protein even if other previous studies suggested that E1B may stimulate E1A expression through an enhancement of the transcription initiation rate of the E1A region, and this transcriptional activation was dependent on a close physical linkage between E1A and E1B gene regions (9). In addition, the observation of a higher level of E1A expression in the tumorous lung of Ad5 E1A+E1B transgenic mice is concordant with the results from in vitro studies, which provided evidence that E1B enhanced E1A expression and led to an increase in the transformation frequency in transformed cell lines and also concluded that in general the transcription of E1A was enhanced by E1B (9, 19). The cellular hyperproliferation in Ad5 E1A transgenic mice is the consequence of the expression of Ad5 E1A 243-aa protein at a moderate level (17). Similarly, the high level of cellular proliferation in Ad5 E1A+E1B transgenic mice, which led to uncontrolled cellular proliferation and neoplasia, is also the consequence of the cooperation of an abnormally high level of E1A 243-aa protein with E1B products such as 58-kDa and 19-kDa proteins.
To fully understand the mechanism by which Ad5 E1A 243-aa protein leads to the LPL, we still need further investigations. However, previous studies implied that the persistent presence of viral proteins and their alternatively upregulated growth cytokines may be responsible for the lung LPL seen in the lungs of transgenic mice (3, 4, 18, 21, 24). Recent studies indicate that some cytokines, such as interleukin-6 and interleukin-15, acting as lymphocyte survival or inflammatory factors, have a fundamental role in homeostatic proliferation and inflammatory filtration (5, 10). Following viral antigen deposition, those cytokines are released by epithelial cells, stromal cells, and activated lymphocytes. They are not required for lymphocyte differentiation but upregulate the proliferation factors such as Bcl2, BAD, and phosphatidylinositol 3-kinase, leading to lymphocyte proliferation and peripheral expansion of lymphocytes. Thus, alternative expression of cytokines and growth factors may have a role in impairment of the immune response for disease malignancy (16, 23).
To rule out the possibility that a lifelong presence of adenovirus E1A and E1B in human lungs may play a role in stimulating lung carcinogenesis requires further clinical investigations, although other previous studies failed to detect the adenoviral genomes in human lung carcinomas and adenocarcinomas except small-cell lung cancers by a low-sensitivity detection method (6, 11). In general, numerous factors exert an effect on stimulating lung carcinogenesis either individually or cooperatively. Thus, if lifelong latency of adenoviral E1A+E1B in human lung functions as one of those factors, the proportion of adenovirus latency-associated lung tumors should be very small among the tumors caused by those numerous factors. Furthermore, if adenoviral E1A+E1B DNA during latency does not integrate into the host genome, the adenoviral genome would be dramatically diluted off during tumor progression (15). Our finding indicated that the E1A transgene product was dramatically increased only at the beginning of tumor formation in the focal adenomas and adenomas and then was dramatically abolished along with tumor progression in adenocarcinomas and carcinomas, suggesting that a high level of E1A 243-aa protein expression and the presence of E1B products are relevant to stimulating carcinogenesis but not necessary for tumor progression. Recent clinical studies demonstrated that relatively high levels of E1A proteins were also detected in the lungs of smokers and patients with respiratory disorders (2, 7), suggesting that the expression of Ad5 E1A can accumulate to a high level under certain conditions during latent adenoviral infection. In addition, the differences in tumor type between the transgenic mice and humans may be due to the species difference, e.g., the lack of small-cell lung cancer in the transgenic mice. The observations that the age of onset of tumor formation is delayed in the transgenic females and that those with tumors appear to have better survival than their male counterparts (Fig. 1B) suggest an effect of sex-associated hormones on tumor promotion.
Overall, our studies may provide clues to understanding the potential in vivo effects of latent Ad5 E1A and E1B on stimulating lung tumorigenesis and inflammation, upon reactivation, through uncontrolled cell proliferation, LPL, deferred p53, or impaired apoptosis response.
Acknowledgments
Colin McKerlie equally contributed to this work with Yongping Yang.
We thank Frank L. Graham for a generous supply of Ad5 E1A and E1B genomic DNA, NCI-CCR principal investigators Shyam Sharan and Ilona Linnoila for their advice and support, and the NCI-CCR Fellows Editorial Board and Maritta P. Grau at NCI-Frederick Scientific Publication for their critical reviews.
This work was supported by grants from Inspiraplex, the Lung Diseases Network of Centres of Excellence, the Hospital for Sick Children Foundation, and the Sunnybrook Trust for Medical Research in Canada.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Adrian, T., G. Schafer, M. K. Cooney, J. P. Fox, and R. Wigand. 1988. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol. Infect. 101503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakrabarti, S., V. Mautner, H. Osman, K. E. Collingham, C. D. Fegan, P. E. Klapper, P. A. Moss, and D. W. Milligan. 2002. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood 1001619-1627. [DOI] [PubMed] [Google Scholar]
- 3.Endter, C., B. Hartl, T. Spruss, J. Hauber, and T. Dobner. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 2455-64. [DOI] [PubMed] [Google Scholar]
- 4.Fischer, R. S., and M. P. Quinlan. 2000. While E1A can facilitate epithelial cell transformation by several dominant oncogenes, the C-terminus seems only to regulate rac and cdc42 function, but in both epithelial and fibroblastic cells. Virology 269404-419. [DOI] [PubMed] [Google Scholar]
- 5.Goldrath, A. W., P. V. Sivakumar, M. Glaccum, M. K. Kennedy, M. J. Bevan, C. Benoist, D. Mathis, and E. A. Butz. 2002. Cytokine requirements for acute and basal homeostatic proliferation of naive and memory CD8+ T cells. J. Exp. Med. 1951515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green, M., W. S. Wold, J. K. Mackey, and P. Rigden. 1979. Analysis of human tonsil and cancer DNAs and RNAs for DNA sequences of group C (serotypes 1, 2, 5, and 6) human adenoviruses. Proc. Natl. Acad. Sci. USA 766606-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi, S. 2002. Latent adenovirus infection in COPD. Chest 121183S-187S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutton, F. G., A. S. Turnell, P. H. Gallimore, and R. J. Grand. 2000. Consequences of disruption of the interaction between p53 and the larger adenovirus early region 1B protein in adenovirus E1 transformed human cells. Oncogene 19452-462. [DOI] [PubMed] [Google Scholar]
- 9.Jochemsen, A. G., L. T. Peltenburg, M. F. te Pas, C. M. de Wit, J. L. Bos, and A. J. van der Eb. 1987. Activation of adenovirus 5 E1A transcription by region E1B in transformed primary rat cells. EMBO J. 63399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalvakolanu, D. V. 1999. Virus interception of cytokine-regulated pathways. Trends Microbiol. 7166-171. [DOI] [PubMed] [Google Scholar]
- 11.Kuwano, K., M. Kawasaki, R. Kunitake, N. Hagimoto, Y. Nomoto, T. Matsuba, Y. Nakanishi, and N. Hara. 1997. Detection of group C adenovirus DNA in small-cell lung cancer with the nested polymerase chain reaction. J. Cancer Res. Clin. Oncol. 123377-382. [DOI] [PubMed] [Google Scholar]
- 12.Louis, N., C. Evelegh, and F. L. Graham. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233423-429. [DOI] [PubMed] [Google Scholar]
- 13.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7535-545. [DOI] [PubMed] [Google Scholar]
- 14.Matsuse, T., S. Hayashi, K. Kuwano, H. Keunecke, W. A. Jefferies, and J. C. Hogg. 1992. Latent adenoviral infection in the pathogenesis of chronic airways obstruction. Am. Rev. Respir. Dis. 146177-184. [DOI] [PubMed] [Google Scholar]
- 15.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 753089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280248-253. [DOI] [PubMed] [Google Scholar]
- 17.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinlan, M. P. 1994. Enhanced proliferation, growth factor induction and immortalization by adenovirus E1A 12S in the absence of E1B. Oncogene 92639-2647. [PubMed] [Google Scholar]
- 19.Senear, A. W., and J. B. Lewis. 1986. Morphological transformation of established rodent cell lines by high-level expression of the adenovirus type 2 E1a gene. Mol. Cell. Biol. 61253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieber, T., and T. Dobner. 2007. Adenovirus type 5 early region 1B 156R protein promotes cell transformation independently of repression of p53-stimulated transcription. J. Virol. 8195-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Templeton, S. P., and S. Perlman. 2007. Pathogenesis of acute and chronic central nervous system infection with variants of mouse hepatitis virus, strain JHM. Immunol. Res. 39160-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Elsen, P., A. Houweling, and A. Van der Eb. 1983. Expression of region E1b of human adenoviruses in the absence of region E1a is not sufficient for complete transformation. Virology 128377-390. [DOI] [PubMed] [Google Scholar]
- 23.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 247686-7696. [DOI] [PubMed] [Google Scholar]
- 24.Yang, Y., C. McKerlie, S. H. Borenstein, Z. Lu, M. Schito, J. W. Chamberlain, and M. Buchwald. 2002. Transgenic expression in mouse lung reveals distinct biological roles for the adenovirus type 5 E1A 243- and 289-amino-acid proteins. J. Virol. 768910-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 35782-85. [DOI] [PubMed] [Google Scholar]