Abstract
In the sera of patients infected with hepatitis B virus (HBV), in addition to infectious particles, there is an excess (typically 1,000- to 100,000-fold) of empty subviral particles (SVP) composed solely of HBV envelope proteins in the form of relatively smaller spheres and filaments of variable length. Hepatitis delta virus (HDV) assembly also uses the envelope proteins of HBV to produce an infectious particle. Rate-zonal sedimentation was used to study the particles released from liver cell lines that produced SVP only, HDV plus SVP, and HBV plus SVP. The SVP made in the absence of HBV or HDV were further examined by electron microscopy. They bound efficiently to heparin columns, consistent with an ability to bind cell surface glycosaminoglycans. However, unlike soluble forms of HBV envelope protein that were potent inhibitors, the SVP did not inhibit the ability of HBV and HDV to infect primary human hepatocytes.
In natural infections with hepatitis B virus (HBV), there is an excess of empty noninfectious subviral particles (SVP) that do not contain the viral capsid. SVP are typically present in a 1,000- to 100,000-fold excess relative to the infectious particles (12, 13). They exist in two main forms: spheres of 25 nm in diameter and filaments of 22 nm in diameter with variable length (15, 17). They can contain all three forms of the HBV envelope proteins: L, M, and S. These share a common C terminus, with M containing the pre-S2 domain relative to S and L containing the pre-S1 domain relative to M (15). There is good evidence that during infection a domain within the pre-S1 of L is what interacts with an as-yet-unidentified host receptor(s) (15). Hepatitis delta virus (HDV), which is assembled using the envelope proteins of HBV, also depends upon this pre-S1 domain (26). HDV can be assembled using only the S protein of HBV, but the particles are noninfectious.
SVP from patients are immunogenic and were used with success in the first HBV vaccine (1). Most current HBV vaccines are prepared in yeast from SVP assembled using just the HBV S protein; such SVP are sufficient to protect individuals against HBV and HDV.
The basis for the excess of SVP detected in patients is unexplained, and the biological function of this excess has been largely ignored. Some authors have suggested that SVP might sop up neutralizing antibodies produced by the host and thus increase the ability of the infectious particles to reach susceptible cells (11, 25). It has also been suggested that SVP contribute to a state of immune tolerance that is a precondition for highly productive persistent infection (13). One study with SVP of duck HBV indicated that for infections at low multiplicity SVP could enhance infection, but when present in large amounts they were inhibitory (3). Another study showed that SVP containing the large envelope protein interfered with duck HBV infection (19).
For the present studies we chose to use SVP as produced by transfection procedures. For the following reasons we consider these more defined than SVP obtained from the sera of infected individuals: (i) the latter contain infectious virus as well as SVP; (ii) they may also contain a spectrum of host antibodies either mixed with the SVP or even directly attached to the SVP; and (iii) since the patients producing SVP are chronically infected, the genetic composition of the SVP will be mixed, with a variety of mutant forms. In contrast, with the in vitro approach, we can assemble SVP that contain just the HBV S protein or those with both the S and L proteins.
As described below, we assembled not just different forms of SVP but also HBV and HDV. We thus found that during HBV and HDV assembly, there was not necessarily a great excess of SVP. In addition, we assembled SVP in the absence of HBV and HDV and found that these SVP were able to bind heparin in vitro yet were not able to interfere with the infection of primary human hepatocytes (PHH) by HBV or HDV.
MATERIALS AND METHODS
Cells and viruses.
Assembly of HDV and SVP was achieved in plasmid-transfected Huh7 cells, as previously described (17). pSVL(D3) was used to initiate HDV genome replication (10). pSV24H and pSVLM−S− were used to express the S and L envelope proteins of HBV, respectively (4, 6). HBV was assembled using HepAD38, a cell line expressing HBV under tetracycline-off control, a gift of Christoph Seeger (20). Alternatively, HBV was produced from Huh7 cells transfected with pRVHBV1.5 (a gift from Volker Bruss), a cloned overlength HBV genome containing only the natural HBV promoters (4). Virus particles and SVP were concentrated 100-fold using polyethylene glycol (PEG) precipitation (17) and then resuspended in STE (0.1 M NaCl, 0.01 M Tris-HCl [pH 8.0], 0.001 M EDTA). Alternatively, SVP were concentrated by ultracentrifugation (with a Beckman SW41 rotor at 40,000 rpm for 16 h at 4°C). PHH plated on rat tail collagen were obtained commercially (Lonza, Cambrex, and Cellzdirect) and infected with virus in the presence of 5% PEG, as previously described (17; N. Chai, H. Chang, E. Nicolas, Z. Han, S. Gudima, and J. Taylor, unpublished data).
RNA extraction and quantitative real-time PCR assays.
At 6 days after infection, total cell RNA was extracted with Tri Reagent (Molecular Research Center). Detection of viral RNAs was by quantitative real-time PCR after a reverse transcription step utilizing random oligonucleotide primers (17; Chai et al., unpublished data).
Rate-zonal sedimentation.
Concentrated sources of HDV, HBV, and SVP were dialyzed against STE and then layered on preformed gradients of 10 to 30% sucrose in STE. For HDV and SVP, centrifugation was in a Beckman SW41 rotor at 40,000 rpm for 4 h at 4°C. Fractions of 0.5 ml were collected from above. For HBV the extent of centrifugation was reduced by using an SW60 rotor at 45,000 rpm for 100 min at 4°C, with fractions of 0.3 ml collected.
Electron microscopy.
Samples from the sucrose gradient shown in Fig. 1B were pooled (pools P to R), diluted in STE, and collected by centrifugation. The pellets were gently washed with STE and again subjected to centrifugation. After careful draining of the supernatant, the pellets were resuspended in 0.1 ml of STE and analyzed by electron microscopy, as previously described (17).
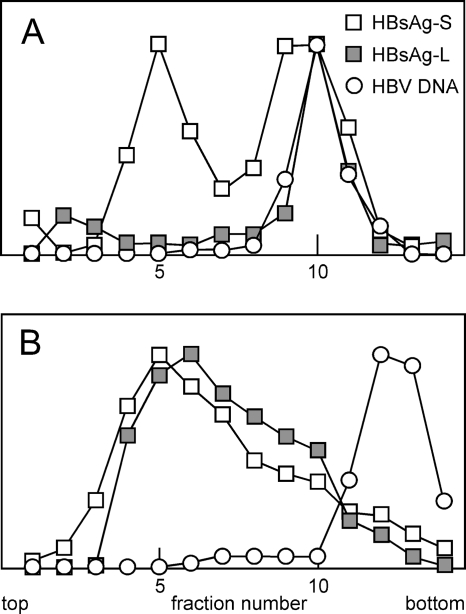
FIG. 1.
Rate-zonal sedimentation of HDV and SVP containing δAg. Huh7 cells were transfected with appropriate plasmids, as described in the text, leading to the assembly and release of particles into the medium. (A) Assembly of HDV with a harvest between days 6 and 9 after transfection. (B) SVP assembly with a harvest between days 0 and 3. In both cases particles were collected by PEG precipitation and analyzed by rate-zonal sedimentation on gradients of 10 to 30% sucrose. Fractions were assayed by immunoblotting for HBV envelope proteins, HBsAg, and δAg and in panel A by real-time PCR for HDV RNA. In both panels the vertical axes use linear scales and arbitrary units. In panel B, pools indicated as P to R were made and assayed by electron microscopy, as shown in Fig. 2.
Heparin-affinity chromatography.
Heparin-agarose beads (Sigma) were washed three times with TN buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl). SVP diluted in TN were applied to 0.1 ml beads with rocking overnight at 4°C. The mixture was transferred to a Micro Bio-Spin column (Bio-Rad) and centrifuged at 1,000 rpm for 20 s, and the flowthrough was collected. The beads in the column were washed three times with TN, then once for each of a series of increasing concentrations of NaCl in TN, and finally with 0.1% sodium dodecyl sulfate. Aliquots of all fractions were assayed by immunoblotting, as described below.
Immunoblotting.
Samples were treated for 10 min at 70°C in Laemmli buffer prior to electrophoresis in precast 10% polyacrylamide gels (NuPage; Invitrogen). After electrotransfer to a nitrocellulose membrane, HBV envelope proteins were detected using as primary antibodies either a rabbit polyclonal antibody specific for the S domain (Fitzgerald) or a rabbit polyclonal antibody specific for a peptide containing the HBV matrix domain, a region spanning the C terminus of pre-S1, and the N terminus of pre-S2 (18). A goat anti-rabbit immunoglobulin G (IgG) coupled to an infrared fluorescent dye was used as the secondary antibody, after which signal was detected with an Odyssey apparatus and quantitated using Odyssey 2.1 software (Li-Cor).
RESULTS
SVP in the presence and absence of HDV and HBV.
As previously described, we are able to assemble HDV using transfection of Huh7 cells with a combination of plasmids (18). Here we used plasmids to express HBV L and S, along with pSVL(D3), a plasmid that initiates HDV genome replication (10). After several days some of the replicating HDV RNA undergoes RNA editing, leading to the synthesis of an altered form of delta antigen (δAg). The original form, the 195-amino-acid small δAg, is essential for the genome replication, while the altered form, the 214-amino-acid large δAg, is essential for the assembly process mediated by the HBV envelope proteins (9, 10).
Figure 1A shows a rate-zonal sedimentation analysis of such particles. The HDV RNA genome and the δAg sedimented together more than halfway down the tube. The smaller particles, in the top half of the gradient, which contained HBV envelope proteins, we refer to as SVP. In terms of HBV envelope content, the small SVP were 5 times more abundant than HDV. We were initially struck by the observation that the SVP did not contain detectable amounts of δAg. However, this actually confirmed an earlier study that used rate-zonal analysis of sera from patients infected with HDV (2).
Others have now shown that the assembly of HDV requires a small cytosolic loop of the HBV S protein (26) and somehow involves an interaction of the large form of δAg and HDV RNA. This together with our result (Fig. 1A) provokes the interpretation that HDV RNA, as a ribonucleoprotein involving large δAg, interacts with the HBV envelope proteins and drives the assembly of HDV particles. As a test of this, we next examined whether any large δAg can be assembled into SVP when HDV RNA is absent.
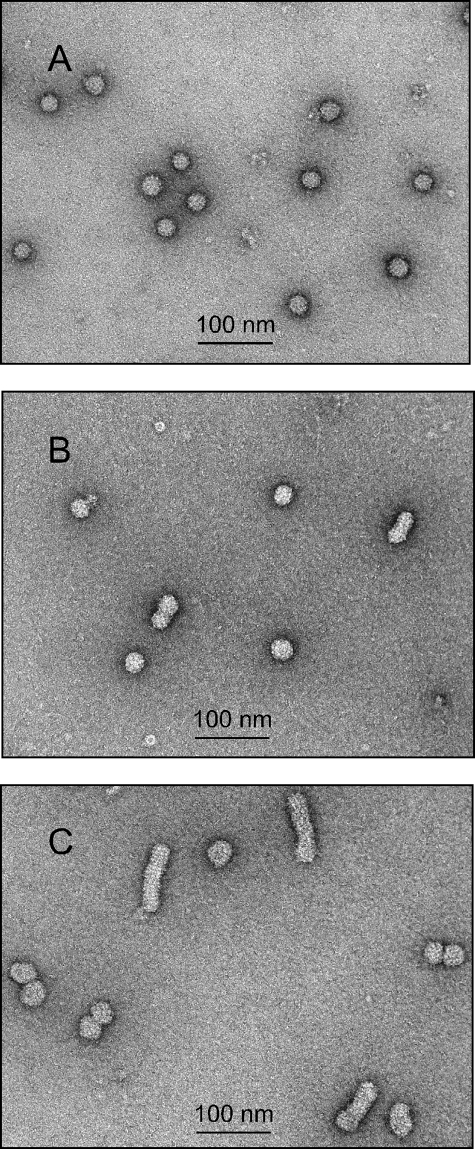
We expressed HBV S together with a plasmid expressing large δAg but in the absence of HDV genome replication. Figure 1B shows that large amounts of small SVP were produced. The δAg was assembled into particles, but the majority had sedimentation values intermediate between those of the small SVP and HDV. Two interpretations are proposed. First, while this assembly was independent of HDV RNA, it might have involved interactions with some forms of host cell RNA species. Second, the assembly might have involved the filamentous forms of SVP that are larger than the small spherical SVP. As a test of this second interpretation, three pools, pools P to R, were made from the gradient in Fig. 1B, after which the particles were collected and examined by electron microscopy; the results are shown in Fig. 2A to C, respectively. As expected, pool P (Fig. 2A) contained largely spherical particles, with <0.1% as filaments. Pool Q (Fig. 2B) contained more filaments (0.8%), while pool R (Fig. 2C) contained many more (11%). This analysis supports but does not establish the second interpretation that most of the assembled δAg was in filaments rather than spheres. The first interpretation, which is not mutually exclusive of the second, nevertheless remains possible. Incidentally, it is worth noting that the filamentous SVP were assembled in the absence of the HBV L protein, a result contrary to the thinking that L protein is needed for such assembly (13). Furthermore, S-only filaments were also observed when large δAg was omitted (data not shown).
FIG. 2.
Electron microscopy of SVP containing δAg. Particles were subjected to rate-zonal sedimentation, fractionation, and the collection of size pools P to R, as shown in Fig. 1B. These materials were concentrated and, after negative staining, examined by electron microscopy, as shown in panels A to C for pools P to R, respectively. For each panel, 10 images were also taken at lower magnification and used to deduce the fractions of filamentous particles as <0.1, 0.8, and 11%, respectively.
Next we tested the assembly of HBV. For this we first used the cell line HepAD38, established by Ladner and colleagues (20). This line expressed an HBV cDNA under the control of a tetracycline-repressed promoter. Particles released from the induced cells were concentrated and then characterized by rate-zonal sedimentation. Since HBV is larger than HDV, the sedimentation conditions were reduced relative to those used for Fig. 1. Figure 3A shows that as expected, the HBV DNA-containing particles sedimented to the bottom half of the gradient. Immunoblot analysis was used to quantitate both HBV S and L envelope proteins. About half of the S protein cosedimented with the DNA, with the remainder in the top half of the gradient, as expected for small spherical SVP. In contrast, the L protein almost all cosedimented with the HBV DNA.
FIG. 3.
Rate-zonal sedimentation of HBV. Culture medium was harvested from HepAD38 cells (A) or Huh7 cells (B) between days 4 and 7 after induction or plasmid transfection, respectively. HBV and associated SVP were concentrated from the medium by PEG precipitation and analyzed by rate-zonal sedimentation. Fractions were assayed by immunoblotting for HBV envelope proteins and by real-time PCR for HBV DNA. From the immunoblot we separately quantitated the L and S forms of the HBV envelope proteins, HBsAg-L and HBsAg-S, respectively. The vertical axes use linear scales and arbitrary units.
In this harvest of media from cells at 4 to 7 days after induction, the SVP represented 45% of the total HBV S envelope protein detected (Fig. 3A). For harvests at 0 to 4 and 7 to 11 days, it was 70 and 23%, respectively (data not shown). Such results with SVP in amounts comparable to HBV are in contrast to citations that HBV-transfected hepatoma cells produce an excess of SVP (13). They are also very different from those for the sera of HBV-infected individuals, which contain huge excesses of SVP (11, 13, 25). Therefore, we performed an assembly experiment in which the cells were of liver origin (Huh7) and transfected using a plasmid where expression of all HBV components was under the control of natural HBV promoters. Sedimentation results for particles harvested between days 4 and 7 are shown in Fig. 3B. It can be seen that there was a major peak of envelope proteins, both S and L, at the fractions expected for SVP. In contrast, in the region of the gradient where HBV DNA was detected, we could not even discern a peak of envelope proteins. That is, in Fig. 3B the SVP were in considerable excess (>94%) relative to the HBV particles. Similar results were obtained for particles harvested at days 7 to 10 (data not shown).
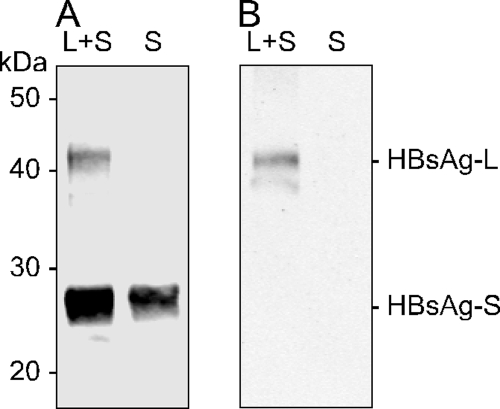
Given the above-described studies of SVP associated with the assembly of HDV and HBV, we next considered the assembly of two kinds of SVP, in the absence of HDV and HBV, with the ultimate aim of testing these for their ability to interfere with HBV and HDV infections. We chose to make forms with S only [SVP(S)] and those from cells expressing equal amounts of L and S [SVP(LS)]. These particles were assembled and analyzed by immunoblotting, and the results are shown in Fig. 4. As expected, the L protein was assembled along with S into SVP, but the relative amount (6%) was less than that of the plasmids expressed in the transfected cells (50%).
FIG. 4.
Immunoblots of SVP(S) and SVP(LS). Huh7 cells were transfected with a plasmid expressing HBV S or with this plasmid plus an equal amount of a plasmid expressing HBV L. SVP released into the medium between days 0 and 3 were concentrated by PEG precipitation, dialyzed against STE, and analyzed by immunoblotting to detect either the HBV S domain (A) or the pre-S domain (B). Molecular mass markers were as indicated.
Such SVP were concentrated 100-fold out of clarified medium either by PEG precipitation or by ultracentrifugation, and then the yield, in terms of the total HBV envelope protein, was determined by immunoblotting relative to a standard of purified HBV SVP, a gift of Dieter Glebe. With such quantitated preparations of SVP(S) and SVP(LS), we next turned our attention to whether they could interfere with the ability of HBV and HDV to infect PHH.
Ability of SVP to interfere with infection of PHH.
In preliminary experiments we observed that such SVP preparations could interfere with the ability of HBV and HDV to infect PHH (data not shown). However, to test the validity of these results, we incorporated three different types of controls.
The first control was to use additional viruses. We used HIV(LS), a human immunodeficiency virus (HIV) pseudotype with the envelope proteins of HBV, and HIV(G), a pseudotype with the envelope protein G of vesicular stomatitis virus (7). These two viruses together with HBV and HDV, can be used to infect PHH, with infection being quantitated 6 days later by specific quantitative real-time PCR assays applied to the total cell RNA (Chai et al., unpublished). As controls for specific inhibition, we would expect HIV(G) to be resistant and HIV(LS) sensitive.
The second control was to produce what we refer to as SVP(−). For this we harvested and concentrated medium from cells that were mock transfected. SVP(−) was used as a negative control for components that might be concentrated from the culture medium along with SVP(S) or SVP(LS).
A third control was to compare SVP with agents that we knew would inhibit HBV and HDV. Specifically, we used S1S2-IA, an immunoadhesin containing both the pre-S1 and pre-S2 domains of HBV L attached to the Fc region of a rabbit IgG (8). Another inhibitor was a chemically synthesized peptide containing part of the HBV pre-S1 sequence (a gift of Stephan Urban) (16).
PHH were treated with either SVP preparations or these compounds for 1 h prior to infection, and then the four-virus mixture, in the presence of 5% PEG and, as indicated, a fresh dose of SVP or the compounds, was added and left for 6 h. After 6 days, total PHH RNA was extracted and assayed by quantitative real-time PCR to detect replication of the four viruses. The results are summarized in Table 1.
TABLE 1.
Ability of potential inhibitors to modulate infection of PHHa
| Potential inhibitor and amt and/or concnb | Relative RNA accumulation (%)c following infection with:
|
|||
|---|---|---|---|---|
| HDV | HBV | HIV(LS) | HIV(G) | |
| None | 100 ± 21 | 100 ± 34 | 100 ± 22 | 100 ± 26 |
| SVP(−) | ||||
| 2 U (0 nM) | 228 ± 206 | 207 ± 94 | 158 ± 56 | 253 ± 77 |
| 50 U (0 nM) | 32 ± 13 | 44 ± 28 | 22 ± 11 | 44 ± 18 |
| SVP(S) | ||||
| 2 U (80 nM) | 220 ± 52 | 170 ± 84 | 123 ± 62 | 183 ± 89 |
| 50 U (2,000 nM) | 13 ± 3 | 36 ± 21 | 18 ± 10 | 27 ± 8 |
| SVP(LS) | ||||
| 2 U (40 nM) | 110 ± 51 | 157 ± 93 | 209 ± 104 | 181 ± 112 |
| 50 U (1,000 nM) | 105 ± 36 | 89 ± 18 | 47 ± 6 | 74 ± 14 |
| S1S2-IA | ||||
| 2 nM | 170 ± 117 | 207 ± 24 | 138 ± 10 | 392 ± 42 |
| 50 nM | <0.6 | 0.6 ± 0.6 | 1.1 ± 0.1 | 117 ± 6 |
| Peptide | ||||
| 2 nM | 25 ± 15 | 187 ± 33 | 139 ± 9 | 409 ± 49 |
| 50 nM | <0.6 | 0.3 ± 0.3 | 0.9 ± 0.4 | 75 ± 18 |
PHH were pretreated for 1 h with the indicated potential inhibitor, and then a mixture of four viruses in 5% PEG, along with the potential inhibitor, was added and left for 6 h. The medium was then replaced and the infection allowed for 6 days, after which total RNA was extracted and quantitative real-time PCR assays were used to determine the amount of replication for each of the four viruses.
The three forms of SVP were prepared from the same volumes of medium harvested between days 0 and 3 after transfection of the cells with either empty plasmid [SVP(−)], HBV S plasmid [SVP(S)], or plasmids expressing HBV L and S [SVP(LS)]. The amounts of the three SVP are expressed in arbitrary units. In addition, for SVP(S) and SVP(LS) we used immunoblots relative to protein standards to determine the final concentrations of HBV envelope proteins in the preparations. For the immunoadhesin S1S2-IA, the concentration was determined as previously described (9), while for the chemically synthesized peptide, the concentration was determined for the purified product (17).
The infections were performed in triplicate, and the values are averages and standard deviations.
Consider first the positive controls. The immunoadhesin S1S2-IA and the synthetic peptide both gave major inhibition of the infection by HDV, HBV, and HIV(LS). This inhibition was achieved at a concentration of 50 nM but not at 2 nM. The inhibition also was specific, in that there was no effect on HIV(G).
The three sources of SVP, however, gave only minor inhibition even for the largest amounts used. The inhibition was also nonspecific, in that SVP(−), which contained no HBV envelope proteins, inhibited all four viruses, including HIV(G). In an attempt to reduce the nonspecific effect of SVP(−), we used an alternative method of SVP preparation, namely, ultracentrifugation. The entire experiment with the results shown in Table 1 was repeated. While we observed some reduction in the nonspecific effects of SVP(−), we were still unable to detect any specific inhibition by either SVP(S) or SVP(LS) (data not shown).
In the two experiments described above, the final concentrations for SVP(S) and SVP(LS) during virus infection were at least 1 μM (25 μg/ml) of HBV S protein sequence. Compared to the immunoadhesin and the synthetic peptide, which strongly blocked infection even at 50 nM, we would conclude that the SVP were at best 20 times less inhibitory. Even if we allow that only 6% of the SVP(LS) envelope proteins were of the pre-S-containing L (Fig. 4), these SVP contained 60 nM of pre-S and yet were still not inhibitory.
A recent cryoelectron microscopy study determined that HBV spherical SVP contain about 48 molecules of envelope protein (14). From this we deduced that the concentration of our SVP(LS) was about 20 nM, yet this was not sufficient to give specific interference with the infection by HDV, HBV, and HIV(LS).
Heparin-affinity chromatography of SVP.
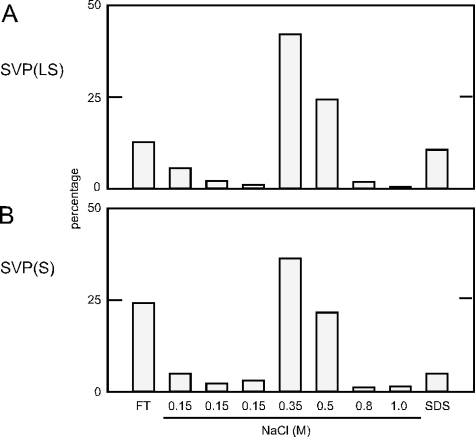
Two recent studies have concluded that HBV initially binds to susceptible cells via cell surface glycosaminoglycans (GAG) and that this interaction is dependent upon the pre-S1 domain of L (21, 24). Also, soluble heparin, which resembles liver GAG, can interfere with infection of hepatocytes by HDV, HBV, and HIV(LS) (Chai et al., unpublished). Therefore, we considered that it would be relevant to determine whether SVP(LS) and SVP(S) would bind to heparin beads, just as has been reported for HBV (27). We performed such affinity chromatography and assayed the fractions via immunoblots. We found that SVP(LS) and SVP(S) bound efficiently and similarly to heparin beads (Fig. 5A and B, respectively). Note that for SVP(LS) we specifically assayed the pre-S domain; that is, we assayed only those SVP that contained the pre-S domain of HBV L.
FIG. 5.
Heparin binding of SVP(S) and SVP(LS). Particles prepared as described for Fig. 4 were subjected to heparin-affinity chromatography. Immunoblots were used to detect particles in the flowthrough (FT), the washes with TN buffer containing 0.15 M NaCl, the eluants with increasing concentrations of NaCl in TN, and a final wash containing sodium dodecyl sulfate (SDS). The results are expressed as percentages of the total recovered material. In panel A, the particles were SVP(LS) and the immunoblotting was to detect the pre-S domain of HBV L. In panel B, the particles were SVP(S) and the immunoblotting was with antibody to detect the HBV S domain.
Since there was no significant difference in the binding and elution of the two forms of SVP (Fig. 5) and since there was no inhibitory effect of such SVP on the infection of PHH by HBV and HDV (Table 1), we are tempted to suggest that heparin binding might not be dependent on the pre-S1 domain and might not even be relevant to the infection of susceptible cells.
DISCUSSION
These studies on the assembly of SVP, either alone or in the context of HDV and HBV assembly, have provided some novel insights. (i) Under our experimental conditions, while HDV genomic RNA and δAg were assembled via HBV envelope proteins into discrete particles, there was only a fivefold excess of SVP produced. Thus, HDV assembly is only somewhat inefficient relative to SVP. (ii) These SVP contained no detectable δAg (Fig. 1A). Apparently, what must have been rate-limiting amounts of δAg were preferentially assembled into HDV particles. (iii) If HDV replication was replaced by expression of large δAg, this protein was assembled by HBV envelope proteins into particles that were larger than 25-nm spherical SVP. Thus, the δAg was probably incorporated into 22-nm filaments (Fig. 1B and 2). (iv) We found that filaments were assembled and released from cells even when only the HBV S protein was expressed (Fig. 2). This is in contrast to studies that concluded that HBV L protein must be present for the formation of filamentous SVP (13). (v) As previously reported, HBV can be assembled from stably transfected HepAD38 cells (20), but we observed that such production was accompanied by only a minor amount of SVP (Fig. 3A). In contrast, when HBV was assembled in Huh7 cells following transfection with a plasmid where all expression was under the control of natural HBV promoters, a great excess of SVP over HBV was produced (Fig. 3B).
There may be many possible reasons why the preparation of HBV assembled from HepAD38 cells contained a relatively small amount of SVP (Fig. 3A). A likely reason is that the HepAD38 cell line contains a cytomegalovirus promoter for the pregenome that is probably much stronger than the downstream promoters of the L mRNA and the shared M and S mRNA (20). Nevertheless, our results show that HBV could be assembled without producing an excess of SVP; that is, HBV assembly is not intrinsically inefficient.
Our study also attempted to address the biological relevance of the SVP. First, we tested the binding of SVP in heparin-affinity chromatography, since it has been reported that HBV binds heparin in vitro (27) and further that HBV attachment to susceptible cells involves binding to cell surface GAG in a manner dependent upon the pre-S1 of the L protein (21, 24). We observed no significant difference between the binding of SVP assembled with HBV S only and the binding of those containing L and S (Fig. 5). Such findings do not support an interpretation that pre-S1 domains are essential for GAG binding. However, they do not exclude the possibility that for infection, GAG binding is needed together with an interaction between pre-S1 and the elusive host cell receptor(s).
Second, we examined whether SVP could interfere with infection of susceptible cells by HBV and HDV. SVP in patient serum can reach 1 mg/ml (13), a concentration we calculate as equivalent to about 40 μM of HBV envelope proteins and a total mass of 5 g in the blood of a patient. With the SVP that we assembled from transfected cells, even after concentration procedures, we were able to achieve only about 1 μM. We found that this did not give specific inhibition of HDV and HBV (Table 1), in contrast to the HBV pre-S peptides, which strongly inhibited infection even at the 20-fold-lower concentration of 50 nM. One possible reason why SVP failed to inhibit is that only 6% of the envelope proteins contained the pre-S domain. We have previously shown that some pre-S is present on the surface of SVP, since we were able to immunoprecipitate SVP with a pre-S antibody (17). However, there are two limitations for such affinity studies. First, they do not determine what fraction of all the pre-S domains is exposed on the surface. Specifically, others have reported that perhaps only 50% of the pre-S domains are exposed (5, 22, 23). Second, the affinity studies cannot exclude that some of the pre-S domains might have been proteolytically removed. Thus, <6% of the total envelope proteins presented pre-S on the surface of the SVP that failed to inhibit infection. Another possible reason for such failure is that SVP are much larger than the soluble peptides and probably have less access to the cell surface receptor(s). Thus, the enigma still remains as to what, if anything, is the function of SVP in natural infections.
Acknowledgments
J.T. was supported by grants AI-26522 and CA-06927 from the NIH and by an appropriation from the Commonwealth of Pennsylvania. N.C. was supported in part by the Elizabeth Knight Patterson Fellowship.
N.C. and H.C. contributed equally to these studies.
The real-time PCR was performed in the Fox Chase Biotechnology Facility and the electron microscopy in the Imaging Facility.
We thank Stephan Urban for the gift of chemically synthesized peptide. We thank Severin Gudima and Volker Bruss for advice. Constructive comments on the manuscript were given by Severin Gudima, Christoph Seeger, and William Mason.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Blumberg, B. S. 1984. Hepatitis B virus and the control of hepatocellular carcinoma. IARC Sci. Publ. 63243-261. [PubMed] [Google Scholar]
- 2.Bonino, F., W. Hoyer, J. W.-K. Shih, M. Rizzetto, R. H. Purcell, and J. L. Gerin. 1984. Delta hepatitis agent: structural and antigenic properties of the delta-associated particles. Infect. Immun. 431000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns, M., S. Miska, S. Chassot, and H. Will. 1998. Enhancement of hepatitis B virus infection by noninfectious subviral particles. J. Virol. 721462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 881059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 132273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V., and K. Vieluf. 1995. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 696652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai, N., H. E. Chang, S. Gudima, J. Chang, and J. Taylor. 2007. Assembly of hepatitis B virus envelope proteins onto a lentivirus pseudotype that infects primary human hepatocytes. J. Virol. 8110897-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai, N., S. Gudima, J. Chang, and J. Taylor. 2007. Immunoadhesins containing pre-S domains of hepatitis B virus large envelope protein are secreted and inhibit virus infection. J. Virol. 814912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, F. L., P. J. Chen, S. J. Tu, M. N. Chiu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis d antigen is crucial for the assembly of hepatitis D virus. Proc. Natl. Acad. Sci. USA 888490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of the hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 645066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem, D. 1991. Assembly of hepadnaviral virions and subviral particles. Curr. Top. Microbiol. Immunol. 16861-83. [DOI] [PubMed] [Google Scholar]
- 12.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 3501118-1129. [DOI] [PubMed] [Google Scholar]
- 13.Gerlich, W. H., and M. Kann. 2005. Hepatitis B, p. 1226-1268. In B. W. J. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 14.Gilbert, R. J., L. Beales, D. Blond, M. N. Simon, B. Y. Lin, F. V. Chisari, D. I. Stuart, and D. J. Rowlands. 2005. Hepatitis B small surface antigen particles are octahedral. Proc. Natl. Acad. Sci. USA 10214783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glebe, D., and S. Urban. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 1322-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gripon, P., I. Cannie, and S. Urban. 2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 791613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudima, S., Y. He, A. Meier, J. Chang, R. Chen, M. Jarnik, E. Nicolas, V. Bruss, and J. Taylor. 2007. Assembly of hepatitis delta virus: particle characterization including ability to infect primary human hepatocytes. J. Virol. 813608-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudima, S., A. Meier, R. Dunbrack, J. Taylor, and V. Bruss. 2007. Two potentially important elements of the hepatitis B virus large envelope protein are dispensable for the infectivity of hepatitis delta virus. J. Virol. 814343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klingmuller, U., and H. Schaller. 1993. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J. Virol. 677414-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladner, S. K., M. J. Otto, C. S. Barker, K. Zaifert, G. H. Wang, J. T. Guo, C. Seeger, and R. W. King. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 411715-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leistner, C. M., S. Gruen-Bernhard, and D. Glebe. 2008. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 10122-133. [DOI] [PubMed] [Google Scholar]
- 22.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 131048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze, A., P. Gripon, and S. Urban. 2007. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 461759-1768. [DOI] [PubMed] [Google Scholar]
- 25.Seeger, C., F. Zoulim, and W. S. Mason. 2007. Hepadnaviruses, p. 2977-3030. In D. M. Knipe (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 26.Sureau, C. 2006. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 307113-131. [DOI] [PubMed] [Google Scholar]
- 27.Zahn, A., and J. P. Allain. 2005. Hepatitis C virus and hepatitis B virus bind to heparin: purification of largely IgG-free virions from infected plasma by heparin chromatography. J. Gen. Virol. 86677-685. [DOI] [PubMed] [Google Scholar]