Recently, Huy et al. described a new hepatitis B virus (HBV) strain isolated in Vietnam (3) and claimed it to be a “new genotype,” “HBV genotype I,” with a complex recombination involving genotypes C, A, and G. We refute both claims.
Using complete genome sequence analysis of their single isolate, VH24 (AB231908), the authors documented an over 98% similarity with three other Vietnamese strains (2). Earlier, Hannoun et al. provided comprehensive information regarding those strains, showing recombination between genotype C and an unknown genotype in the pre-S/S region (2). Mean genetic divergence from genotype C of <8% in the entire genome and evidence of recombination had prevented the authors from assigning the strains to a new genotype. The same conclusion for the strains was reached by a later study using a new methodological approach (10). By providing neither additional information nor a new analytical approach, Huy et al. (3) surprisingly conclude that their strain, with those previously reported, represent a new genotype.
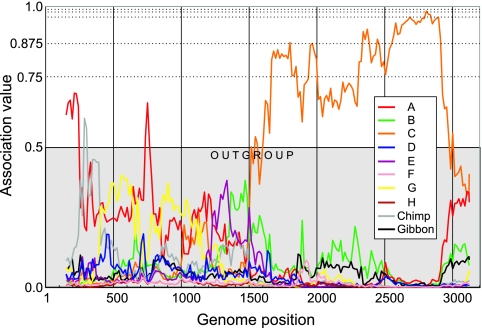
First, phylogenetic analysis of the complete genome of the four Vietnamese HBV isolates shows them to cluster with subgenotypes of C (C1 to C5) and to differ from genotype C by a mean nucleotide distance of only 7.0% ± 0.4%, which falls within the range of intragenotype and not intergenotype divergence (4). Furthermore, their conclusion of a “complex A/G/C recombination” arose from the use of Simplot software that has methodological limitations, which can be overcome by using GroupScanning (10). Reanalyzing AB231908 by using GroupScanning provides no strong evidence for recombination with known human or ape HBV genotypes in the pre-S/S regions (apart from two restricted regions, with association values of >0.5), in contrast to its consistent penetration into the genotype C clade from position 1600 (Fig. 1). In the pre-S/S regions, AB231908 formed variable, inconsistent outgroup associations with a range of genotypes, including A and G (originally identified as recombination partners by Huy et al. [3], using SimPlot) and with chimpanzee variants (Fig. 1, gray line; not included in the original analysis), a recombination partner even more improbable geographically than genotype A or G.
FIG. 1.
GroupScanning analysis (10) of VH24 against reference groups of nonrecombinant HBV sequences of human genotypes A to H and nonhuman ape-derived variants (chimpanzee/gibbon) (n = 288), incorporating all HBV sequences used for recombination detection in the Huy et al. study (3). Association values of approximately 0.5 or lower indicate an outgroup position or no phylogenetic clustering with a reference group. Analysis of previously described Vietnamese variants (AF241407 to AF241409) produced almost identical results (data not shown).
Finally, Huy et al. (3) “justified” assigning the four Vietnamese strains into a new genotype on the basis of seven “unique” conserved amino acids: His56, Ala60, Asn87, Val90, Val91, Ile136, and Lys198. From the databases, it is evident that His56 is present in subgenotype B1 and genotype C; Ala60 is the consensus for genotype D and present in subgenotypes C2 to C4; Val90, found in only three of the four Vietnamese sequences, is present in subgenotype C2; Val91 is common in genotype A; and Lys198 is found in subgenotypes B1 to B4, C3, F1, and F2 and genotypes E and H. Ile136 and Asn87 are therefore the only amino acids unique to the four Vietnamese strains, a far-from-recognized criterion of HBV genotyping.
Since 1988, when nucleotide diversity of >8% in the entire genome was first proposed for genotyping (9), eight genotypes have been described and named A to H (1, 7, 8, 11), and their geographical distribution and clinical relevance have been extensively reported (5, 6). In addition to the eight currently recognized genotypes, intergenotype recombination generates novel HBV variants, with over 24 phylogenetically independent recombinant variants described (10, 13). These recombinants can spread in humans and develop specific distributions and epidemiology as shown for the B/C recombinant, which accounts for the majority of genotype B strains in mainland Asia (12). Since sequencing and phylogenetic analyses are widely available, numerous further reports on HBV variation can be expected. If every new recombinant is assigned to a new genotype, we would soon be running out of alphabet letters. Principles of HBV classification must be established and accepted by the international community of experts in the field in order to ensure that genotyping is consistent, relevant, and significant.
Ed. Note: The author of the published article declined to reply.
REFERENCES
- 1.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 832059-2073. [DOI] [PubMed] [Google Scholar]
- 2.Hannoun, C., H. Norder, and M. Lindh. 2000. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol. 812267-2272. [DOI] [PubMed] [Google Scholar]
- 3.Huy, T. T. T., T. T. Ngoc, and K. Abe. 2008. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J. Virol. 825657-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramvis, A., K. Arakawa, M. C. Yu, R. Nogueira, D. O. Stram, and M. C. Kew. 2008. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J. Med. Virol. 8027-46. [DOI] [PubMed] [Google Scholar]
- 5.Kramvis, A., M. Kew, and G. Francois. 2005. Hepatitis B virus genotypes. Vaccine 232409-2423. [DOI] [PubMed] [Google Scholar]
- 6.Miyakawa, Y., and M. Mizokami. 2003. Classifying hepatitis B virus genotypes. Intervirology 46329-338. [DOI] [PubMed] [Google Scholar]
- 7.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198489-503. [DOI] [PubMed] [Google Scholar]
- 8.Norder, H., A. M. Courouce, and L. O. Magnius. 1992. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J. Gen. Virol. 733141-3145. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastrosoewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 692575-2583. [DOI] [PubMed] [Google Scholar]
- 10.Simmonds, P., and S. Midgley. 2005. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 7915467-15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 8167-74. [DOI] [PubMed] [Google Scholar]
- 12.Sugauchi, F., E. Orito, T. Ichida, H. Kato, H. Sakugawa, S. Kakumu, T. Ishida, A. Chutaputti, C. L. Lai, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J. Virol. 765985-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suwannakarn, K., P. Tangkijvanich, A. Theamboonlers, K. Abe, and Y. Poovorawan. 2005. A novel recombinant of hepatitis B virus genotypes G and C isolated from a Thai patient with hepatocellular carcinoma. J. Gen. Virol. 863027-3030. [DOI] [PubMed] [Google Scholar]