Abstract
The pathogenic mechanism by which parvovirus B19 may induce inflammatory cardiomyopathy (iCMP) is complex but is known to involve inflammatory processes, possibly including activation of JAK/STAT signaling. The nonstructural B19 protein NS1 acts as a transactivator triggering signaling cascades that eventually lead to activation of interleukin 6 (IL-6). We examined the impact of NS1 on modulation of STAT signaling in human endothelial cells (HMEC-1). The NS1 sequences were identified from B19 DNA isolated from the myocardia of patients with fatal iCMP. B19 infection as well as NS1 overexpression in HMEC-1 cells produced a significant upregulation in the phosphorylation of both tyrosine705 and serine727 STAT3 (P < 0.05). The increased STAT3 phosphorylation was accompanied by dimerization, nuclear translocation, and DNA binding of pSTAT3. In contrast, NS1 expression did not result in increased STAT1 activation. Notably, the expression levels of the negative regulators of STAT activation, SOCS1 and SOCS3, were not altered by NS1. However, the level of PIAS3 was upregulated in NS1-expressing HMEC-1 cells. Analysis of the transcriptional activation of target genes revealed that NS1-induced STAT3 signaling was associated with upregulation of genes involved in immune response (e.g., the IFNAR1 and IL-2 genes) and downregulation of genes associated with viral defense (e.g., the OAS1 and TYK2 genes). Our results demonstrate that B19 NS1 modulates the STAT/PIAS pathway. The NS1-induced upregulation of STAT3/PIAS3 in the absence of STAT1 phosphorylation and the lack of SOCS1/SOCS3 activation may contribute to the mechanisms by which B19 evades the immune response and establishes persistent infection in human endothelial cells. Thus, NS1 may play a critical role in the mechanism of viral pathogenesis in B19-associated iCMP.
Human parvovirus B19 is emerging as an important pathogenic agent in the etiology of inflammatory cardiomyopathy (iCMP). Recent studies have indicated an association between infection with B19 and acute myocarditis in both children and adults (5, 44). However, the role of B19 infection in the development of chronic iCMP patients is still unclear. We have recently demonstrated that endothelial cells but not cardiac myocytes are B19-specific target cells in patients with B19-associated myocarditis (21). Furthermore, B19 could be detected frequently in patients with unexplained isolated diastolic dysfunction (2, 49).
The B19 single-stranded DNA genome of 5,600 base pairs contains two open reading frames encoding the nonstructural protein NS1 and two structural capsid proteins, VP1 and VP2, by a combination of alternative splicing (15). A functional phospholipase A2-like activity has been demonstrated recently in the VP1 region which is involved in intracellular Ca2+ regulation (28). In addition, three small viral proteins of unknown function have been described previously (39, 55).
The main function of NS1 includes transactivation of the viral P6 promoter, which is important for viral replication in a process that involves the direct binding of NS1 to specific promoter regions and through protein-protein interaction with p21/WAF (37, 55). However, NS1 can also transactivate a variety of cellular genes, including interleukin 6 (IL-6) and tumor necrosis factor alpha as well as unrelated viral promoters like the long terminal repeat of human immunodeficiency virus (11, 18, 46). In addition to its transactivator functions, NS1 is also cytotoxic in vitro (34, 45) and can initiate proapoptotic processes through activation of caspases 3 and 9 (43). The NS1 protein contains a nucleoside triphosphate (NTP)-binding motif which is involved in the cytotoxicity of the protein. The cytotoxic effects of NS1 can be abolished by mutations within this NTP-binding domain and thereby rescue cells from NS1-induced apoptosis without having an effect on the NS1-induced activation of IL-6 expression (34, 43).
Signal transducer and activator of transcription (STAT) proteins represent a family of latent transcription factors that have a critical immune regulatory role in the transcriptional activation of cytokine response genes (54). These proteins are activated upon phosphorylation in response to extracellular signals such as cytokines (interferons [IFNs]) and growth factors (42). STAT3 has been shown to suppress the endothelial cell activation and transmigration of leukocytes (9, 42). Dysregulated STAT3 signaling in T lymphocytes and macrophages can lead to chronic inflammation through uncontrolled IFN production and the expression of proinflammatory cytokines (20). Constitutive STAT3 activation also contributes to oncogenic transformation (6) and promotes apoptosis (27). This ambivalent role of STAT3 signaling contributes to maintaining the balance between cytoprotection and programmed cell death (12).
Activated STAT1 is crucial for the innate immune response (48). However, numerous viruses, including paramyxoviruses (52), hepatitis C virus (25), and coronaviruses (e.g., severe acute respiratory syndrome coronavirus) (10), have evolved mechanisms to circumvent the host defense by inhibition of STAT1 signaling, which may contribute to establishing chronic disease (13). Suppressor of cytokine signaling (SOCS) proteins are negative regulators of the JAK/STAT signaling. SOCS1 and SOCS3 have been shown to directly inhibit JAK phosphorylation activity and activation of cytokine-induced STAT proteins (23). Protein inhibitor of activated STAT3 (PIAS3) also acts as a negative regulator of phosphorylated STAT3 in IL-6-stimulated M1 cells (7). The constitutive expression of PIAS implies that their physiological function differs from SOCS proteins. Consequently, PIAS proteins may act like a buffer that titrates the concentration of active STAT dimers within the cell (14).
Understanding the underlying molecular mechanisms by which B19 may cause iCMP remains a challenge. A feature of the B19 infection of myocardial muscle is through the sole involvement of cardiac endothelial cells, resulting in a combination of inflammatory events and endothelial cell dysfunction. The present investigation explored the effect of NS1 on the interaction and regulation of the STAT/SOCS/PIAS signaling pathway using patient-specific NS1 constructs in functional human endothelial cell culture systems.
MATERIALS AND METHODS
Cell culture, transfection, and infection.
The immortalized human microvascular endothelial cell line (HMEC-1) was kindly provided by Wieder (University of Tuebingen, Germany). HMEC-1 cells were grown in Medium 199 (Invitrogen, Karlsruhe, Germany) supplemented with 1% penicillin-streptomycin, 10 ng/ml endothelial cell growth factor (Invitrogen, Karlsruhe, Germany), 1 μg/ml hydrocortisone, and 15% fetal calf serum. A total of 2.5 × 105 HMEC-1 cells were seeded in six-well cell culture dishes (Nunc, Wiesbaden, Germany). Transient and stable transfection was performed with FuGENE 6 (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. All experiments were performed at least in triplicate.
The B19-susceptible UT7/Epo-S1 cells were propagated in RPMI 1640 medium containing 10% fetal calf serum and 2 U/ml of recombinant erythropoietin (Epo) (Hofmann-LaRoche, Basel, Switzerland) (36). B19 virions were purified from high-titer B19-positive serum using ultracentrifugation through a 30% sucrose cushion in phosphate-buffered saline (100,000 rpm for 24 h at room temperature). Purified virus stock solution was adjusted to 5.5 × 1012 viral particles/ml by quantitative real-time PCR (qPCR). UT7/Epo-S1 cells were infected with a multiplicity of infection of 1,000 in Isocove modified Dulbecco medium and incubated for 2 h at 4°C. After adsorption of B19, the cells were maintained in Isocove modified Dulbecco medium containing 10% fetal calf serum and 2 U/ml Epo at 37°C and 5% CO2.
Plasmids and site-directed mutagenesis.
B19 DNA was isolated from the deparaffinized myocardial tissues of patients with fatal B19-associated iCMP as described previously (accession no. AY768535 and AF162273) (5). The NS1 region was amplified by PCR using high-fidelity polymerase (Pwo SuperYield DNA polymerase; Hofmann-LaRoche, Basel, Switzerland). The resulting NS1-specific amplicons were subcloned with ApaI into the pcDNA3.1 expression vector (Invitrogen, Karlsruhe, Germany). SOCS1 and SOCS3 expression constructs were generated using PCR amplification of genomic DNA isolated from HMEC-1 cells. The PCR fragments were then subcloned into pcDNA3.1. Newly generated constructs were verified by DNA sequencing for correct cloning (BigDye Terminator cycle sequencing; PE Applied Biosystems, Foster City, CA).
An NTP-binding motif mutant within the NS1 coding region (pcDNA-NS1K334E) was constructed as described previously using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions (37). K334E mutants were confirmed by DNA sequencing. Primer sequences for cloning can be supplied upon request.
A replication-competent B19 plasmid (pB19-M20, kindly provided by Kevin E. Brown and Susan Wong, Hematology Branch, National Heart Lung and Blood Institute, Bethesda, MD) (56) was used for the generation of stable B19-expressing HMEC-1 cells. Stable transfection was performed with FuGENE 6 in a 1:20 ratio of pB19-M20 to pCMV-Hygromycin and selected in Medium 199 containing 300 U/ml of hygromycin B. Individual colonies were picked and expanded. Infectivity of selected stable B19-expressing HMEC-1 cells was demonstrated by infection of HEMC-1 cells with medium supernatant of the stable cells, followed by reverse transcription-PCR (RT-PCR) for NS1 and VP1 genes (data not shown).
Control cell extracts and Western blot analysis.
IFN-γ- and IL-6-stimulated cells (each 100 ng/ml for 30 min at 37°C; Cell Signaling, Beverly, MS) were used as positive controls for STAT1 and STAT3 phosphorylation, respectively. Nuclear extracts of HeLa cells known to express high levels of PIAS3 were used as positive controls (53). Western blot analysis was performed as described previously (16). In brief, HMEC-1 cells transfected with pcDNA-NS1 and pcDNA3.1 control constructs as well as B19-transfected and mock-transfected UT7/Epo-S1 cells were harvested at indicated time points and lysed (3, 4). Twenty-five micrograms of protein extract was subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE), followed by transfer to a polyvinylidene difluoride (PVDF) membrane (Perkin Elmer, Wellesley, MA) as described previously (3, 4). The following antibodies were used: anti-B19 (NS1), anti-STAT1, anti-STAT3, anti-PIAS3, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-pSTAT1Tyr701, anti-pSTAT3Tyr705, and anti-pSTAT3Ser727 (Cell Signaling, Danvers, MA). Anti-SOCS1 and anti-SOCS3 antibodies were kindly provided by H. Zentgraf (German Cancer Research Center, Heidelberg, Germany). Anti-B19 NS1 antibodies for immunofluorescence were provided by Nobuyuki Tanaka. Membranes were probed with anti-goat antibodies conjugated to horseradish peroxidase (Dianova, Hamburg, Germany) and visualized using an enhanced chemiluminescence detection assay according to the manufacturer's instructions (Applied Biosystems, Darmstadt, Germany).
EMSA.
In order to determine pSTAT3 binding to STAT3-specific binding sites (cis-inducible element [CIE]) in response to NS1 expression, electrophoretic mobility shift assays (EMSAs) were performed as previously described (16). HMEC-1 cells were transfected with pcDNA3.1-NS1 and pcDNA3.1 control constructs and harvested 48 h after transfection. Nuclear extracts were obtained as described previously (16). A double-stranded oligonucleotide containing the CIE consensus sequence was labeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas, St. Leon-Rot, Germany). The binding reaction was performed at room temperature for 30 min as described previously (16). The samples were separated on 5% Tris-borate-EDTA polyacrylamide gels for 2 h and exposed to X-ray films. Supershift assays were performed as described previously using nuclear extracts and CIE probes incubated with anti-STAT3 and anti-GAPDH antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) (16).
BN-PAGE.
In order to determine STAT3 dimer formation, pcDNA-NS1-transfected HMEC-1 cells were harvested 12, 24, and 36 h posttransfection and lysed in blue native PAGE (BN-PAGE) sample buffer (50 nM Bis-Tris, 6 N HCl, 50 mM NaCl, 10% [wt/vol], 0.001% Ponceau S, 1% digitonin, pH 7.2) and analyzed following the manufacturer's instructions. A total of 30 μg of cell extracts was separated on BN-PAGE Novex 4% to 16% Bis-Tris gels (Invitrogen, Karlsruhe, Germany). Proteins were transferred to a PVDF membrane (Perkin Elmer, Wellesley, MA) as described previously (16) and probed with anti-STAT3 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and secondary goat anti-rabbit antibodies conjugated to horseradish peroxidase (Dianova, Hamburg, Germany), followed by detection using an enhanced chemiluminescence assay (Applied Biosystems, Darmstadt, Germany).
RNA extraction, semiquantitative RT-PCR, real-time PCR, and the RT2 profiler assay.
Total cellular RNA was isolated using the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions 24 h after transfection of HMEC-1 cells with pcDNA-NS1 and pcDNA3.1. Semiquantitative RT-PCR was performed using the one-step RT-PCR kit (Qiagen, Hilden, Germany) with B19 NS1 primers B19 NS1-s (5′-ATGGAGCTATTTAGAGGG-3′) and B19 NS1-as (5′-AAGTAGCAGAAATACAGGT-3′) (cycle conditions were as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s for a total of 45 cycles) and GAPDH primers GAPDH-s (5′-CATGTTCGTCATGGGTGTGA-3′) and GAPDH-as (5′-AGTGAGCTTCCCGTTCAGCTC-3′) (cycle conditions were as follows: 95°C for 60 s, 55°C for 45 s, and 72°C for 45 s for a total of 25 cycles). qPCR to determine SOCS1 and SOCS3 expression was performed using the TaqMan gene expression assay mix (Applied Biosystems) after reverse transcription of RNA with the high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA). NS1 mRNA expression of stable B19-expressing and B19-infected HMEC-1 cells was determined by real-time RT-qPCR as described previously (49). ATP synthase 6 (ATP-S-6) served as an internal control to standardize the system. The probes were as follows: NS1, ACCTCCAAACCACCCCAATTGTCACA; NS1-s, TTCCTGGAATTAATGCAGATGC; NS1-as, CACTGCTGCTGATACTGGTGTCT; ATP-S-6, 5′-AGCCCACCTCTTACCACAAGGCACA-3′; ATP-S-6 forward primer, 5′-CAGTGATTATAGGCTTTCGCTCTAA-3′; ATP-S-6 reverse primer, 5′-CAGGGCTATTGGTTGAATGAGTA-3′. Cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min for a total of 45 cycles. To avoid DNA contamination in RT-PCR assays, samples were extensively processed using DNase 1 digestion. The lack of DNA was monitored using NS1-specific nested PCR (data not shown).
Gene expression profiling for JAK/STAT target genes was performed using RT2 profiler PCR arrays (SuperArray, Frederick, MD) following the manufacturer's instructions. In brief, 1 μg RNA was reverse transcribed with the RT2 profiler PCR array first strand synthesis assay (SuperArray, Frederick, MD) followed by real-time PCR with RT2 real-time PCR master mix Sybr green (SuperArray, Frederick, MD).
Luciferase assay promoter studies.
Luciferase assays were performed as described previously (3, 50). SOCS3 promoter activity was analyzed using pLucSOCS3Min (a generous gift from Hiroshi Nakajima, Chiba University School of Medicine, Chiba, Japan). The CIE (hSIEm67) containing the construct pLucTK-SIE was a generous gift from Richard Jove (H. Lee Moffitt Cancer Center and Research Institute). Luciferase activity was monitored using the Steady-Glo luciferase assay system (Promega, Mannheim, Germany) according to the manufacturer's instructions.
Statistical analysis.
The densitometric evaluation of the Western blot analysis was performed using Aida software version 3.24 (Raytest; Straubenhardt, Germany). Statistical analysis and graphing was performed using Prism software version 4.0 (GraphPad Software Inc., San Diego, CA). All quantitative assays were performed at least in triplicate. The results were expressed as means ± standard deviations. For the significance test, a one-way analysis of variance followed by post hoc testing and Tukey's multiple comparison test were used.
RESULTS
Expression of NS1 in HMEC-1 cells.
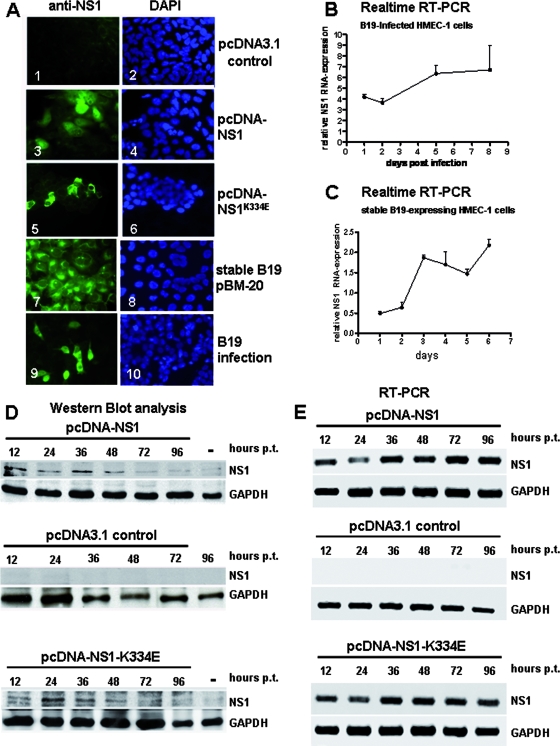
In order to determine the effects of B19 infection and especially of the B19 NS1 protein on STAT signaling patient-derived purified B19 virions, replication-competent B19 constructs (pB19-M20), pcDNA-NS1, and pcDNA-NS1K334E constructs were used to infect and transfect HMEC-1 cells, respectively. In contrast to a previously described study, we were successful in obtaining B19 stable cell clones that survived and produced B19 proteins, including NS1 (Fig. 1A and B), using the recently described B19 infectious clone pB19-M20 (40, 56). This construct has been shown to be B19 replication competent, producing infectious virions as early as 72 h after transfection in B19-permissive UT7/Epo-S1 cells. The production of NS1 proteins as well as NS1K334E proteins and transcripts in infected, transfected, and stably transfected HMEC-1 cells was demonstrated by immunofluorescence (Fig. 1A), Western blot analysis (Fig. 1D), RT-qPCR (Fig. 1B and C), and RT-PCR (Fig. 1E). NS1 and NS1K334E proteins of the correct size were confirmed by Western blot analysis (Fig. 1D). NS1 protein expression was maximal between 12 h and 48 h, whereas NS1 mRNA expression was constantly present throughout the analyzed time periods. The NS1K334E mutant showed comparable expression levels to the wild-type (WT) NS1. The constant level of NS1 mRNA in comparison to the declining expression of NS1 protein over time in the transfection experiments may be due to the more sensitive RT-PCR technique. However, the NS1 protein is detectable throughout all time points. Notably, in immunofluorescence experiments, a predominant nuclear and perinuclear distribution of NS1 proteins was observed (Fig. 1A). The perinuclear localization was more obvious in NS1K334E mutant proteins than in the WT NS1 (Fig. 1A).
FIG. 1.
Expression of NS1 in HMEC-1. (A) Immunofluorescence experiments demonstrating NS1 protein expression of transfected HMEC-1 cells with pcDNA-NS1 (panel 3), mutant NS1K334E (panel 5), stable B19 pBM-20 HMEC-1 cells (panel 7), and B19-infected cells (panel 9). The NS1 protein was detected with monoclonal anti-NS1 antibodies 48 h after transfection. Corresponding images of DAPI (4′,6-diamidino-2-phenylindole) staining are shown in blue (panels 2, 4, 6, 8, and 10). pcDNA3.1 control transfection showed no NS1-specific staining (panel 1). (B) Real-time RT-PCR analysis of NS1 mRNA expression demonstrated a time-dependent increase in NS1 mRNA after B19 infection of HMEC-1 cells. (C) Stable B19-expressing HMEC-1 cells showed increased NS1 mRNA expression by real-time RT-PCR. (D) Western blot analyses demonstrating correct expression of NS1 polypeptides (71 kDa). HMEC-1 cells were transfected with pcDNA-NS1 (top panel), control constructs pcDNA3.1 (middle panel), and mutant pcDNA-NS1K334E (bottom panel), and harvested 12, 24, 36, 48, 72, and 96 h posttransfection (p.t.). Cell lysates of the transfected HMEC-1 cells were applied to 7.5% sodium dodecyl sulfate-PAGE, transferred to PVDF membranes, and probed with monoclonal anti-NS1 antibodies. pcDNA3.1 control transfectants showed no NS1-specific signals. The housekeeping gene GAPDH served as an internal control for the equal loading of proteins. Mock-transfected cells harvested 36 h after transfection were used as negative controls (−). (E) RT-PCR analyses showing B19 NS1 mRNA expression in HMEC-1 cells. HMEC-1 cells were transfected with pcDNA-NS1 (top panel), pcDNA3.1 control constructs (middle panel), and mutant pcDNA-NS1K334E (bottom panel). Total RNA was extracted 12, 24, 36, 48, 72, and 96 h posttransfection (p.t.) and subjected to RT-PCR analyses using gene-specific primers for B19 NS1. GAPDH-specific primers were used to demonstrate correct RNA synthesis. NS1- and GAPDH-specific mRNA amplicons were separated on 2% agarose gel electrophoreses and visualized by ethidium bromide staining.
NS1 induces STAT3 but not STAT1 activation in human endothelial cells.
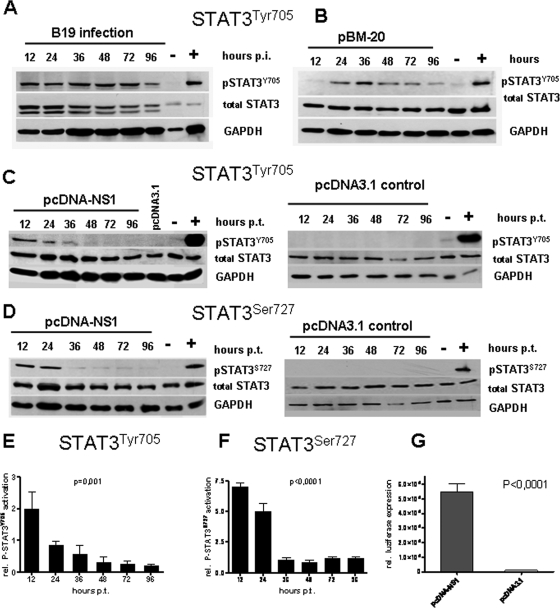
It has been reported that NS1 can activate IL-6 production, which can be responsible for STAT3 activation (18, 33). In order to investigate the influence of B19 NS1 on the activation of STAT signaling in human endothelial cells, we performed cell culture experiments. First, the UT7/Epo-S1 cells highly susceptible for B19 were infected with purified B19 virions to determine STAT activation. The induction of P-STAT3 (pSTAT3Tyr705) that significantly increased throughout all time points up to 96 h after infection could be shown (Fig. 2A). In addition, compared to that of mock-transfected cells, STAT3 activation could be demonstrated in stable B19-expressing HMEC-1 cells. The greatest activation of STAT3 was detectable between 24 h and 72 h following the induction of NS1 expression (Fig. 2B). Western blot analyses of transiently transfected HMEC-1 cell lysates revealed that STAT3 tyrosine phosphorylation (pSTAT3Tyr705) was upregulated 12 h after transfection with pcDNA-NS1 in contrast to that of control plasmid pcDNA3.1 (Fig. 2C). Coincident with the decline in the intracellular NS1 protein, expression in the signal intensity of pSTAT3Tyr705 also declined with time, returning to the background level by 96 h posttransfection.
FIG. 2.
Expression of NS1 leads to activation of p-STAT3 in HMEC-1 cells. Western blot analyses of STAT3 activation by phosphorylation of STAT3Tyr705 (A to C) and STAT3Ser727 (D) in HMEC-1 cells at indicated time points after B19 infection (A), stable B19 expression (B), and transfection of NS1 (C and D). Corresponding pcDNA3.1 controls not expressing NS1 are shown (C and D, right panels). IL-6-stimulated HMEC-1 cells served as positive controls (+), and mock-transfected HMEC-1 cells served as negative controls (−) for STAT3 activation. Notably, the expression level of total STAT3 was not altered by NS1 (middle lanes). The housekeeping gene GAPDH showed that an equal amount of protein was loaded in each lane (A to D, bottom lanes). (E to F) Densitometric evaluation of STAT3 activation. The data represent a minimum of five independent experiments (P = 0.001 for STAT3Tyr705 [left panel]; P < 0.0001 for STATSer727 [right panel]). (G) Luciferase assays demonstrating P-STAT3 binding to the STAT3-specific CIE by NS1 expression 24 h after transfection of pcDNA-NS1 and the pcDNA3.1 control construct, respectively. The statistical analysis (E to F) was performed using a one-way analysis of variance and Tukey's multiple comparison posttest.
Similar Western blot analyses following pcDNA-NS1 transfection of HMEC-1 cells failed to show any upregulation of STAT1 phosphorylation (pSTAT1) (data not shown). In contrast, the total STAT3 and STAT1 were not significantly altered by B19 NS1 expression (Fig. 2).
Similar to that of cells with the WT NS1 protein, transfection of HMEC-1 cells with the NS1 NTP mutant (pcDNA-NS1K334E) demonstrated a comparable level of upregulation of pSTAT3Tyr705, while there was no activation of STAT1, indicating that the NTP-binding motif with its ATPase, nickase, and helicase enzymatic activities is not necessary for the regulation of STAT signaling by NS1 (data not shown).
NS1 induces STAT3 serine phosphorylation in HMEC-1 cells.
Tyrosine phosphorylation of STAT3 (pSTAT3Tyr705) is known to be necessary for dimer formation and DNA binding (42). STAT3 serine phosphorylation (pSTATSer727), on the other hand, potentiates the transcriptional activity of STAT3 (1). We analyzed the effect of B19 NS1 on STAT3Ser727 phosphorylation in HMEC-1 cells. Western blot analyses using a pSTAT3Ser727-specific antibody revealed significant upregulation of STAT3Ser727 phosphorylation 12 h to 36 h after transfection with pcDNA-NS1. Also, accompanying the decline in NS1 expression, this was followed by a decline in pSTAT3Ser727 and a return to baseline by 48 h posttransfection (Fig. 2D).
NS1 activates STAT3 dimer formation, translocation of pSTAT3 to the nucleus, and DNA binding.
Having shown that B19 NS1 can modulate STAT3 phosphorylation, we next wanted to determine if this effect influences pSTAT3 dimerization, nuclear translocation, and DNA binding, all of which are essential steps associated with the induction of genes involved in inflammatory responses.
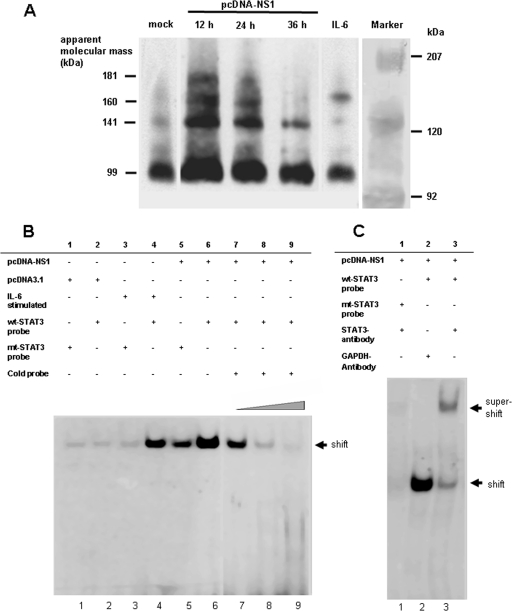
To demonstrate STAT3 dimer formation, we performed STAT3-specific BN-PAGE. The STAT3 dimer formation of pcDNA-NS1-transfected HMEC-1 cells was detectable at 12 h to 36 h after transfection, declining in the time course (Fig. 3A). This result was in accordance with the pattern of STAT3 activation in NS1-transfected HMEC cells (Fig. 2).
FIG. 3.
NS1-induced STAT3 dimer formation. (A) BN-PAGEs showing P-STAT3 dimer formation after transfection of HMEC-1 cells with pcDNA-NS1, mock-transfected (mock), and IL-6-stimulated cells. Cells were harvested at indicated time points after transfection. Membranes were probed with anti-STAT3 antibodies. STAT3 monomers (94 kDa) and putative STAT3 dimers (141 kDa, 160 kDa, and 181 kDA) are shown. (B) EMSAs showing binding of P-STAT3 to the STAT3-specific CIE in B19 NS1-expressing cells. HMEC-1 cells were transfected with pcDNA-NS1 (lanes 5 to 9) and control vector pcDNA3.1 (lanes 1 and 2). IL-6-stimulated HMEC-1 cells (lanes 3 and 4) were used as positive controls for STAT3 activation. Cell extracts were either incubated with WT (lanes 2, 4, and 6 to 9) or mutant CIE oligonucleotides (lanes 1, 3, and 5). Competition experiments showed increasing amounts of nonradioactive WT CIE oligonucleotides (1 μg, lane 7; 3 μg, lane 8; and 5 μg, lane 9) demonstrating specificity of STAT3 binding. (C) Supershift experiments were performed using anti-STAT3 (lane 3) and control GAPDH antibodies (lane 2). Mutated CIE oligonucleotides showed no supershift (lane 1).
By using an EMSA, we could also show that pSTAT3 molecules were present in nuclear extracts of pcDNA-NS1-transfected HMEC-1 cells 24 h after transfection and that these proteins bound to the STAT3-specific binding motif of the CIE probe, which results in a shift of this complex (Fig. 3B, lane 6). In contrast, in HMEC-1 cells transfected with pcDNA-NS1 and incubated with a mutant CIE probe representing a defective STAT3-binding motif, only a faint shift in the EMSA was observed compared to that with the WT probe (Fig. 3B, lanes 1, 3, and 5). Additionally, by titrating increasing amounts of nonradioactive WT CIE oligonucleotides, the complex formation could be driven to completion (Fig. 3, lanes 7 to 9). These results were also confirmed by luciferase reporter gene assays demonstrating binding of activated STAT3 dimers to the CIEs of the pLucTK-SIE construct (Fig. 2G). NS1-expressing HMEC-1 cells showed upregulation of luciferase activity of pLucTK-SIE 24 h after transfection of pcDNA-NS1 constructs compared to pcDNA3.1-transfected control cells.
NS1 activates PIAS3 but not SOCS1 and SOCS3 in HMEC-1 cells.
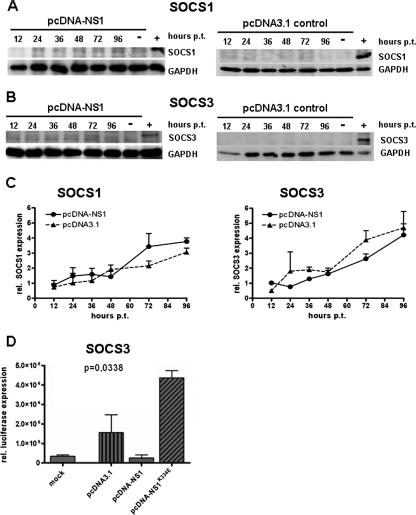
SOCS and PIAS proteins are the negative regulators of the JAK/STAT pathway. To determine if SOCS1 and SOCS3 expression were increased following STAT3 activation, we performed SOCS1- and SOCS3-specific real-time qPCR and Western blot analyses. These experiments revealed that neither SOCS1 nor SOCS3 expression was significantly upregulated after transfection of HMEC-1 cells with pcDNA-NS1 (Fig. 4).
FIG. 4.
NS1 expression does not activate SOCS1 and SOCS3 expression in HMEC-1 cells. Western blot analyses of SOCS1 (A) and SOCS3 (B) expression after transfection of pcDNA-NS1 (A and B, left panels) and pcDNA3.1 control constructs (A and B, right panels) of HMEC-1 cells. Cells were harvested at indicated time points after transfection and subjected to Western blot analyses. Cells transfected with pcDNA-SOCS1 (A) and pcDNA-SOCS3 (B) expression plasmids served as positive controls (+); mock-transfected cells were used as negative controls (-). (C) Real-time (TaqMan) qPCR of SOCS1 (left panel) and SOCS3 mRNA (right panel) after transfection of HMEC-1 cells with pcDNA-NS1 and pcDNA3.1, demonstrating that SOCS1 and SOCS3 were not upregulated. (D) Luciferase gene reporter experiments demonstrating suppression of SOCS3 promoter activity 24 h after transfection of pcDNA-NS1. Mock-transfected cells served as the negative control. pcDNA3.1 and pcDNA-NS1K334E NTP mutant constructs showed upregulation of SOCS3 promoter activity.
In addition, in order to determine if NS1 has a direct interaction with the SOCS3 promoter element, we performed luciferase gene reporter assays using the pLucSOCS3Min plasmid harboring a SOCS3 promoter element that controls luciferase gene activity. HMEC-1 cells were transfected with pcDNA-NS1, mutant pcDNA-NS1K334E, and pcDNA3.1 control constructs, and 48 h later, transfection cells were harvested and luciferase activity was measured. Interestingly, NS1-expressing cells showed suppression of SOCS3 promoter activity compared to pcDNA3.1 control cells (Fig. 4D). In contrast, compared to that of WT NS1, the SOCS3 promoter activity was upregulated in cells expressing the mutant NS1K334E protein. These results suggest that a direct negative interaction may exist between the NTP-binding motif of NS1 and the SOCS3 promoter element.
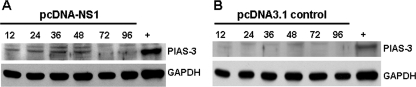
As SOCS3 was not involved in STAT3 regulation in the NS1-transfected HMEC cells, we investigated the possible role of PIAS3 activation as an alternative STAT3 negative regulator. In NS1-expressing cells, we demonstrated that the levels of PIAS3 increased 12 to 48 h after transfection, followed by a gradual decline (Fig. 5). This was in accordance with the pattern of intracellular NS1 protein expression and of STAT3 activation after transfection of pcDNA-NS1 (Fig. 2).
FIG. 5.
PIAS3 is upregulated by NS1 expression. Western blot analyses of (A) pcDNA-NS1 and (B) pcDNA3.1 control plasmid-transfected HMEC-1 cell extracts were analyzed at indicated time points. HeLa cell extracts served as the positive control (+). GAPDH showed that an equal amount of protein was loaded in each lane (A and B, bottom lanes).
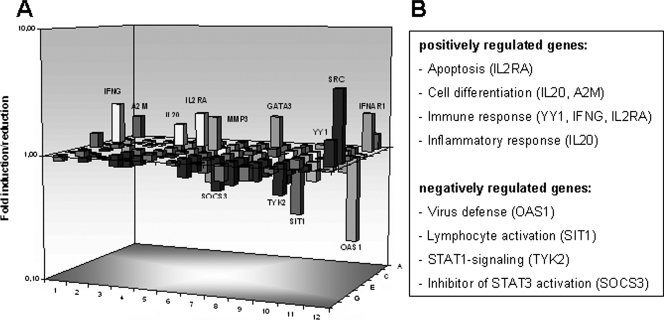
A negative regulation of SOCS3 mRNA expression after B19 NS1 expression could also be shown using RT2 profiler array assays of JAK/STAT gene regulation. In comparison to that in controls, SOCS3 expression was reduced to 1.86-fold in NS1-transfected HMEC-1 cells 24 h after transfection (Fig. 6).
FIG. 6.
STAT3 target gene transcriptional expression by NS1 expression. (A) Gene expression profiling for JAK/STAT target genes was performed using RT2 profiler PCR arrays 24 h after transfection of HMEC-1 cells with pcDNA-NS1, showing upregulation of genes of immune response (e.g., the IL-20 and IL2R genes) and downregulation of genes involved in virus defense (e.g., the OAS1, TYK2, and SIT1 genes). (B) The products of the most abundant regulated genes, with induction greater than twofold, are shown.
NS1-induced regulation of STAT-dependent target gene expression.
Having shown that NS1 upregulates pSTAT3, we wanted to determine if this was associated with the upregulation of STAT-induced target gene expression. We performed RT2 profiler array assays to analyze important STAT-activated genes (84 target genes were included in the RT2 profiler array) which could be modulated by JAK/STAT-signaling. Twenty-four hours after transfection of HMEC-1 cells with pcDNA-NS1, the expression profiling showed a significant induction of nine genes compared to that of mock-transfected cells (IFNG, IFNAR1, IL-20, IL2RA, SRC, MMP3, GATA3, YY1, and A2M) (Fig. 6). All of these have important roles in immune response and activation of transcription. Most notably, the negative regulators of STAT signaling, PIAS2 and PTPN1 (protein tyrosine phosphatase N1), were also upregulated. Four genes (OAS1, TYK2, SOCS3, and SIT) important for virus defense and STAT1- and STAT3-related signal transduction were downregulated. These findings were in accordance with the STAT3 activation and absence of STAT1 activation we observed 12 h after transfection/infection with B19.
DISCUSSION
The association between B19 infection and iCMP has been described recently with the identification of myocardial endothelial cells as target cells of B19 (2, 5, 24). Furthermore, analyses of consecutive endomyocardial biopsies of patients with progressive left ventricular systolic dysfunction demonstrated the persistence of B19 infection in heart tissue (24, 38). Besides enteroviruses and human herpesvirus 6, B19 has been described as being the most common agent of biopsy-proven viral myocarditis, which may lead to the development of endothelial cell and isolated left ventricular diastolic dysfunction (29, 49).
It has been proposed that the B19 NS1 protein acts as a transcriptional transactivator that regulates a variety of viral and cellular promoters (11, 18, 31, 55), including the expression levels and activities of cellular factors like IL-6, AP-1, SP1, NF-kB, tumor necrosis factor alpha, and the tumor suppressor p53 (11, 37, 45). This led us to hypothesize that during B19 infection of myocardial endothelial cells, NS1 may modulate the expression of inflammatory genes, including STAT signaling. We have shown that B19 NS1 expression in infected and transfected human endothelial cells leads to the activation and dimerization of STAT3 and that the STAT3 dimers are translocated into the nucleus where they bind to specific recognition sites of a CIE. These observations are consistent with earlier reports which showed that NS1 is capable of inducing IL-6 expression, which is an important factor of STAT3 activation (11, 18, 31).
STAT3 serine phosphorylation (pSTAT3Ser727) is necessary to achieve maximal transcriptional activation of STAT3 target genes. STAT3Ser727 phosphorylation can negatively modulate tyrosine phosphorylation (8, 19). Our experiments demonstrate that NS1-induced pSTAT3Ser727 activation follows pSTAT3Tyr705 activation. These findings are consistent with the expected sequential pattern of STAT3 tyrosine705/serine727 phosphorylation.
The B19 NS1-induced STAT3 activation was accompanied by modulation of STAT target gene expression. The upregulation of genes involved in immune response against viral infections, including those of IL-10 receptor α, IL-2 receptor α, and IFN receptor proteins (IFNG and IFNAR1), could be detected using RT profiler arrays. Downregulation of genes involved with virus defense, e.g., suppression of STAT1 activation by TYK2 downregulation, and STAT3 signaling (OAS1, TYK2, SOCS3, and SIT1) were also shown.
While the NS1 gene contains an NTP-binding motif that supports ATPase, nickase, and helicase enzymatic activities, all of which have been reported to be linked to viral replication, cytotoxicity, and proapoptotic effects (34, 35, 43, 45), we have demonstrated that this motif is not involved in STAT3 activation.
Our experiments have also indicated that B19 NS1 expression does not result in the upregulation of SOCS1 and SOCS3 expression as would have been expected following the induction of STAT3. In fact, we have shown that NS1 may directly suppress SOCS3 promoter activity and consequently suppress SOCS3 gene expression. However, our findings support the hypothesis that NS1-activated PIAS3 expression may participate in the negative regulation of NS1-induced STAT3 activation and gene expression. PIAS3 exhibits E3 SUMO (small ubiquitin-related modifier) ligase activity if overexpressed in the nucleus, where it blocks the DNA binding of activated STAT3 as well as STAT3-mediated gene regulation (26, 47).
STAT3 activation has been described as being involved in anti-inflammatory processes like suppression of vascular permeability and transendothelial migration of leukocytes (9). By using STAT3 knockout mice, it has been shown that chronic inflammation originates from increased IFN and proinflammatory cytokine production (e.g., IL-10) (20). The time-dependent regulation of STAT3 and PIAS3 activation after NS1 expression we have shown may play a role in virus-host interaction to determine inflammatory, cytoprotective, and/or proapoptotic effects associated with B19 infection. STAT3 deregulation by NS1 followed by PIAS3 activation in the absence of a strong immune response can therefore be beneficial for viral replication and promote viral persistence and chronic inflammation. Similar mechanisms involving deregulation of anti-inflammatory signaling have also been recently described for Vaccinia virus (30).
In contrast to STAT3 activation in response to NS1 expression, STAT1 is not activated by NS1, and so it might be expected that this would result in a reduced IFN response to B19 infection. This could be one mechanism by which B19 exerts its pathogenic effect on the host endothelial cells. Other viruses, like mumps virus, measles virus, and paramyxovirus, also block activation of STAT1 by promoting the degradation of the unphosphorylated protein, thereby enabling these viruses to evade the host anti-inflammatory response (17, 41, 51). However, in the case of B19 infection, it cannot be assumed that increased STAT1 degradation has occurred since total STAT1 expression was not reduced. Hypothetically, NS1 could be able to inhibit phosphorylation of STAT1 by interacting with the Janus kinases JAK1, JAK2, and TYK2. On the other hand, NS1 may cause early dephosphorylation of STAT1, as has been shown for severe acute respiratory syndrome coronavirus, coronavirus, and Sendai virus infections (22, 32). We found that TYK2 mRNA expression was reduced, indicating that NS1 inhibits STAT1 activation by suppressing TYK2, which is essential for type I IFN response. More detailed analyses of STAT1 regulation in response to NS1 expression are needed to elucidate the exact mechanisms by which NS1 modulates STAT1 activation.
In conclusion, the present observations revealed NS1-activated STAT3/PIAS3 signaling which may contribute to the pathogenesis of B19-associated iCMP through heightened expression of inflammatory cytokines. This could be accompanied by a disruption of endothelial cell barrier function and ultimately lead to endothelial dysfunction. Accordingly, the altered regulation of STAT3/PIAS3 signaling by NS1 is likely to exert a profound effect on the function of host cells.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich-Transregio 19 (project B5; C.-T.B, R.K., F.L., and C.T.).
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Aznar, S., P. F. Valeron, S. V. del Rincon, L. F. Perez, R. Perona, and J. C. Lacal. 2001. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol. Biol. Cell 123282-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock, C. T., K. Klingel, S. Aberle, A. Duechting, A. Lupescu, F. Lang, and R. Kandolf. 2005. Human parvovirus B19: a new emerging pathogen of inflammatory cardiomyopathy. J. Vet. Med. B 52340-343. [DOI] [PubMed] [Google Scholar]
- 3.Bock, C. T., S. Kubicka, M. P. Manns, and C. Trautwein. 1999. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology 291236-1247. [DOI] [PubMed] [Google Scholar]
- 4.Bock, C. T., N. P. Malek, H. L. Tillmann, M. P. Manns, and C. Trautwein. 2000. The enhancer I core region contributes to the replication level of hepatitis B virus in vivo and in vitro. J. Virol. 742193-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bultmann, B. D., K. Klingel, K. Sotlar, C. T. Bock, H. A. Baba, M. Sauter, and R. Kandolf. 2003. Fatal parvovirus B19-associated myocarditis clinically mimicking ischemic heart disease: an endothelial cell-mediated disease. Hum. Pathol. 3492-95. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. L., A. Loy, L. Cen, C. Chan, F. C. Hsieh, G. Cheng, B. Wu, S. J. Qualman, K. Kunisada, K. Yamauchi-Takihara, and J. Lin. 2007. Signal transducer and activator of transcription 3 is involved in cell growth and survival of human rhabdomyosarcoma and osteosarcoma cells. BMC Cancer 7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 2781803-1805. [DOI] [PubMed] [Google Scholar]
- 8.Chung, J., E. Uchida, T. C. Grammer, and J. Blenis. 1997. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol. Cell. Biol. 176508-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahmy, R. G., A. Waldman, G. Zhang, A. Mitchell, N. Tedla, H. Cai, C. R. Geczy, C. N. Chesterman, M. Perry, and L. M. Khachigian. 2006. Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat. Biotechnol. 24856-863. [DOI] [PubMed] [Google Scholar]
- 10.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, Y., K. K. Ishii, Y. Munakata, T. Saitoh, M. Kaku, and T. Sasaki. 2002. Regulation of tumor necrosis factor alpha promoter by human parvovirus B19 NS1 through activation of AP-1 and AP-2. J. Virol. 765395-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujio, Y., K. Kunisada, H. Hirota, K. Yamauchi-Takihara, and T. Kishimoto. 1997. Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J. Clin. Investig. 992898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 812341-2364. [DOI] [PubMed] [Google Scholar]
- 14.Greenhalgh, C. J., and D. J. Hilton. 2001. Negative regulation of cytokine signaling. J. Leukoc. Biol. 70348-356. [PubMed] [Google Scholar]
- 15.Heegaard, E. D., and K. E. Brown. 2002. Human parvovirus B19. Clin. Microbiol. Rev. 15485-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heegaard, E. D., H. Hasle, L. Skibsted, J. Bock, and K. E. Brown. 2000. Congenital anemia caused by parvovirus B19 infection. Pediatr. Infect. Dis. J. 191216-1218. [DOI] [PubMed] [Google Scholar]
- 17.Horvath, C. M. 2004. Weapons of STAT destruction: interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2714621-4628. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, T. C., B. S. Tzang, C. N. Huang, Y. J. Lee, G. Y. Liu, M. C. Chen, and G. J. Tsay. 2006. Increased expression and secretion of interleukin-6 in human parvovirus B19 non-structural protein (NS1) transfected COS-7 epithelial cells. Clin. Exp. Immunol. 144152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain, N., T. Zhang, S. L. Fong, C. P. Lim, and X. Cao. 1998. Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene 173157-3167. [DOI] [PubMed] [Google Scholar]
- 20.Kano, A., M. J. Wolfgang, Q. Gao, J. Jacoby, G. X. Chai, W. Hansen, Y. Iwamoto, J. S. Pober, R. A. Flavell, and X. Y. Fu. 2003. Endothelial cells require STAT3 for protection against endotoxin-induced inflammation. J. Exp. Med. 1981517-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klingel, K., M. Sauter, C. T. Bock, G. Szalay, J. J. Schnorr, and R. Kandolf. 2004. Molecular pathology of inflammatory cardiomyopathy. Med. Microbiol. Immunol. 193101-107. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511139-144. [DOI] [PubMed] [Google Scholar]
- 23.Krebs, D. L., and D. J. Hilton. 2001. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19378-387. [DOI] [PubMed] [Google Scholar]
- 24.Kuhl, U., M. Pauschinger, M. Noutsias, B. Seeberg, T. Bock, D. Lassner, W. Poller, R. Kandolf, and H. P. Schultheiss. 2005. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation 111887-893. [DOI] [PubMed] [Google Scholar]
- 25.Lan, K. H., K. L. Lan, W. P. Lee, M. L. Sheu, M. Y. Chen, Y. L. Lee, S. H. Yen, F. Y. Chang, and S. D. Lee. 2007. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J. Hepatol. 46759-767. [DOI] [PubMed] [Google Scholar]
- 26.Levy, C., Y. N. Lee, H. Nechushtan, O. Schueler-Furman, A. Sonnenblick, S. Hacohen, and E. Razin. 2006. Identifying a common molecular mechanism for inhibition of MITF and STAT3 by PIAS3. Blood 1072839-2845. [DOI] [PubMed] [Google Scholar]
- 27.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3651-662. [DOI] [PubMed] [Google Scholar]
- 28.Lupescu, A., C. T. Bock, P. A. Lang, S. Aberle, H. Kaiser, R. Kandolf, and F. Lang. 2006. Phospholipase A2 activity-dependent stimulation of Ca2+ entry by human parvovirus B19 capsid protein VP1. J. Virol. 8011370-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahrholdt, H., A. Wagner, C. C. Deluigi, E. Kispert, S. Hager, G. Meinhardt, H. Vogelsberg, P. Fritz, J. Dippon, C. T. Bock, K. Klingel, R. Kandolf, and U. Sechtem. 2006. Presentation, patterns of myocardial damage, and clinical course of viral myocarditis. Circulation 1141581-1590. [DOI] [PubMed] [Google Scholar]
- 30.Maloney, G., M. Schroder, and A. G. Bowie. 2005. Vaccinia virus protein A52R activates p38 mitogen-activated protein kinase and potentiates lipopolysaccharide-induced interleukin-10. J. Biol. Chem. 28030838-30844. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, L. A. 2002. Parvovirus B19 nonstructural (NS1) protein as a transactivator of interleukin-6 synthesis: common pathway in inflammatory sequelae of human parvovirus infections? J. Med. Virol. 67267-274. [DOI] [PubMed] [Google Scholar]
- 32.Mizutani, T., S. Fukushi, M. Murakami, T. Hirano, M. Saijo, I. Kurane, and S. Morikawa. 2004. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett. 577187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffatt, S., N. Tanaka, K. Tada, M. Nose, M. Nakamura, O. Muraoka, T. Hirano, and K. Sugamura. 1996. A cytotoxic nonstructural protein, NS1, of human parvovirus B19 induces activation of interleukin-6 gene expression. J. Virol. 708485-8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffatt, S., N. Yaegashi, K. Tada, N. Tanaka, and K. Sugamura. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J. Virol. 723018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momoeda, M., S. Wong, M. Kawase, N. S. Young, and S. Kajigaya. 1994. A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity. J. Virol. 688443-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita, E., K. Tada, H. Chisaka, H. Asao, H. Sato, N. Yaegashi, and K. Sugamura. 2001. Human parvovirus B19 induces cell cycle arrest at G2 phase with accumulation of mitotic cyclins. J. Virol. 757555-7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakashima, A., E. Morita, S. Saito, and K. Sugamura. 2004. Human parvovirus B19 nonstructural protein transactivates the p21/WAF1 through Sp1. Virology 329493-504. [DOI] [PubMed] [Google Scholar]
- 38.Nigro, G., V. Bastianon, V. Colloridi, F. Ventriglia, P. Gallo, G. D'Amati, W. C. Koch, and S. P. Adler. 2000. Human parvovirus B19 infection in infancy associated with acute and chronic lymphocytic myocarditis and high cytokine levels: report of 3 cases and review. Clin. Infect. Dis. 3165-69. [DOI] [PubMed] [Google Scholar]
- 39.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 612395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozawa, K., J. Ayub, S. Kajigaya, T. Shimada, and N. Young. 1988. The gene encoding the nonstructural protein of B19 (human) parvovirus may be lethal in transfected cells. J. Virol. 622884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 777635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5375-386. [DOI] [PubMed] [Google Scholar]
- 43.Poole, B. D., J. Zhou, A. Grote, A. Schiffenbauer, and S. J. Naides. 2006. Apoptosis of liver-derived cells induced by parvovirus B19 nonstructural protein. J. Virol. 804114-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schowengerdt, K. O., J. Ni, S. W. Denfield, R. J. Gajarski, N. E. Bowles, G. Rosenthal, D. L. Kearney, J. K. Price, B. B. Rogers, G. M. Schauer, R. E. Chinnock, and J. A. Towbin. 1997. Association of parvovirus B19 genome in children with myocarditis and cardiac allograft rejection: diagnosis using the polymerase chain reaction. Circulation 963549-3554. [DOI] [PubMed] [Google Scholar]
- 45.Sol, N., J. Le Junter, I. Vassias, J. M. Freyssinier, A. Thomas, A. F. Prigent, B. B. Rudkin, S. Fichelson, and F. Morinet. 1999. Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J. Virol. 738762-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sol, N., F. Morinet, M. Alizon, and U. Hazan. 1993. Trans-activation of the long terminal repeat of human immunodeficiency virus type 1 by the parvovirus B19 NS1 gene product. J. Gen. Virol. 742011-2014. [DOI] [PubMed] [Google Scholar]
- 47.Sonnenblick, A., C. Levy, and E. Razin. 2004. Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes. Mol. Cell. Biol. 2410584-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suarez Gonzalez, A., L. Otero Guerra, G. V. De La Guerra, P. De La Iglesia Martinez, G. Solis Sanchez, and A. Rodriguez Fernandez. 2002. Varicella and parvovirus B19 immunity among pregnant women in Gijon, Spain. Med. Clin. (Barcelona) 119171-173. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 49.Tschope, C., C. T. Bock, M. Kasner, M. Noutsias, D. Westermann, P. L. Schwimmbeck, M. Pauschinger, W. C. Poller, U. Kuhl, R. Kandolf, and H. P. Schultheiss. 2005. High prevalence of cardiac parvovirus B19 infection in patients with isolated left ventricular diastolic dysfunction. Circulation 111879-886. [DOI] [PubMed] [Google Scholar]
- 50.Turkson, J., T. Bowman, R. Garcia, E. Caldenhoven, R. P. De Groot, and R. Jove. 1998. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol. Cell. Biol. 182545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 776385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vag, T., E. Sonkoly, B. Kemeny, S. Karpati, A. Horvath, and J. Ongradi. 2004. Familiar occurrence of papular-purpuric ‘gloves and socks’ syndrome with human herpes virus-7 and human parvovirus B19 infection. J. Eur. Acad. Dermatol. Venereol. 18639-641. [DOI] [PubMed] [Google Scholar]
- 53.Wang, X., G. Zhang, F. Liu, M. Han, D. Xu, and Y. Zang. 2004. Prevalence of human parvovirus B19 DNA in cardiac tissues of patients with congenital heart diseases indicated by nested PCR and in situ hybridization. J. Clin. Virol. 3120-24. [DOI] [PubMed] [Google Scholar]
- 54.Wen, Z., and J. E. Darnell, Jr. 1997. Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 252062-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhi, N., I. P. Mills, J. Lu, S. Wong, C. Filippone, and K. E. Brown. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 805941-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318142-152. [DOI] [PubMed] [Google Scholar]