Abstract
Little is known about the in vivo development of resistance to human immunodeficiency virus type 1 (HIV-1) CCR5 antagonists. We studied 29 subjects with virologic failure from a phase IIb study of the CCR5 antagonist vicriviroc (VCV) and identified one individual with HIV-1 subtype C who developed VCV resistance. Studies with chimeric envelopes demonstrated that changes within the V3 loop were sufficient to confer VCV resistance. Resistant virus showed VCV-enhanced replication, cross-resistance to another CCR5 antagonist, TAK779, and increased sensitivity to aminooxypentane-RANTES and the CCR5 monoclonal antibody HGS004. Pretreatment V3 loop sequences reemerged following VCV discontinuation, implying that VCV resistance has associated fitness costs.
The human immunodeficiency virus type 1 (HIV-1) envelope third variable loop (V3) is the major structural element of gp120 that determines coreceptor recognition and specificity (9, 15, 16). Vicriviroc (VCV; Schering-Plough) and maraviroc (Selzentry; Pfizer) are allosteric noncompetitive antagonists that bind to similar sites on CCR5 and antagonize the gp120-CCR5 interaction (19). To date, data on resistance to these agents have come largely from in vitro selection studies. Phenotypically, resistance manifests as a plateau in the maximum achievable suppression of viral replication (19). This plateau, referred to as the percent maximal inhibition, correlates with viral adaptation to the use of the inhibitor-bound form of CCR5 for entry (13, 21). Genotypically, VCV resistance has been associated with a variety of amino-acid-changing mutations throughout the envelope gene (env) that most often involve V3 but whose effect on drug susceptibility depends on the env backbone into which they are introduced (5). In vitro data suggest that resistance to the closely related CCR5 antagonist AD101 does not confer a significant loss of viral fitness (1, 8). In vivo resistance to the CCR5 antagonists remains poorly defined.
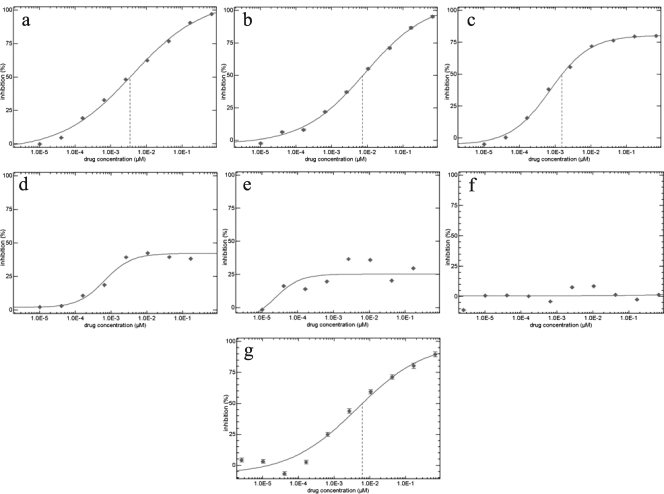
To study the emergence of VCV resistance in vivo, we monitored subjects enrolled in ACTG 5211, a 48-week study of VCV in 118 HIV-1-infected, treatment-experienced subjects (3). Among the 90 subjects receiving VCV, we studied all 29 who experienced protocol-defined virologic failure. We amplified full-length HIV-1 env from plasma samples collected during the period from study entry through week 48. These env sequences were used to generate pseudovirions for examining VCV susceptibility and coreceptor usage in the PhenoSense entry susceptibility and Trofile assays (Monogram Biosciences), respectively (20, 22). In 28 of 29 subjects analyzed, no evidence of decreased VCV susceptibility was observed (data not shown). Samples from the remaining subject demonstrated increasing VCV resistance over 28 weeks (Fig. 1). This subject, randomly assigned to receive 10 mg of VCV daily, experienced protocol-defined virologic failure at week 16 but continued VCV treatment through week 28 (see Fig. S1 in the supplemental material). Samples from 13 of the 29 subjects showed the emergence of CXCR4-using virus at the time of virologic failure. Virologic failure in the remaining 15 subjects could not be explained by coreceptor switching or VCV resistance.
FIG. 1.
VCV susceptibility of HIV-1 from a subject for whom VCV-containing antiretroviral therapy failed. VCV susceptibility was examined at week 0 (study entry) (a), week 2 (b), week 8 (c), week 19 (d), week 24 (e), week 28 (f), and week 48 (20 weeks after VCV discontinuation) (g) by using the PhenoSense entry assay (Monogram Biosciences, South San Francisco, CA) (21). Susceptibilities were plotted as micromolar drug concentrations versus the percent viral inhibition relative to the infection level in the absence of the drug. The vertical dashed lines indicate the 50% inhibitory concentration for VCV.
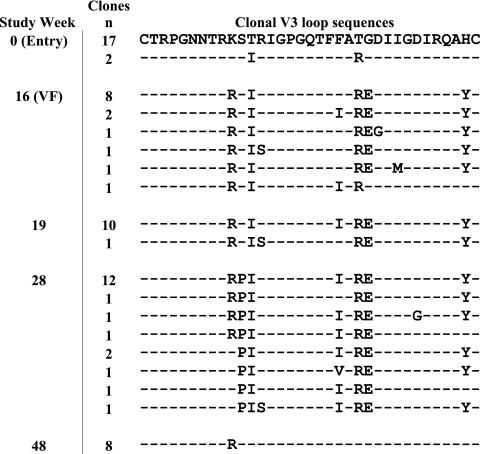
We assessed the genotypic changes occurring within env over this same time period by isolating and sequencing multiple independent full-length clones of env at time points from week 0 through week 48 (Fig. 2). Viral RNA was extracted from the subjects' plasma samples by using a QIAamp viral RNA mini kit (Qiagen), and env amplicons encoding gp160 were generated by nested PCR as described previously (5). Sequence and phylogenetic analyses showed that the envelope gene from the subject with VCV resistance clustered with HIV-1 subtype C genotypes (data not shown). At weeks 16 and 19, when partial VCV resistance was observed by the PhenoSense assay, the majority of envelope sequences showed amino acid substitutions K305R, T307I, F316I, T318R, and G319E in the V3 loop stem (numbering is based upon the HxB2 envelope sequence. The development of complete phenotypic VCV resistance at week 28 corresponded to the appearance of the S306P mutation in the V3 loops of all clones. This observation suggests that the V3 loop mutations present at weeks 16 and 19 confer partial resistance to VCV, with the addition of S306P leading to complete resistance. The discontinuation of VCV treatment at week 28 was associated with a shift toward a VCV-susceptible wild-type virus phenotype by week 48.
FIG. 2.
Alignment of V3 loop sequences from independent clones obtained at weeks 0, 16, 19, 28, and 48. The predominant clone at baseline (week 0) was designated the reference clone. Predicted amino acid differences are shown, and similarities are indicated with dashes. The number of independent clones with the same sequence is indicated to the left of each sequence. VF, virologic failure.
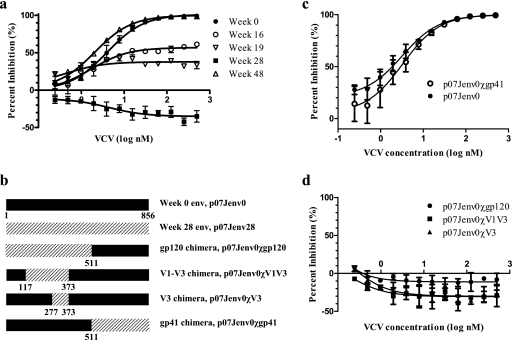
We next generated replication-competent recombinant HIV-1 clones to confirm the findings of the population-based data generated by the PhenoSense assay. We modified a Saccharomyces cerevisiae gap repair homologous recombination system to create infectious molecular clones of HIV-1 that carried the predominant full-length env sequence observed at each time point in an NL4-3 backbone (7) (M. Sagar, unpublished data). Viral stocks were generated by transfecting 293T cells. The susceptibilities of the resulting viruses to various HIV-1 inhibitors were assessed on TZM-bl cells (4, 23). Recombinant viruses expressing the cloned baseline env sequence were fully susceptible to VCV (Fig. 3a), whereas those expressing the cloned envelope genes from weeks 16 and 19 demonstrated a progressive decrease in VCV susceptibility. Recombinant virus expressing the cloned week 28 env showed no inhibition by VCV; increasing VCV concentrations resulted in increasing virus replication. This observation suggested that fully VCV-resistant virus had adapted to enter cells preferentially via the VCV-bound form of CCR5 and is consistent with similar findings with a VCV-resistant isolate selected in vitro (13). Recombinant virus with the cloned week 28 envelope gene did not infect CXCR4-expressing cell lines (see Table S1 in the supplemental material), demonstrating that the VCV-resistant virus continued to use CCR5 exclusively. Viruses expressing env cloned from the week 48 plasma sample, obtained 20 weeks after VCV discontinuation, showed restored sensitivity to VCV (Fig. 3a); the week 48 envelopes had near-wild-type V3 sequences.
FIG. 3.
VCV susceptibilities of recombinant viruses with full-length and chimeric envelopes. In each graph, the percentages of inhibition relative to the viral level in the no-drug control at various inhibitor concentrations are shown. (a) VCV susceptibilities of recombinant viruses with the predominant full-length envelope at sequential time points after VCV initiation. (b) Schematic representation of the chimeric envelopes; amino acid numbering is based on the HxB2 reference sequence. (c) VCV susceptibility of recombinant virus expressing a chimeric envelope with week 28 gp41 substituting within the week 0 envelope. (d) VCV susceptibilities of recombinant viruses expressing chimeric envelopes with week 28 envelope segments substituting for week 0 envelope regions. Error bars represent the standard errors of the means of results from two to four experiments, each performed in triplicate. Nonlinear regression was used to estimate a fitted curve.
We next constructed chimeric envelopes by incorporating portions of env from the VCV-resistant week 28 virus into a VCV-sensitive env backbone (week 0) to ascertain which envelope domain(s) determined VCV resistance (Fig. 3b). Different env segments encoding gp120, gp41, V1 to V3, or V3 were amplified using specific primers. These amplified segments were substituted into the week 0 env by using the modified yeast gap repair homologous recombination method described above. The substitution of the week 28 gp41 sequence into the week 0 envelope did not alter VCV sensitivity (Fig. 3c). By contrast, recombinant viruses incorporating gp120, the V1-V3 segment, or the V3 loop from the week 28 envelope demonstrated complete VCV resistance and enhanced viral replication in the presence of VCV (Fig. 3d). The substitution of the V3 loop alone, without V1 or V2, into the VCV-sensitive week 0 envelope was sufficient to confer complete VCV resistance. Although the V3 loop chimeras included some additional env sequences from the resistant virus immediately adjacent to both sides of the sequence encoding V3 (Fig. 3b), no consistent substitutions in these flanking regions were noted. Therefore, these data support the conclusion that the V3 loop is the principal determinant of VCV resistance in this virus.
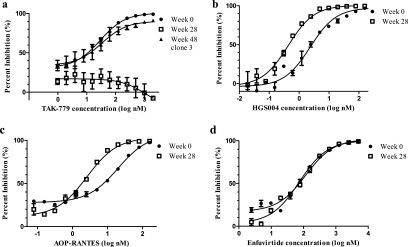
To explore the mechanism of VCV resistance, we tested the susceptibilities of VCV-resistant virus to other entry inhibitors. Recombinant viruses expressing the week 28 env showed cross-resistance to another small-molecule CCR5 inhibitor, TAK779 (2); recombinants expressing the week 48 env showed the restoration of TAK779 susceptibility (Fig. 4a). Cross-resistance to TAK779 and VCV may occur because the two compounds occupy similar binding sites on CCR5 (19) and likely lead to similar allosteric changes in the receptor. By contrast, VCV resistance sensitized virus to inhibition by the CCR5 monoclonal antibody HGS004 (Human Genome Sciences) and by aminooxypentane (AOP)-RANTES, resulting in 5.4- and 7.8-fold decreases in 50% inhibitory concentrations, respectively (Fig. 4b and c). HGS004 binds CCR5, competitively antagonizing gp120 binding, whereas AOP-RANTES triggers CCR5 internalization (6, 10). Thus, both compounds reduce the number of CCR5 molecules available on the cell surface. Increased susceptibilities to HGS004 and AOP-RANTES provided additional evidence that VCV-resistant virus has a decreased capacity to utilize the VCV-free form of CCR5. This observation, coupled with the VCV-enhanced replication of VCV-resistant virus, suggested that VCV-resistant envelope has decreased affinity for the non-drug-bound form of the receptor, but formal binding studies are needed to confirm our interpretation of these data. Previous studies suggested a correlation between CCR5 affinity and sensitivity to the fusion inhibitor enfuvirtide (ENF) (14). Interestingly, recombinant viruses with week 0 and week 28 envelopes showed no difference in ENF sensitivity (Fig. 4d), suggesting that ENF sensitivity and CCR5 affinity were not correlated for this viral envelope.
FIG. 4.
Resistance to VCV modulates sensitivities to other entry inhibitors. In each graph, percentages of inhibition relative to the viral level in the no-drug control at various TAK779 (a), HGS004 (b), AOP-RANTES (c), and ENF (d) concentrations are shown. Error bars represent the standard errors of the means of results from two to four experiments, each performed in triplicate. Nonlinear regression was used to estimate a fitted curve.
Our results show that in this HIV-1 subtype C isolate from a VCV-treated subject, increasing VCV resistance was conferred by the stepwise accumulation of mutations on both sides of the V3 loop stem. The complete loss of VCV susceptibility correlated with the emergence of S306P at position 11 of the V3 loop. The presence of a positively charged amino acid residue at this position is a determinant of CXCR4 usage, but substitutions such as S306K, S306L, and S306H, which confer CXCR4 usage, were not found in the samples we analyzed. It is likely that the introduction of a proline residue at position 11 significantly alters the conformation of the V3 loop. The presence of a 306P substitution as a naturally occurring polymorphism has been reported previously for only two subtype C viruses (0.05%) and eight subtype B viruses (0.02%) (Los Alamos Sequence Database [www.hiv.lanl.gov/content/index; accessed 8 December 2007]). Preliminary data from other subjects experiencing the virologic failure of maraviroc or VCV show mutations in the V3 loop stem that differ from subject to subject, although virus from one subject who experienced the failure of an initial VCV-containing regimen developed K305R, as seen in the samples from our subject (11, 17). No signature resistance mutations for the CCR5 antagonists have been identified to date.
A change in coreceptor usage accounted for the majority of virologic failures in the phase 3 trials of maraviroc (18) and in 13 of 29 subjects in the present study. The use of CXCR4 by subtype C viruses appears to be relatively uncommon for reasons that are not yet understood (12). Only two other subjects enrolled in the parent clinical trial (ACTG A5211) were infected with HIV-1 subtype C virus. One subject was randomly assigned to the 5-mg VCV arm but discontinued use of the study drug by week 2 of the study; the other subject was randomly assigned to the placebo group. Virus from both subjects remained exclusively R5 and VCV susceptible (data not shown). It is possible that viral or host factors that limit the emergence of CXCR4-using variants of HIV-1 subtype C constrain the pathways available to the virus to escape from VCV inhibition, thereby favoring the emergence of VCV-resistant mutants that have adapted to use the VCV-bound form of CCR5.
The loss of virtually all V3 loop mutations by week 48 implies that VCV resistance incurs a fitness cost relative to the wild-type phenotype in the absence of the drug. This finding contrasts with published in vitro data (1). Possible explanations for the disparate results include the limited genetic diversity of viruses in culture and the absence of immune selective pressure in vitro.
The emergence of VCV resistance in ACTG A5211 was uncommon, being identified in just 1 of 29 subjects with virologic failure. Although the causal role of V3 loop mutations in VCV resistance has been demonstrated for this subtype C virus, additional examples of clinically derived CCR5 antagonist-resistant viruses should be studied to elucidate the effects of the viral subtype on the emergence of VCV resistance and to describe the full range of changes in env that can confer CCR5 antagonist resistance.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited under GenBank accession numbers EU664612 to EU664683.
Supplementary Material
Acknowledgments
This research was supported by funds from the Clinical Investigator Training Program, Harvard/MIT Health Sciences and Technology-Beth Israel Deaconess Medical Center, in collaboration with Pfizer Inc. and Merck and Co., to A.M.N.T.; NIH grants R37 AI553537 and K24 RR016482 to D.R.K. and K24 AI-51966 to R.M.G.; and the AIDS Clinical Trials Group (U01 AI068636), the Harvard Virology Specialty Laboratory of the AIDS Clinical Trials Group, and the Harvard University Center for AIDS Research (P30 AI060354).
We thank B. M. Baroudy (Schering-Plough) for VCV and the entire A5211 protocol team for their efforts during this trial. ENF, TAK779, TZM-bl cells, and pNL4-3 were obtained from the NIH AIDS Reference Repository.
Footnotes
Published ahead of print on 21 May 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anastassopoulou, C. G., A. J. Marozsan, A. Matet, A. D. Snyder, E. J. Arts, S. E. Kuhmann, and J. P. Moore. 2007. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS Pathog. 3e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 965698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gulick, R. M., Z. Su, C. Flexner, M. D. Hughes, P. R. Skolnik, T. J. Wilkin, R. Gross, A. Krambrink, E. Coakley, W. L. Greaves, A. Zolopa, R. Reichman, C. Godfrey, M. Hirsch, and D. R. Kuritzkes. 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 196304-312. [DOI] [PubMed] [Google Scholar]
- 4.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 662232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 782790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalezari, J., G. K. Yadavalli, M. Para, G. Richmond, E. Dejesus, S. J. Brown, W. Cai, C. Chen, J. Zhong, L. A. Novello, M. M. Lederman, and G. M. Subramanian. 2008. Safety, pharmacokinetics, and antiviral activity of HGS004, a novel fully human IgG4 monoclonal antibody against CCR5, in HIV-1-infected patients. J. Infect. Dis. 197721-727. [DOI] [PubMed] [Google Scholar]
- 7.Marozsan, A. J., and E. J. Arts. 2003. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods 111111-120. [DOI] [PubMed] [Google Scholar]
- 8.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338182-199. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien, W. A., Y. Koyanagi, A. Namazie, J. Q. Zhao, A. Diagne, K. Idler, J. A. Zack, and I. S. Chen. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 34869-73. [DOI] [PubMed] [Google Scholar]
- 10.Pastore, C., G. R. Picchio, F. Galimi, R. Fish, O. Hartley, R. E. Offord, and D. E. Mosier. 2003. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob. Agents Chemother. 47509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfizer Inc. 24 April 2007, posting date. Maraviroc tablets NDA 22-128. Antiviral Drugs Advisory Committee (AVDAC) briefing document, p. 103. Pfizer Inc., New York, NY. http://www.fda.gov/OHRMS/DOCKETS/AC/07/briefing/2007-4283b1-01-Pfizer.pdf.
- 12.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 736271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 9916249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 2801949-1953. [DOI] [PubMed] [Google Scholar]
- 16.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349167-169. [DOI] [PubMed] [Google Scholar]
- 17.Strizki, J. M., P. Qiu, N. Murgolo, W. Greaves, R. Landovitz, and J. Whitcomb. 2006. 7th Annu. Symp. Antivir. Drug Resist., Chantilly, VA, 12 to 15 November, 2006, abstr. 36.
- 18.van der Ryst, E., and M. Westby. 2007. Changes in HIV-1 co-receptor tropism for patients participating in the maraviroc MOTIVATE 1 and 2 clinical trials, abstr. H-715. Abstr. 47th Annu. Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL, 17 to 20 September 2007.
- 19.Watson, C., S. Jenkinson, W. Kazmierski, and T. Kenakin. 2005. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 671268-1282. [DOI] [PubMed] [Google Scholar]
- 20.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 804909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 812359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitcomb, J. M., W. Huang, S. Fransen, K. Limoli, J. Toma, T. Wrin, C. Chappey, L. D. Kiss, E. E. Paxinos, and C. J. Petropoulos. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, X., A. B. Parast, B. A. Richardson, R. Nduati, G. John-Stewart, D. Mbori-Ngacha, S. M. Rainwater, and J. Overbaugh. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.