Abstract
Duck hepatitis B virus (DHBV) is a model virus for human hepatitis B virus (HBV), which infects approximately 360 million individuals worldwide. Nucleoside analogs can decrease virus production by inhibiting the viral polymerase; however, complete clearance by these drugs is not common because of the persistence of the HBV episome. HBV DNA is present in the nucleus as a covalently closed circular (cccDNA) form, where it drives viral transcription and progeny virus production. cccDNA is not the direct target of antiviral nucleoside analogs and is the source of HBV reemergence when antiviral therapy is stopped. To target cccDNA, six different zinc finger proteins (ZFP) were designed to bind DNA sequences in the DHBV enhancer region. After the binding kinetics were assessed by using electrophoretic mobility shift assays and surface plasmon resonance, two candidates with dissociation constants of 12.3 and 40.2 nM were focused on for further study. The ZFPs were cloned into a eukaryotic expression vector and cotransfected into longhorn male hepatoma cells with the plasmid pDHBV1.3, which replicates the DHBV life cycle. In the presence of each ZFP, viral RNA was significantly reduced, and protein levels were dramatically decreased. As a result, intracellular viral particle production was also significantly decreased. In summary, designed ZFPs are able to bind to the DHBV enhancer and interfere with viral transcription, resulting in decreased production of viral products and progeny virus genomes.
Hepatitis B virus (HBV) causes a significant global health burden with an estimated 360 million people persistently infected and 500,000 to 700,000 deaths annually from HBV-associated liver disease (42). A total of 5% of adults and 95% of neonates exposed to the virus become persistently infected (42). Persistent infection with HBV may lead to liver cirrhosis and/or hepatocellular carcinoma, the latter of which has a 5-year survival rate of only 9% (38). Therapeutics such as nucleoside analogs are effective at clearing the infection in ca. 20 to 30% of treated patients (27); however, resistance to nucleoside analogs is an increasing problem, with 70% of patients becoming resistant to lamivudine and 18% becoming resistant to adefovir or tenofovir after 4 years of treatment (27). Thus, there is need for improved treatment options that can target the HBV episome, which is the primary source of HBV persistence.
HBV is a member of the Hepadnaviridae family and has a small double-stranded DNA (dsDNA) genome of approximately 3,200 bp and a strict tropism for hepatocytes (27, 38). Duck hepatitis B virus (DHBV) is a model virus for HBV with a comparable tropism for avian hepatocytes and a common viral structure, life cycle, and genome organization (15). Upon infection, the viral genome is converted from a relaxed circular form to a covalently closed circular (cccDNA) form in the nucleus of hepatocytes (15). This cccDNA form associates with several proteins to form a “minichromosome” structure (31) and is the reservoir from which transcription of viral genes and progeny genomes occur. cccDNA is highly stable with 3 to 50 copies per nucleus and a half-life of approximately 50 days (2, 43). When treatment with nucleoside analogs is stopped in infected patients, the long-lived cccDNA reservoir frequently results in the resurgence of viral production. There are currently no therapeutics available that specifically target the cccDNA of HBV.
Zinc finger proteins (ZFPs) are Cys2His2 DNA-binding proteins that can be designed to target novel DNA sequences with high specificity and affinity (3, 10, 11, 12, 26, 36). Each zinc finger is approximately 30 amino acids in length and is composed of two beta sheets and an alpha helix that are coordinated by a zinc ion. The alpha helix lies within the major groove of dsDNA and makes specific contact with 3 bp of DNA. By stringing zinc fingers in tandem, a unique DNA sequence of 18 bp can be specifically recognized (17).
We have designed six ZFPs targeting the enhancer region of DHBV, which is an accessible region of the cccDNA minichromosome and controls the core and surface promoters (7, 23, 26). After assessment of binding kinetics by using electrophoretic mobility shift assays (EMSAs) and surface plasmon resonance (SPR), we selected two candidate ZFPs with dissociation constants (Kd) in the nanomolar range. We have found that production of viral RNA, protein, and virus progeny was decreased in the presence of each ZFP after transfection into a DHBV tissue culture system, indicating that ZFPs binding the DHBV enhancer are capable of inhibiting the viral replicative process at the transcriptional level.
MATERIALS AND METHODS
Design of DHBV-specific ZFPs.
ZFPs were designed to target DHBV Canada isolate (AF047045) using the program Zinc Finger Tools (28). ZFPs were designed with flanking XhoI and SpeI restriction endonuclease sites, and zinc fingers was linked in tandem by the canonical TGEKP linker. All ZFPs were designed to bind to target sites within the enhancer region of DHBV (positions 2170 to 2361) (Fig. 1). One control ZFP (ZFP1) was constructed by scrambling the order of the fingers of ZFPa. ZFP target sites and the corresponding zinc finger amino acid sequences that mediate binding are shown in Table 1.
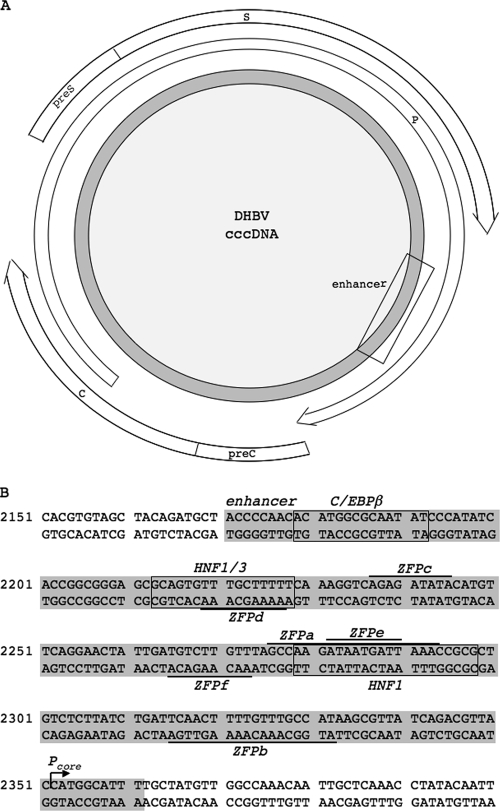
FIG. 1.
Schematic representation of the DHBV genome and detailed map of DHBV enhancer including ZFP and other TF binding sites. (A) The gray circle represents DHBV cccDNA. The open arrows represent open reading frames for core (C) and precore (preC), Pol (P), and surface (large [preS] and small [S]). The square approximates the location of the DHBV enhancer. (B) The enhancer region (gray) with the binding sites for ZFPs (overlined and underlined), and transcription factors C/EBPβ, HNF1, and HNF3 (boxed). ZFPa and ZFPb both bind 18-bp sequences, while ZFPc, ZFPd, ZFPe, and ZFPf each bind 9-bp sequences.
TABLE 1.
DNA binding sites and corresponding amino acid sequences of ZFPs
| ZFP | DNA target
|
Finger design | |
|---|---|---|---|
| Sequence (5′-3′)a | Subsites (5′-3′)b | ||
| ZFPa | GCCAAGATAATGATTAAAc | GCCa | DCRDLAR |
| AAGa | RKDNLKN | ||
| ATAa | QKSSLIA | ||
| ATGa | RRDELNV | ||
| ATTa | HKNALQN | ||
| AAAc | QRANLRA | ||
| ZFPb | ATGGCAAACAAAAGTTGAa | ATGg | RRDELNV |
| GCAa | QSGDLRR | ||
| AACa | DSGNLRV | ||
| AAAa | QRANLRA | ||
| AGTt | HRTTLTN | ||
| TGAa | QAGHLAS | ||
| ZFPc | AGAGATATAc | AGAg | QLAHLRA |
| GATa | TSGNLVR | ||
| ATAc | QKSSLIA | ||
| ZFPd | AAAAGCAAAc | AAAa | QRANLRA |
| AGCa | ERSHLRE | ||
| AAAc | QRANLRA | ||
| ZFPe | ATAATGATTa | ATAa | QKSSLIA |
| ATGa | RRDELNV | ||
| ATTa | HKNALQN | ||
| ZFPf | AACAAGACAt | AACa | DSGNLRV |
| AAGa | RKDNLKN | ||
| ACAt | SPADLTR | ||
| ZFP1 | ATAAAAATGAAGGCCATTa | ATAa | QKSSLIA |
| AAAa | QRANLRA | ||
| ATGa | RRDELNV | ||
| AAGg | RKDNLKN | ||
| GCCa | DCRDLAR | ||
| ATTa | HKNALQN | ||
The entire DNA-binding site sequence is shown from 5′ to 3′.
Each subsite is shown with its corresponding zinc finger amino acid sequence displayed (in the last column), with amino acid positions from −1 up to +6 representing the amino acids of the alpha helix that make site-specific contacts with the DNA (5).
Cloning, expression, and purification of ZFPs.
ZFPs were codon optimized for Anas platyrhyncos using a database from Blue Heron Biotechnology (Bothell, WA) and then synthesized and cloned into pUC19 vectors by Blue Heron Biotechnology. ZFPs were transferred to pMAL (E8000S NEB) using the flanking XhoI and SpeI sites, creating maltose-binding protein (MBP) fusion proteins. These constructs were expressed in Escherichia coli BL21(DE3) and purified on amylose resin (E8021L NEB) according to the manufacturer's specifications, with the addition of 15% glycerol to the elution buffer. Proteins were quantified by using the Micro-BCA protein assay (catalog no. 23235; Pierce) and stored at −80°C.
EMSA.
dsDNA oligonucleotides (2.5 μM) were incubated for 1 h at room temperature in gel-shift buffer [25 mM Tris-HCl (pH 8), 100 mM NaCl, 2 mM dithiothreitol, 100 μM ZnCl2, 10% glycerol, 50 μg of bovine serum albumin/ml, 4 μg of poly(I-C)/ml, and 0.01% bromophenol blue] with serial dilutions of ZFP from 150 nM down to 9.5 nM (30, 33, 40). The oligonucleotide sequence for the ZFPa target is (AGTACTGCCA AGATAATGAT TAAAAGTACT) and its complement and for the ZFPb target is (AGTACTATGG CAAACAAAAG TTGAAGTACT) and its complement. Reactions were run on 7% nondenaturing polyacrylamide gels at 100 V for 1 h, stained by using a Molecular Probes EMSA kit (E33075; Invitrogen), and then scanned by using a Fujifilm FLA-5100 phosphorimager. EMSAs were quantified by using Fujifilm ImageGauge v4.22 software. Nonlinear regression plots were produced from these data by using the program Enzyme Kinetics (v1.11; Trinity Software).
Radioactive 32P-labeled dsDNA probes for competition EMSAs were made using T4 polynucleotide kinase (catalog no. 18004-010; Invitrogen) according to the manufacturer's specifications, and unincorporated [γ-32P]ATP was removed by using a Qiagen QIAquick nucleotide removal kit (catalog no. 28304) (40). Then, 150 nM ZFP was incubated with 10,000 cpm of radioactive dsDNA probe in gel shift buffer for 1 h at room temperature. Nonradioactive competitor oligonucleotides were added at increasing concentrations prior to the addition of ZFPs. Reactions were run as described above, and then gels were exposed to an image plate overnight at room temperature. Image plates were scanned by using the Fujifilm FLA-5100 phosphorimager.
SPR.
SPR using a Sensor Chip SA (BR-1003-98; BIAcore) was performed by using a BIAcore 3000 as previously described (14), except that 1× HBS-EP (BR-1001-88; BIAcore) was used as the running and sample buffers and 0.5% sodium dodecyl sulfate (SDS) was used as the regeneration buffer. Oligonucleotides were produced by Operon Biotechnologies (Huntsville, AL) and were biotinylated at the 5′ end of the sense strand only. Sequences were as described for the EMSA. Analysis was done on the BIAeval program by fitting the data to the 1:1 binding with a drifting baseline model.
Cloning of ZFPs into eukaryotic expression vector.
Primers encoding a simian virus 40 nuclear localization signal and a His6 tag at the 5′ end were used to amplify each ZFP by PCR. ZFPa was amplified with the primers ZFPa.fw (GGATCCATGC ATCATCACCA TCACCATCCC AAGAAAAAGC GTAAGGTCCT CGAACCCGGC GAAAAGCCTT AT) and ZFPa.rv (GAATTCACTT GTCTTCTTAC CTGTGTGG), ZFPb with ZFPb.fw (GGATCCATGC ATCATCACCA TCACCATCCC AAGAAAAAGC GTAAGGTCCT CGAACCAGGT GAAAAACCCT) and ZFPb.rv (GAATTCTGAA GTCTTCTTTC CTGTGTGA) and ZFP1 with ZFP1.fw (GGATCCATGC ATCATCACCA TCACCATCCC AAGAAAAAGC GTAAGGTCCT GGAACCCGGC GAGAAAC) and ZFP1.rv (GAATTCGGAG GTCTTTTTTC CGGTGTG). PCR products were cloned into pCR4 using the TOPO TA cloning kit (K4530-20; Invitrogen) and then transferred into the eukaryotic expression vector pcDNA3.1(+) (V790-20; Invitrogen) using BamHI and EcoRI restriction endonuclease sites. Enhanced green fluorescent protein (EGFP) was cloned into pCR4 as described above following PCR amplification from pAdTrack-CMV with these primers: EGFP.fw (GAATTCGCCA CAATGGTGAG CAAGGGCGAG G) and EGFP.rv (GTCTGACAGA ACATCAAAGA ACCC). EGFP was then fused in frame to ZFPa and ZFPb by EcoRI/NotI digestion of pcDNA3.1(+)-ZFPa and pcDNA3.1(+)-ZFPb and ligation of EGFP.
Cell lines and culture conditions.
Longhorn male hepatoma (LMH) cells were maintained in 1:1 MEM-F-12 medium (MEM, catalog no. 11700-077 Gibco; F-12, catalog no. 21700-026, Gibco) supplemented with 10% fetal calf serum (catalog no. 12483-020; Gibco), 50 IU of penicillin/ml, 10 μg of streptomycin/ml, and 1 mM glutamine. LMH cells were cotransfected with 1 μg of pDHBV1.3 and 3 μg of pcDNA3.1(+) or pcDNA3.1(+)-ZFPa, -ZFPb, or -ZFP1 using Lipofectamine 2000 (catalog no. 11668-027; Invitrogen) according to the manufacturer's specifications with a DNA/Lipofectamine 2000 ratio of 2:1.
Confocal microscopy.
LMH cells were transfected with 4 μg of pcDNA3.1(+)-ZFPa-EGFP or pcDNA3.1(+)-ZFPb-EGFP in 32-mm dishes with glass coverslips affixed. After 24 h, 10 μl/ml of 0.1-mg/ml Hoechst 33342 (catalog no. 14533; Biochemika) was added to the medium, and the cells were incubated at 37°C in 5% CO2 for 15 min. The medium was replaced, and live cells were visualized by using the Zeiss NLO510 multiphoton microscope. The emission and excitation wavelengths were, respectively, 488 and 509 nm for EGFP and 355 and 465 nm for Hoechst 33342.
Isolation of viral nucleic acids.
Intracellular viral (ICV) DNA was isolated 48 h posttransfection as previously described (41). Briefly, cells were lysed in 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.3% Triton X-100, and 8% sucrose. Nuclei and cellular debris were pelleted by centrifugation at 13,000 × g for 10 min, and then the supernatants were incubated at 37°C for 30 min with 6 mM MgCl2, 100 μg of DNase I/ml, and 10 μg of RNase A/ml to digest the cellular nucleic acids. Samples were centrifuged again as described above, and virus was precipitated from the supernatants with 0.3 volumes of 26% polyethylene glycol 8000, 1.4 M NaCl, and 10 mM EDTA overnight at 4°C. Virus was pelleted by centrifugation as described above and resuspended in 100 μl of 50 mM Tris-HCl (pH 8), 150 mM NaCl, and 10 mM EDTA. Samples were incubated overnight at 42°C with 800 μg of proteinase K/ml and 0.1% SDS to digest capsid and polymerase and then phenol-chloroform extracted. DNA was precipitated with 10 μg of yeast tRNA as carrier, a 0.1 volume of 3 M sodium acetate, and a 2× volume 95% ethanol. After centrifugation at 13,000 × g for 10 min, viral DNA was resuspended in 15 μl of DNA loading buffer, and the entire sample was used for Southern analysis.
RNA isolation and quantitative PCR.
RNA was isolated 24 h posttransfection by using TRIzol reagent (catalog no. 15596-018; Invitrogen) according to the manufacturer's specifications. cDNA was produced from 1 μg of total RNA by using oligo(dT)20 (catalog no. 18418-020; Invitrogen) and SuperScript II reverse transcriptase (catalog no. 18064-022; Invitrogen) according to the manufacturer's specifications. Quantitative PCR was performed on a Roche LightCycler using a LightCycler FastStart DNA MasterPLUS Sybr Green I kit (catalog no. 3515885001; Roche) and the following primer pairs: primers DHBV.2553.fw (AGCTGCTTGC CAAGGTATCT TT) and DHBV.2752.rv and primers Chick.GAPDH.25.fw (GTTGACGTGC AGCAGGAACA CT) and Chick.GAPDH.222.rv (CTTGAAGTGT CCGTGTGTAG AATC). The plasmid pDHBV1.3 was used as the standard for the pregenomic assessment, and chicken GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was cloned into the pCR4 vector as described above, which was used as the standard for GAPDH.
Northern blot.
Total RNA was heated to 50°C for 60 min in the presence of dimethyl sulfoxide, 6 M glyoxal, and 1× morpholinepropanesulfonic acid buffer. The denatured RNA was run on a 1% 1× morpholinepropanesulfonic acid agarose gel and transferred to Hybond-XL by Northern transfer overnight in 7.5 mM NaOH (RPN303S; Amersham Biosciences). Membranes were prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt reagent, and 0.1% SDS for 4 h at 65°C, and then DHBV or GAPDH specific DNA probe was added overnight. Membranes were washed twice with 1× SSC-0.1% SDS and twice with 0.1× SSC-0.1% SDS and then imaged with an image plate on a Fujifilm FLA-5100 phosphorimager.
SDS-polyacrylamide gel electrophoresis and Western blot.
LMH cells were washed with phosphate-buffered saline (PBS) and lysed in RIPA buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholic acid, 0.025% NaN3) at 24 h posttransfection. Proteins were separated by SDS-10% polyacrylamide gel electrophoresis and then transferred to Hybond-ECL nitrocellulose membranes (RPN303D; Amersham Biosciences). Primary antibodies to DHBV core (J112) and DHBV preS (1H1) were provided by Jesse Summers (University of New Mexico) and Pat Nakajima (Fox Chase Institute), respectively. The horseradish peroxidase (HRP)-labeled goat anti-rabbit (catalog no. 1706515; Bio-Rad) and goat anti-mouse (catalog no. 115-035-174; Jackson Immunoresearch) antibodies, respectively, were used as secondary antibodies. Actin was assessed by using an anti-actin antibody (MAB1501; Chemicon) and HRP-labeled goat anti-mouse secondary antibody. EGFP was assessed by using an anti-GFP antibody (catalog no. 33-2600; Zymed) and the HRP-labeled goat anti-mouse secondary described above. Proteins were visualized by chemiluminescence detection.
Viability assay.
LMH cells were plated at 2 × 104 cells/well in 96-well plates and transfected as described above. At 24 h after transfection, 10 μl of 3-(4, 5-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide) (MTT) at 5 mg/ml in PBS was added to the cells for 2 h, followed by incubation at 37°C in 5% CO2. Cells were washed once with PBS, and then 100 μl of isopropanol supplemented with 0.1 N HCl was added to each well for 5 min before measurement at 570 nm on a Spectramax Plus plate reader (Molecular Devices).
Statistical analysis.
Data from the MTT assay, quantitative PCR, and Southern blots were analyzed in Microsoft Excel 2004 for Mac (v11.3.6) by using two-tailed paired t tests for two samples for means.
RESULTS
Design of ZFPs targeted to the DHBV enhancer region.
To interfere with the binding of transcription factors and the movement of the transcriptional machinery along the DHBV episome, we designed ZFPs to bind the DHBV enhancer region where other cis-acting transcription factors, such as hepatocyte nuclear factor 1 (HNF1), HNF3, and CCAAT/enhancer binding protein beta (C/EBPβ), are known to bind (Fig. 1B) (23, 26). To ensure that off-target binding did not occur, we performed BLAST searches of the DNA target sites against the chicken genome and found no matches. The chicken genome was searched because the duck genome is not sequenced, and our DHBV tissue culture system uses chicken hepatoma cells.
Assessment of dissociation constants and binding affinities using EMSA.
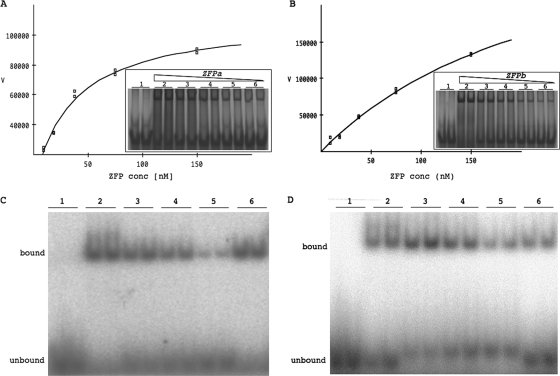
To determine the dissociation constants of the designed ZFPs, ZFPs were first purified, and then DNA binding was assessed by EMSA. Two of the six ZFPs caused a shift in the mobility of their cognate dsDNA oligonucleotide, indicating binding by the ZFP to the target DNA (Fig. 2A and B). The dissociation constants (Kd), calculated by nonlinear regression, were 37.0 nM for ZFPa and 179.0 nM for ZFPb, indicating that their affinities for their respective target sites are strong. The control ZFP, ZFP1, did not bind the ZFPa binding site (data not shown).
FIG. 2.
Determination of affinity and specificity of ZFPa and ZFPb. The nonlinear regression plots of ZFPa (A) and ZFPb (B) are derived from quantifying the EMSA (inset). The inset EMSA shows the unbound probe in the absence of ZFP (lane 1) and the mobility shift in the presence of ZFP at 150 nM (lane 2), serial diluted 1 in 2 (lanes 3 to 5) down to 9.5 nM (lane 6). Each sample is in duplicate and was repeated three times. A representative experiment is shown. The x axis (V) represents quantification of ZFP binding to oligonucleotide using arbitrary units. Competition EMSAs for ZFPa (C) and ZFPb (D). Lane 1, 32P-labeled specific oligonucleotides without ZFP; lane 2, 150 nM ZFP with labeled specific oligonucleotides; lanes 3 to 5, 150 nM ZFP with labeled specific oligonucleotides and 5, 10, or 50 μM (respectively) of unlabeled specific oligonucleotides; lane 6, 150 nM ZFP with labeled specific oligonucleotides and 50 μM concentrations of unlabeled nonspecific oligonucleotides. Each samples is in duplicate and was performed three times. A representative experiment is shown.
The specificity of the designed ZFPs to their target sequence was assessed by competition EMSA (32, 39). By adding excess unlabeled (nonradioactive) oligonucleotides, we were able to see competition by specific unlabeled oligonucleotides (Fig. 2C and D, lanes 3 to 5) but not with nonspecific unlabeled oligonucleotides (Fig. 2C and D, lane 6), indicating our ZFPs bound specifically to their target oligonucleotides.
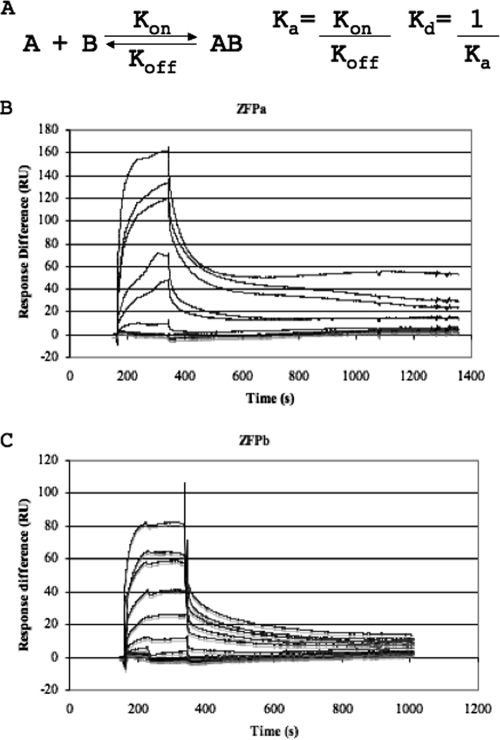
Assessment of dissociation constants using SPR.
We used SPR analysis to further model the binding kinetics of the ZFPs. SPR measures real-time interactions between a ligand anchored to a detection surface and an analyte that flows over the detection surface. The general kinetic equation for the binding of ZFP to its target DNA describes a 1:1 binding scenario (Fig. 3A). We produced kinetic graphs of ZFP binding target oligonucleotides (Fig. 3B and C) and calculated the Kd for each ZFP using 1:1 binding models on the BIA evaluation software. The Kds calculated by SPR were 12.3 nM for ZFPa and 40.2 nM for ZFPb, confirming the nanomolar range dissociation constants obtained by EMSA.
FIG. 3.
Kinetic analysis of ZFPa and ZFPb. (A) The kinetic equation describing the binding relationship of a ZFP (A) to its target DNA (B), with the dissociation (Kd) and association constants (Ka) shown. (B and C) SPR analysis of ZFPa (B) and ZFPb (C) was performed. Each line represents duplicate analysis of different concentrations of ZFP binding to surface-bound oligonucleotides over time. Response difference is measured in resonance units (RU) and represents the binding of the ZFP to the anchored oligonucleotides. Three blanks were also performed in duplicate.
Viral RNA production in the presence of ZFPs.
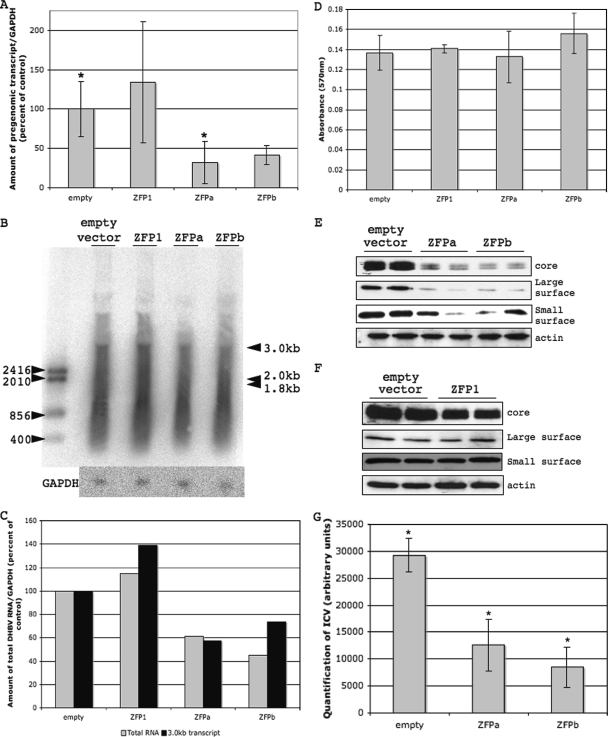
To assess the effects of ZFPs on viral transcription, we expressed each ZFP in a DHBV tissue culture system. In order to ensure ZFP localization in the nucleus of cells, where the plasmid driving DHBV transcription is located, we engineered an simian virus 40 nuclear localization signal onto each ZFP. After cotransfection, production of viral pregenomic RNA was significantly decreased in the presence of ZFPa compared to empty vector control (31.9% of control) and substantially decreased by ZFPb compared to the empty vector (41.6% of control), as determined by quantitative PCR (Fig. 4A). In addition, cotransfection of a ZFP with scrambled zinc finger domains (ZFP1) did not cause any decrease in viral RNA production (Fig. 4A). DHBV produces three transcripts: the pregenomic transcript at 3.0 kb and the large and small surface antigen transcripts at 2.0 and 1.8 kb, respectively (7). The two smaller transcripts are entirely overlapped by the pregenomic transcript and cannot be assayed directly using quantitative PCR; therefore, we performed Northern blots on total RNA. We saw a slight trend for decreased DHBV transcript production in the presence of ZFPa and ZFPb (Fig. 4B), which is highlighted further by quantification of the Northern blot and normalization to the GAPDH loading control (Fig. 4C). Normalization shows total RNA and pregenomic RNA both decreased in the presence of ZFPa (61.2 and 57.2% of control, respectively) and ZFPb (45.3 and 73.5% of control, respectively) but increased slightly in the presence of the control ZFP, ZFP1, which mimics the trend seen by quantitative PCR. To ensure the difference in protein expression was not due to ZFP toxicity, we performed MTT assays on transfected cells and found no difference in cell viability in cells expressing any ZFP compared to cells transfected with the empty vector (Fig. 4D).
FIG. 4.
Determination of the effects of ZFPs on the DHBV life cycle in tissue culture cells. LMH cells were cotransfected with pDHBV1.3 and pcDNA3.1(+)-ZFPa, -ZFPb, and -ZFP1 or empty vector. (A) Total RNA was collected and reverse transcribed into cDNA, upon which quantitative PCR was performed with DHBV-specific or chicken GAPDH-specific primers. The amount of DHBV was normalized to the amount of GAPDH in each sample. (B) Total RNA was also run on a Northern blot, which was probed in parallel with either a DHBV-specific (top) or a chicken GAPDH-specific (bottom) probe. The DHBV transcript sizes are shown by the black arrows on the right. (C) Total DHBV RNA (░⃞) or the 3.0-kb transcript (▪) were quantified from the Northern blot, normalized to GAPDH, and plotted as a percentage of the control. (D) The toxicity of ZFPs in LMH cells was measured by using an MTT assay after 24 h. Uptake of dye by cells indicates cell viability. There was no statistically significant difference between empty vector and ZFP-transfected cells. This was repeated three times. (E and F) LMH cells were harvested after 24 h, and total cell lysates were assessed for DHBV core, large, and small surface levels, with actin as a loading control. This was repeated three times. (G) LMH cells were harvested after 48 h for ICV DNA. ICV DNA was analyzed by Southern blot and quantified by using a Fujifilm FLA-5100 phosphorimager. The chart is the quantification by the phosphorimager of duplicates. *, P < 0.05 (two-tailed paired t test [mean of two samples]).
Viral protein expression in the presence of ZFPs.
To further characterize the effects of ZFPs on viral transcription, we assessed viral core and surface protein expression in the presence of each ZFP and found a dramatic reduction in the amount of all viral proteins (Fig. 4E). Further, transfection with a ZFP with scrambled zinc finger domains, ZFP1, did not cause any decrease in surface protein production and only a minor decrease in core protein production (Fig. 4F). The extent of decreased core protein production with ZFPa and ZFPb was dramatically higher than the decrease seen with ZFP1. Peak activity of ZFPa and ZFPb was detected 24 h posttransfection, and by 72 to 96 h posttransfection there was no effect of the ZFPs on DHBV protein production (data not shown).
ICV production in the presence of ZFPs.
To determine whether decreased viral RNA and protein production had an impact on viral progeny production, we assessed the effects of ZFPs on ICV DNA. We found that ICV DNA production was significantly decreased in the presence of ZFPa and ZFPb (Fig. 4G). This suggests that the reduction in viral RNA and proteins results in the decreased production of virus progeny, suggesting the binding of ZFPs to DHBV DNA in this tissue culture system can specifically reduce the virus's ability to replicate.
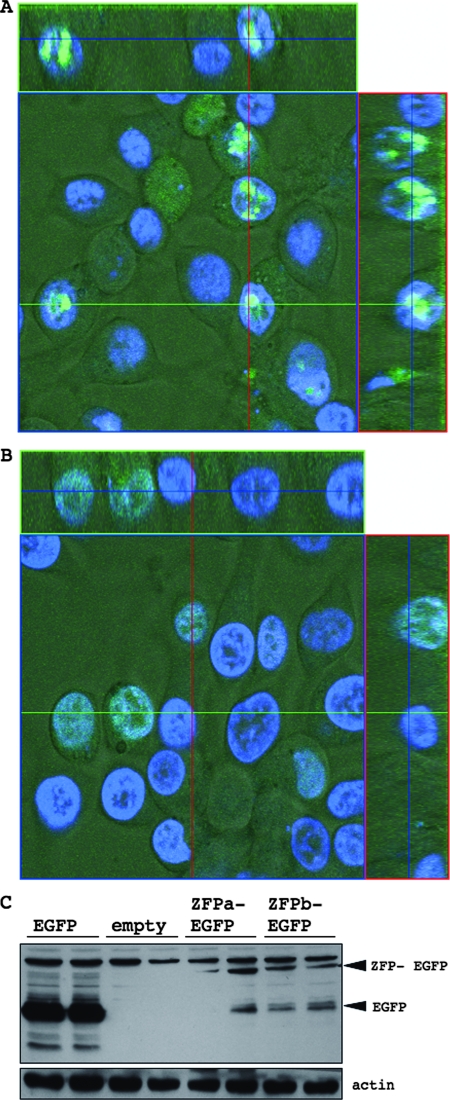
Expression and localization of ZFPs.
To confirm the expression of the designed ZFPs, we transfected LMH with ZFPs fused to EGFP and visualized the ZFPs by using confocal microscopy. Cells were costained with Hoeschst 33342 to visualize the nucleus. Both ZFPa and ZFPb are found predominantly in the nucleus of cells (Fig. 5A and B), although the distribution within the nucleus differed between the two ZFPs. ZFPb was distributed homogeneously throughout the nucleus, whereas ZFPa appeared to collect into focused regions in the nucleus. Expression of the ZFPs were also confirmed by Western blotting on total cell lysates using an anti-GFP antibody (Fig. 5C).
FIG. 5.
Expression of ZFPa- and ZFPb-EGFP fusion proteins in LMH cells. LMH cells were transfected with pcDNA3.1(+)-ZFPa-EGFP (A) or -ZFPb-EGFP (B) and visualized by confocal microscopy of live cells after 24 h. Cells were costained with Hoechst 33342 (blue). (C) Total cell lysates of LMH cells transfected as described above were harvested after 24 h and assessed for EGFP levels, with actin as a loading control. The positive control was pcDNA3.1(+)-EGFP and the empty vector was pcDNA3.1(+) alone.
DISCUSSION
HBV infection frequently results in serious liver complications, including cirrhosis, end-stage liver disease, and hepatocellular carcinoma. Therapy with nucleoside analogs can significantly reduce the viral load, normalize liver enzymes, and improve liver histopathology in many patients. However, the “cure” rate by treatment with nucleoside analogs is low, and many patients experience a relapse once antiviral therapy is stopped.
The first generation of nucleoside analog therapeutics for HBV was lamivudine, which is a deoxycytidine analog (37). Many patients treated with lamivudine can achieve undetectable HBV DNA in the serum within 2 to 3 months of treatment, and 49 to 56% have improved liver histology after 1 year of therapy (39). However, 20 to 50% of HBeAg-positive patients and 90% of HBeAg-negative patients experience a resurgence of viremia, as measured by HBV DNA in sera (39). In addition, the occurrence of HBV resistance to lamivudine is 15% after 1 year of treatment and up to 70% after 5 years of treatment (35, 39). Newer nucleoside analogues such as adefovir, dipivoxil, and entecavir are treatment options for lamivudine-resistant patients, but resistance to these drugs is still occurring, albeit at a lower rate (39).
The persistence of HBV is due to its highly stable cccDNA, which has a calculated half-life of approximately 33 to 50 days (2, 43). While nucleoside analogs function by inhibiting the virion-associated viral polymerase, which suppresses viral DNA replication (35, 39), only entecavir has shown some evidence of directly impacting the amount of cccDNA in animal models of HBV. In models of chronic HBV infection, such as woodchuck hepatitis B virus and DHBV, entecavir was able to reduce the amount of cccDNA in the liver; however, in both models, viral rebound occurred in some animals 2 to 12 weeks after treatment was stopped (16, 29). In humans, entecavir treatment has not shown any significant improvement compared to lamivudine treatment in the key serological markers that would be expected to correlate with a sustained virological response after therapy was stopped (8). The development of new nucleoside analogs has offered limited therapeutic advantages with respect to directly decreasing the cccDNA of HBV infection. We believe that specifically targeting cccDNA is an important direction to improve the treatment and sustained virological response rates for chronic HBV carriers.
We have designed ZFPs that specifically target the cccDNA of DHBV infection and inhibit viral transcription and replication. Compared to nucleoside analogs, which interfere with the viral feedback loop to replenish cccDNA, ZFPs are a unique and direct method to target cccDNA. We have demonstrated that ZFPs bind to their target sequence with nanomolar affinities, ranges that are required for further development of therapeutics. Expression of the ZFPs in LMH cells undergoing the DHBV viral life cycle resulted in decreased expression of viral RNA and protein expression compared to the empty vector control, without any apparent toxicity effects. In addition, the production of viral particles was also decreased in the presence of the expressed ZFPs.
The designed ZFPs target the DHBV enhancer, which is known to control the core and small surface promoters, but not the large surface promoter (7, 9, 25, 34). Our data show that the designed ZFPs inhibit not only core and small surface protein production but also large surface protein production. All three DHBV transcripts include 3′-untranslated regions that span the enhancer region prior to the polyadenylation signal (7). It is possible that the bound ZFPs are sterically hindering the RNA polymerase and preventing readthrough across the enhancer region. This would result in incomplete transcripts being produced that lack the stability of the poly(A) tail. In this way, the ZFPs may be having dual effects on DHBV—(i) inhibition of enhancer activity on core and small surface promoters and (ii) steric hindrance of RNA polymerase across the enhancer—resulting in a reduction of stable complete transcripts.
ZFP therapeutics can be delivered by gene therapy or in protein form by using a number of different delivery methods. Replication-incompetent adenoviruses offer a gene therapy approach for delivering ZFPs, since they can hold up to 10 kb of exogenous DNA, induce little in the way of immune responses, and have relatively small risk of integration into the host genome because they do not replicate within host cells (18, 21). Our lab has shown that adenoviruses are efficient at transducing genetic material into primary duck hepatocytes isolated from congenitally DHBV-infected Pekin ducks (41), and other work has shown that adenoviruses are hepatotropic in mouse models (4, 22). As well, adenoviruses have been used to deliver ZFPs in a mouse model of wound healing (32) and have been shown to maintain the liver expression of delivered genes for 2.5 years in mice (19). ZFPs as protein therapeutics can be delivered by using liposomes, nanoparticles, or synthetic polymers. Liposomes are small vesicles that surround an aqueous phase with one or more phospholipids bilayers (21). Hydrophilic drugs can be encapsulated within the aqueous phase, while hydrophobic drugs can be associated with the bilayer (21). Liposomes can be taken up by a number of cell types, including monocytes and macrophages, spleen cells and, importantly, liver cells. Incorporation of polyethylene glycol into the liposome bilayer can enhance uptake by the spleen and liver because it prevents liposome uptake by phagocytic cells. In addition, cationic liposomes can be used to deliver DNA as a gene therapy approach (21); thus, liposomes may be an option to deliver ZFPs either in protein format or through gene delivery. Nanoparticles, which are solid, polymeric particles, are another option for delivery to the lungs, spleen, bone marrow, and liver (21). Natural or synthetic polymers can be used, and drugs can be incorporated into or bound to the polymer (21). Another approach to drug delivery includes erythrocyte ghosts or bacterial ghosts. In both cases, the cells are nonliving yet intact and can be filled with drugs, protein therapeutics, or DNA. Although bacterial ghosts are more likely targeted to phagocytic cells, erythrocyte ghosts are effectively targeted to the liver and could be used to deliver ZFPs (21).
We expect little immune response against therapeutic ZFPs because the liver is known to be an immune tolerant organ (reviewed in references 6 and 13). The liver is continually exposed to dietary and commensal antigens via the portal vein. Without a tolerogenic status, inflammatory responses and immune activation would constantly be occurring in the liver (13). In addition to oral tolerance, the tolerance of the liver is further highlighted by the high rate of allograft acceptance in liver transplants, which is thought to be due to soluble major histocompatibility complex class I secretion, donor leukocytes, hepatic dendritic cells (DC), and hepatocytes themselves (6). Antigen-presenting cells in the liver, such as liver sinusoidal endothelial cells, Kupffer cells, and hepatic DCs, poorly activate inflammatory responses and Th1 T-cell differentiation in vivo (13, 20). They also secrete molecules that encourage a tolerant environment (13, 20), such as interleukin-10, transforming growth factor β, and prostanoids (13). Hepatic DCs, in particular, have been found to induce liver tolerance in vivo (13) and are weakly immunogenic compared to spleen-derived DCs (1). The tolerance of the liver is further demonstrated in a transgenic mouse model displaying peripheral tolerance of a liver-specific antigen. When naive T cells recognizing this antigen were adoptively transferred into the mice, the specific CD8+ T cells did not infiltrate the liver, even if they were activated against a skin graft bearing the antigen (24), indicating immune exclusion from the liver. Thus, the immune exclusion and tolerance of the liver should allow little immune response against ZFPs if used as therapeutics.
In summary, we offer evidence of a novel approach to directly target the cccDNA of DHBV using designed ZFPs that specifically bind sequences within the DHBV enhancer.
Acknowledgments
This study was supported by grant MOP 57824 from the Canadian Institute for Health Research, by studentship funding from the Alberta Heritage Foundation for Medical Research and the Natural Sciences and Engineering Research Council of Canada, and by an infrastruture grant from the Canadian Foundation for Innovation.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Abe, M., F. Akbar, N. Horiike, and M. Onji. 2001. Induction of cytokine production and proliferation of memory lymphocytes by murine liver dendritic cell progenitors: role of these progenitors as immunogenic resident antigen-presenting cells in the liver. J. Hepatol. 3461-67. [DOI] [PubMed] [Google Scholar]
- 2.Addison, W. R., W. W. Wong, K. P. Fischer, and D. L. J. Tyrrell. 2000. A quantitative competitive PCR assay for covalently closed circular form of the duck hepatitis B virus. Antivir. Res. 4827-37. [DOI] [PubMed] [Google Scholar]
- 3.Blancafort P., D. J. Segal, and C. F. Barbas III. 2004. Designing transcription factors architectures for drug discovery. Mol. Pharmacol. 661361-1371. [DOI] [PubMed] [Google Scholar]
- 4.Brave, A., K. Ljungberg, B. Wahren, and M. A. Liu. 2007. Vaccine delivery methods using viral vectors. Mol. Pharm. 418-32. [DOI] [PubMed] [Google Scholar]
- 5.Beerli, R. R., D. J. Segal, B. Dreier, and C. F. Barbas III. 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 9514628-14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benseler, V., G. W. McCaughan, H. J. Schlitt, G. A. Bishop, D. G. Bowen, and P. Bertolino. 2007. The liver: a special case of transplantation tolerance. Semin. Liver Dis. 27194-213. [DOI] [PubMed] [Google Scholar]
- 7.Buscher, M., W. Reiser, H. Will, and H. Schaller. 1985. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell 40717-724. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. T., R. G. Gish, R. de Man, A. Gadana, J. Sollan, Y. C. Chao, A. S. Lok, K. H. Han, Z. Goodman, J. Zhu, A. Cross, D. DeHertogh, R. Wilber, R. Colonno, and D. Apelian. 2006. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 3541001-1010. [DOI] [PubMed] [Google Scholar]
- 9.Crescenzo-Chaigne, B., J. Pillot, A. Lilienbaum, M. Levrero, and E. Elfassi. 1991. Identification of a strong enhancer element upstream from the pregenomic RNA start site of the duck hepatitis B virus genome. J. Virol. 653882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreier, B., D. J. Segal, and C. F. Barbas III. 2000. Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J. Mol. Biol. 303489-502. [DOI] [PubMed] [Google Scholar]
- 11.Dreier, B., R. R. Beerli, D. J. Segal, J. D. Flippin, and C. F. Barbas III. 2001. Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 27629466-29478. [DOI] [PubMed] [Google Scholar]
- 12.Dreier, B., R. P. Fuller, D. J. Segal, C. Lund, P. Blancafort, A. Huber, B. Koksch, and C. F. Barbas III. 2005. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 28035588-35597. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, W., S. A. Shu, E. Gershwin, and Z. X. Lian. 2007. The current immune function of hepatic dendritic cells. Cell Mol. Immunol. 4321-328. [PubMed] [Google Scholar]
- 14.Galio, L., S. Briquet, and C. Vaquero. 1999. Real-time study of interactions between a composite DNA regulatory region (HIV-1 LTR NRE) and several transcription factors of nuclear extracts. Biochem. Biophys. Res. Commun. 2646-13. [DOI] [PubMed] [Google Scholar]
- 15.Ganem, D., and R. Schneider. 2005. Hepadnaviridae, p. 2977-3030. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 16.Genovisi, E. V., L. Lamb, I. Medina, D. Taylor, M. Seifer, S. Innaimo, R. J. Colonno, D. N. Standring, and J. M. Clark. 1998. Efficacy of the carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob. Agents Chemother. 423209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson, A. C., J. C. Miller, and C. O. Pabo. 2003. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug Discov. 2361-368. [DOI] [PubMed] [Google Scholar]
- 18.Jozkowicz, A., and J. Dulak. 2005. Helper-dependent adenoviral vectors in experimental gene therapy. Acta Biochim. Pol. 52589-599. [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, I. H., A. Jozkowicz, P. A. Piedra, K. Oka, and L. Chan. 2001. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. USA 9813282-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knolle, P. A., E. Schmitt, S. Jin, T. Germann, R. Duchmann, S. Hegenbarth, G. Gerken, and A. W. Lohse. 1999. Induction of cytokine production in naive CD4+ T cells by antigen-presenting murine liver sinusoidal endothelial cells but failure to induce differentiation toward Th1 cells. Gastroenterology 1161428-1440. [DOI] [PubMed] [Google Scholar]
- 21.Lanao, J. M., E. Briones, and C. I. Colino. 2007. Recent advances in delivery systems for anti-HIV1 therapy. J. Drug Target 1521-36. [DOI] [PubMed] [Google Scholar]
- 22.Lelliott, C. J., A. Ljungberg, A. Ahnmark, L. William-Olsson, K. Ekroos, A. Elmgren, G. Arnerup, C. C. Shoulders, J. Oscarsson, and D. Linden. 2007. Hepatic PGC-1β overexpression induces combined hyperlipidemia and modulates the response to PPARα activation. Arterioscler. Thromb. Vasc. Biol. 272707-2713. [DOI] [PubMed] [Google Scholar]
- 23.Lilienbaum, A., B. Crescenzo-Chaigne, A. A. Sall, J. Pillot, and E. Elfassi. 1993. Binding of nuclear factors to functional domains of the duck hepatitis B virus enhancer. J. Virol. 676192-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limmer, A., T. Sacher, J. Alferink, M. Kretschmar, G. Schonrich, T. Nichterlein, B. Arnold, and G. J. Hammerling. 1998. Failure to induce organ-specific autoimmunity by breaking of tolerance: importance of the microenvironment. Eur. J. Immunol. 282395-2406. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C., L. D. Condreay, J. B. E. Burch, and W. Mason. 1991. Characterization of the core promoter and enhancer of duck hepatitis B virus. Virology 184242-252. [DOI] [PubMed] [Google Scholar]
- 26.Liu, C., W. S. Mason, and J. B. E. Burch. 1994. Identification of factor-binding sites in the duck hepatitis B virus enhancer and in vivo effects of enhancer mutations. J. Virol. 682286-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locarnini, S., and M. Omata. 2006. Molecular virology of hepatitis B virus and the development of antiviral drug resistance. Liver Int. 2611-22. [Google Scholar]
- 28.Mandell, J. G., and C. F. Barbas III. 2006. Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 34(Web Server issue)W516-W523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marion, P. L., F. H. Salazar, M. A. Winters, and R. J. Colonno. 2002. Potent efficacy of entecavir (BMS-200475) in a duck model of hepatitis B virus replication. Antimicrob. Agents Chemother. 4682-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, M., Y. Choo, and A. Klug. 2001. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl. Acad. Sci. USA 981432-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollicino, T., L. Belloni, G. Raffa, N. Pediconi, G. Squadrito, G. Raimondo, and M. Levero. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130823-837. [DOI] [PubMed] [Google Scholar]
- 32.Rebar, E. J., Y. Huang, R. Hickey, A. K. Nath, D. Meoli, S. Nath, B. Chen, L. Xu, Y. Liang, A. C. Jamieson, L Zhang, S. K. Spratt, C. C. Case, A. Wolffe, and F. J. Giordano. 2002. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat. Med. 81427-1432. [DOI] [PubMed] [Google Scholar]
- 33.Reidling, J. C., and H. M. Said. 2007. Regulation of the human biotin transporter hSMVT promoter by KLF4 and AP-2: confirmation of promoter activity in vivo. Am. J. Physiol. Cell Physiol. 2921305-1312. [DOI] [PubMed] [Google Scholar]
- 34.Schneider, R., and H. Will. 1991. Regulatory sequences of duck hepatitis B virus C gene transcription. J. Virol. 655693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreibman, I. R., and E. R. Schiff. 2006. Prevention and treatment of recurrent hepatitis B after liver transplantation: the current role of nucleoside and nucleotide analogues. Ann. Clin. Microbiol. Antimicrob. 58-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal, D. J., B. Dreier, R. R. Beerli, and C. F. Barbas III. 1999. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl. Acad. Sci. USA 962758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severini, A., X. Liu, J. S. Wilson, and D. L. J. Tyrrell. 1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 391430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepard, C. W., E. P. Simard, L. Finelli, A. E. Fiore, and B. P. Bell. 2006. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol. Rev. 28112-125. [DOI] [PubMed] [Google Scholar]
- 39.Sims, K. A., and A. M. Woodland. 2006. Entecavir: a new nucleoside analog for the treatment of chronic hepatitis B infection. Pharmacotherapy 261745-1757. [DOI] [PubMed] [Google Scholar]
- 40.Smith, J., J. M. Berg, and S. Chandrasegaran. 1999. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 27674-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters, K. A., M. A. Joyce, W. R. Addison, K. P. Fischer, and D. L. J. Tyrrell. 2004. Superinfection exclusion in duck hepatitis B virus infection is mediated by the large surface antigen. J. Virol. 787925-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2002. Hepatitis B virus. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/hepatitis/whocdscsrlyo20022/en/index3.html.
- 43.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]