Abstract
Promyelocytic Leukemia nuclear body (PML NB) proteins mediate an intrinsic cellular host defense response against virus infections. Herpesviruses express proteins that modulate PML or PML-associated proteins by a variety of strategies, including degradation of PML or relocalization of PML NB proteins. The consequences of PML-herpesvirus interactions during infection in vivo have yet to be investigated in detail, largely because of the species-specific tropism of many human herpesviruses. Murine gammaherpesvirus 68 (γHV68) is emerging as a suitable model to study basic biological questions of virus-host interactions because it naturally infects mice. Therefore, we sought to determine whether γHV68 targets PML NBs as part of its natural life cycle. We found that γHV68 induces PML degradation through a proteasome-dependent mechanism and that loss of PML results in more robust virus replication in mouse fibroblasts. Surprisingly, γHV68-mediated PML degradation was mediated by the virion tegument protein ORF75c, which shares homology with the cellular formylglycinamide ribotide amidotransferase enzyme. In addition, we show that ORF75c is essential for production of infectious virus. ORF75 homologs are conserved in all rhadinoviruses but so far have no assigned functions. Our studies shed light on a potential role for this unusual protein in rhadinovirus biology and suggest that γHV68 will be a useful model for investigation of PML-herpesvirus interactions in vivo.
Promyelocytic leukemia nuclear bodies (PML NBs; also known as nuclear dot 10 or ND10) are 0.2- to 1-μm nuclear organelles that vary in frequency from 2 to 30 per cell, depending on the cell type and its status (19, 41). PML NBs are defined by the presence of the promyelocytic leukemia protein PML, which is required for assembly of PML NBs (33). Several cellular proteins are found constitutively in PML NBs, including Sp100 (speckled protein 100), Daxx, and SUMO-1 (small ubiquitin-like modifier 1) (19, 41, 44). Other cellular factors such as p53, regulators of p53 (homeodomain-interacting protein kinase, CREB binding protein, and mdm2 [mouse double-minute protein]), heterochromatin protein 1, and DNA damage response proteins (RAD50, nbs1, and mre11) can be partially or transiently found in PML NBs (19, 41, 44). While PML NBs do not move substantially within the nucleus, several studies have demonstrated that some of the constituent proteins like PML, Sp100, CREB binding protein, and Daxx exchange rapidly between PML NB structures and the surrounding nucleoplasm over a period of minutes to seconds (9, 27, 68). Thus, PML NBs are rather dynamic structures.
It is not surprising that, due to the vast array of diverse functions associated with PML NBs, they have been linked to a wide variety of cellular functions including the regulation of gene expression, chromatin dynamics, protein modification, apoptosis, p53 pathways, senescence, DNA repair, the interferon response, and viral infection (8, 19, 23, 25, 41, 44). Our laboratory is particularly interested in the interactions between PML NBs and herpesviruses. An emerging theme is that PML NBs carry out an intrinsic cellular defense mechanism against viruses (25, 28, 37, 55, 61). Herpesviruses from all classes (alpha-, beta-, and gammaherpesviruses) have evolved mechanisms to counteract this host defense (25). Thus, modulation of PML NB function(s) appears to be a general feature of most herpesviruses, which suggests that this modulation is necessary for successful persistence of these viruses in their natural hosts.
A great deal of what is known concerning interactions between PML NBs and herpesviruses has been derived from investigations of herpes simplex virus type 1 (HSV-1). Following infection with HSV-1, viral genomes enter the nucleus and appear to localize at or near PML NBs (32, 40). A recent study suggested that this association occurs through the migration of PML components (i.e., PML and Sp100) from existing PML NBs to incoming viral genomes to form “new” PML NB-like structures (27). However, the integrity of these structures is destroyed shortly after infection following the synthesis of the viral immediate-early protein ICP0. The association of viral genomes with PML NBs has also been noted with cytomegalovirus (CMV), varicella-zoster virus, Epstein-Barr virus (EBV), and other DNA viruses including adenoviruses, simian virus 40, polyomavirus, and the parvovirus adeno-associated virus (22).
Several herpesvirus proteins have potent effects on PML NBs. One of the best characterized is the HSV-1 ICP0 protein, which is a RING finger E3 ubiquitin ligase that disrupts PML NBs by initiating a proteasome-dependent degradation of PML (13, 26). This results in the transient association of PML NBs that are first found associated with incoming genomes. HSV-1 viruses expressing ICP0 mutants unable to induce PML degradation have a much lower probability of progressing into productive lytic infection at low multiplicities of infection (MOIs) (24, 54, 59). In addition, extensive depletion of PML in human fibroblast cells significantly increases gene expression and plaque formation of ICP0-null viruses (28). Moreover, viruses with mutations that inactivate ICP0 do not enter productive replication after infection of human fibroblasts at low MOIs but, instead, are retained in a repressed quiescent state that in some respects resembles latency (24, 51, 52). The CMV immediate-early protein IE1 appears to induce the loss of SUMO-lated forms of PML, resulting in the destruction of PML NBs (3, 35). Like HSV-1, CMV mutants impaired for inducing PML NB destruction are less efficient at establishing productive infection at low MOIs, and these mutants can be complemented in human fibroblast cells depleted of PML protein (35, 61). CMV also encodes a tegument protein known as pp71 that induces the degradation of the PML NB-associated protein human Daxx (hDaxx) (55). Depletion of hDaxx enhances human CMV gene expression, especially by pp71-deficient viruses (62).
In contrast to alpha- and betaherpesviruses, EBV, a gammaherpesvirus, typically establishes a latent infection in human B cells, which involves the expression of several EBV proteins that drive B-cell proliferation. Two of the first proteins made following EBV infection are EBNA2 and EBV nuclear antigen leader protein (EBNA-LP) (6, 7). EBNA2 is a transcriptional regulator of viral latent genes and cellular genes (11), while EBNA-LP is a coactivator of EBNA2 (31, 45, 48, 49). Recent work from our laboratory showed that EBNA-LP interacts with Sp100 and displaces it from PML NBs, and this correlates with a coactivation function (37). We have speculated that incoming viral genomes are subject to intrinsic gene silencing mechanisms regardless of whether the virus enters a productive infection or initiates nonproductive infection, as is the case for EBV. EBNA-LP may function like an immediate-early gene to counteract these effects through interactions with Sp100. Latently infected cells also reactivate occasionally, which switches on the viral genetic program for productive lytic infection. The virally encoded regulatory protein Zta, which is critical for productive lytic infection, has also been observed to induce the disruption of PML NBs (1, 14). Collectively, the data suggest that PML NB-associated proteins have a role in inhibiting virus gene expression during productive lytic infection. In addition, some of these studies suggest that PML NBs may play a role in establishment or maintenance of latency.
Our understanding of PML-herpesvirus interactions and their consequences in vivo has been somewhat limited due to the species specificity of most human herpesviruses. Murid herpesvirus 4, also known as murine gammaherpesvirus 68 (γHV68), naturally infects mice and establishes chronic infection in them (58, 65). The virus shares many genetic and biological features with other gammaherpesviruses including EBV and Kaposi's sarcoma-associated herpesvirus (43, 66). We considered that γHV68 might be a good model system for investigating PML-herpesvirus interactions although it was unknown whether γHV68 modulated PML. Thus, we asked whether γHV68 had any effects on PML or its associated proteins and, if so, which viral protein(s) mediated this activity. We found that infection of mouse fibroblast cells with γHV68 induced the rapid degradation of PML by a proteasome-dependent mechanism. Furthermore, γHV68 replication was more robust in PML-negative fibroblasts than in control PML-positive fibroblasts. Unexpectedly, γHV68-mediated PML degradation was regulated by the open reading frame 75c (ORF75c) virion tegument protein. ORF75 homologs are found in all rhadinoviruses and have homology to the cellular formylglycinamide ribotide amidotransferase (FGARAT) enzyme, which carries out the fourth step in de novo purine biosynthesis. Our results reveal a novel function for this family of unusual viral proteins and implicate γHV68 as a unique model system to investigate the role of PML in modulation of herpesvirus infections in a natural animal host.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T12, 3T3, and 293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and grown in 5% CO2 tissue culture incubators at 37°C. Virus stocks were generated by transfecting γHV68-bacterial artificial chromosome (BAC) DNA into 3T12 cells or Vero-Cre cells to excise the BAC sequence from the virus, as described previously (42). Mouse embryo fibroblast (MEF) cells derived from C57BLK/6 mice (passage 2) were a generous gift from Lawrence Donehower. PML+/+ and PML−/− cells have been described previously (67) and were generously provided by Gerd Maul. PML+/+ and PML−/− MEFs were maintained and passed under conditions described for 3T12 cells. PML−/− cells grow somewhat faster than PML+/+ cells. For infection experiments, PML−/− cells were plated out at a lower density 24 h before infection, so at the time of infection they were approximately equal in number to PML+/+ cells. At 24 h postinfection, the PML−/− cell number never exceeded the number of PML+/+ cells by more than 10%. PML−/− cells were also converted to PML-positive cells by retrovirus transduction. Plasmid pJT222 (see “Plasmids” below) was transfected into the Ecopack 2-293 packaging cell line (Clontech), and the supernatant containing virus was used to transduce PML−/− cells. At 24 h posttransduction, the cells were placed in medium containing 10 μg/ml puromycin, and cell pools surviving under selection conditions were amplified for experiments. Similar experiments were done with retroviruses generated from empty murine stem cell virus vector. PML−/− cells converted to express PML are designated PML1, while PML−/− cells transduced with the empty vector are PML0. Cycloheximide and phosphonoacetic acid (PAA) were used in some experiments at concentrations of 50 μg/ml and 200 μg/ml as described previously (38, 39). UV-inactivated virus was generated by exposing 4 ml of γHV68 virus (1 × 107 PFU/ml) to UV light for 5 min. UV-treated virus had no plaque-forming activity following this method of treatment (data not shown).
Plasmids.
Plasmids encoding γHV68 ORFs 45, 48, 50 (Rta), 52, 57, 73 (LANA), 75a, 75b, and 75c were generated by PCR using the γHV68-BAC as a template. All cDNAs were fused with the hemagglutinin (HA) epitope tag encoding sequence at the 3′ end. The cDNAs were cloned into pCR-Blunt (Invitrogen), sequenced to confirm that no mutations were introduced during PCR, and then cloned into the eukaryotic expression vector pCI (Promega). An ORF75c cDNA was also generated, as described above for the HA epitope-tagged cDNAs, to express the AU1 (Covance) epitope tag at its carboxy terminus (75c.AU1). A mouse PML (mPML) cDNA (MMM1013-64912; Open Biosystems) encoding the equivalent of human PML isoform I was PCR amplified to encode an HA epitope tag and cloned into the pCI expression vector. A second version of mPML isoform I was PCR amplified to encode a carboxy-terminal Flag epitope tag and cloned into the murine stem cell virus (Clontech) retroviral vector (pJT222) for transduction of MEF cells.
Immunofluorescence, transfection, and immunoblot assays.
For immunofluorescence assays, 3 × 104 3T12, 3T3, or MEF cells were plated onto coverslips in 1 ml of medium in 24-well tissue culture plates 24 h prior to transfection or infection. Transfection was performed using Superfect (Qiagen) according to the manufacturer's protocol. At 24 h posttransfection or at designated times following virus infection, the cells were washed with phosphate-buffered saline (PBS) and fixed at 4°C with 4% paraformaldehyde. Cells were then permeabilized using 1% Triton, washed, and then blocked for either 1 h at room temperature or at 4°C overnight with 5% dried milk in Tris-buffered saline. Following incubation with primary and secondary antibodies in blocking buffer diluted 1:10 in Tris-buffered saline, the cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and visualized on a Zeiss Axioplan II upright microscope. Captured images were imported and processed in Adobe Photoshop. 293 cells were plated at 5 × 105 cells per well in six-well plates 1 day before transfection. Plasmids were transfected into the 293 cells using Superfect. In some experiments, the proteasome inhibitor MG132 was added to a concentration of 500 nM at 24 h posttransfection for an additional 24 h, whereupon the cells were harvested and analyzed by immunoblotting. Control experiments indicated that cell viability began to decrease significantly at concentrations above 500 nM over the same time period (data not shown). MG132 and lactacystin were also used in some virus infection experiments. 3T12 cells were plated at 4 × 106 cells/10-cm plate. After 24 h the cells were treated with MG132 (10 μM) or lactacystin (10 μM) for 45 min, followed by virus infection for another 4 h. The cells were then harvested for immunoblot analysis. For immunoblot assays, nitrocellulose membranes were blocked in PBS, 5% nonfat dried milk, and 0.01% Tween 20 for 1 h at room temperature. Following blocking, primary antibody was incubated in blocking buffer diluted 1:10 in PBS overnight at 4°C. The blots were then washed in PBS and then incubated with secondary antibody in blocking buffer diluted 1:10 for 1 h at room temperature. The blots were washed again in PBS several times and then developed using a SuperSignal West Pico Chemiluminescent substrate kit (Pierce).
Quantitative reverse transcription-PCR (RT-PCR).
RNA was extracted from mock-infected and γHV68-infected 3T12 cells and reverse transcribed as described previously (49). Oligonucleotide primers and probes for detection of murine β-actin, PML, and γHV68 ORF57 were purchased from Applied Biosystems. Primers were designed to amplify between exons, and probes were designed to hybridize across exon-exon junctions to avoid detection of genomic DNA. In addition, RNA samples that were not reverse transcribed were analyzed to control for potential DNA contamination. Real-time PCR was performed with an Applied Biosystems Step-One instrument using the comparative threshold cycle method as described by the manufacturer.
Antibodies.
Primary antibodies used in this study were against mPML (Upstate), α-tubulin (Sigma), Daxx (Santa Cruz), CBF-B (the B subunit of the CCAAT-binding factor; Santa Cruz), Akt (Covance), AU1 (Covance), and HA (Covance). Polyclonal antibody against γHV68 ORF57 was generated by immunizing rabbits with a recombinant His-tagged ORF57 protein that was produced in Escherichia coli. A mouse monoclonal antibody (9C7/A6) against γHV68 ORF4 was generously provided by Philip Stevenson. Secondary antibodies to mouse and rabbit conjugated with either Alexa Fluor 488 or 594 were purchased from Molecular Probes. Secondary antibodies against mouse and rabbit conjugated with horseradish peroxidase were purchased from Jackson Immunolaboratories.
Generation of virus recombinants by allelic exchange.
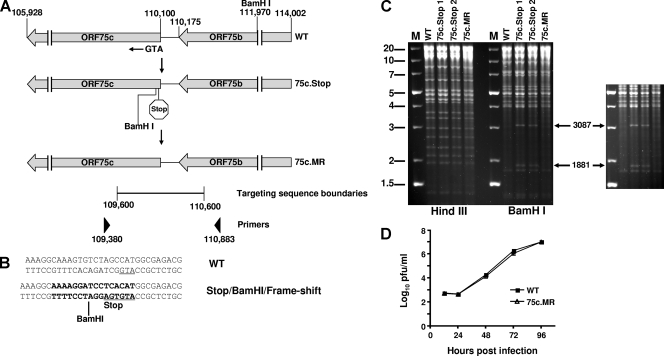
The γHV68 genome cloned into a BAC was a kind gift from Ulrich Koszinowski (2). An ORF75c-null virus was generated by using allelic exchange in RecA-positive E. coli as described previously (42, 56). Briefly, a PCR product was generated from nucleotides 109600 to 110600 that introduced a stop codon following the first methionine codon for ORF75c. A BamHI restriction site was also introduced following the stop codon as well as a single base insertion to give a frameshift in the ORF75c coding sequence (see Fig. 8B). The targeting sequence was cloned into pCR-Blunt and checked by sequencing. The targeting sequence was subcloned into the suicide vector pGS284, and allelic exchange was performed with this vector and the wild-type γHV68-BAC in the S17λpir E. coli strain. 75c.Stop1 and 75c.Stop2 are two BACs selected from two independent allelic exchange experiments. 75c.Stop1 was rescued to the wild-type sequence (marker-rescued 75c[75c.MR]) using the same methodology described for generation of the mutants except that the targeting sequence was identical to wild-type γHV68. PCR was used to amplify between nucleotides 109380 and 110883 (which are outside of the original targeting sequence boundaries) of the 75c.Stop and 75c.MR genomes, and the DNA was sequenced to verify that the mutant and rescued sequences were correct. Restriction enzyme digestion of γHV68-BAC DNA isolated from bacteria was used to compare the ORF75c mutant and MR genomes to the wild-type DNA to confirm that no overt deletions had occurred during the recombination process.
FIG. 8.
ORF75c is required for production of infectious virus. (A) Schematic diagram of the wild-type γHV68 genome that encodes ORF75c. The nucleotide positions are indicated above the diagram as well as the ORF75 initiation methionine (ATG) and a BamHI restriction site. Below the wild-type genome is a schematic of the approximate location of the introduced stop codon and novel BamHI restriction site (75c.Stop). The mutant sequence was converted to wild type and is shown below the 75c.Stop (75c.MR). The locations and boundaries of the targeting sequence used to generate recombinant viruses and the PCR primers used to analyze the recombinants are also shown. (B) Wild-type and mutant sequences corresponding to the 75c.Stop virus are compared. The initiation methionine on the antisense strand is underlined. (C) Wild-type, ORF 75c.Stop, and ORF75c.MR BACs were analyzed by restriction enzyme digestion and resolved on 0.7% agarose gels stained with ethidium bromide. Molecular weight markers (M) are shown on the left side of the panel. Enzymes used to digest the BAC DNAs are indicated below the panel. Novel BamHI digestion products released from the mutant viral DNAs are indicated by the arrows on the right-hand side of the panel. A darker exposure of the section containing the novel BamHI fragments is shown on the right. (D) Multistep growth curve comparing wild-type (WT) and ORF 75c.MR viruses in 3T12 cells. The data are representative of three independent experiments. WT, wild type.
Plaque assays.
NIH 3T12 cells were plated into 12-well tissue culture dishes at a density of 5 × 104 cells/well. The following day, virus was absorbed to the monolayer for 1 h in a volume of 0.2 ml. The samples were overlaid with a mixture of 0.75% methyl-cellulose, 5% fetal bovine serum, 1 × minimal essential medium, and antibiotics. Monolayers were stained with neutral red after 5 to 7 days, and plaques were counted. Plaque-forming efficiency assays were carried out using normal plaque assay conditions; only PML0 and PML1 cells were used instead of 3T12 cells.
RESULTS
γHV68 infection induces the disruption of PML NBs and degradation of PML.
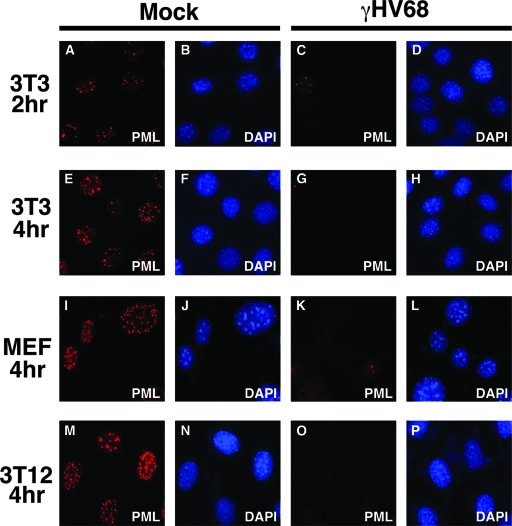
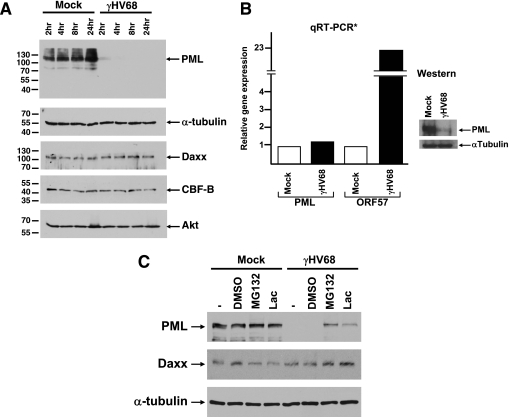
To examine γHV68 effects on PML NBs, mouse 3T3, MEF, and 3T12 cells were infected at an MOI of 5. At 2 or 4 h postinfection, PML was analyzed by immunofluorescence. Staining with anti-PML detected numerous punctate spots in the nucleus of mock-infected cells, characteristic of PML and its localization in PML NBs (Fig. 1A, E, I, and M). In contrast, as early as 2 h postinfection, PML was almost undetectable in γHV68-infected cells (Fig. 1C, G, K, and O). To determine whether the disappearance of PML was due to the absence of protein or if it was merely being redistributed, we analyzed infected cell extracts by Western blotting. PML protein levels were substantially reduced as early as 2 h postinfection and were undetectable at 4 h postinfection (Fig. 2A). To see if this was a global effect induced by the virus, we probed the same infected cell lysates with antibodies to several other cellular proteins. In contrast to PML, α-tubulin and Akt protein levels remained unaltered throughout the 24-h time period following γHV68 infection. CBF-B, a nuclear transcription factor, and Daxx, which resides in PML NBs, were not degraded, suggesting that the virus does not induce global destruction of nuclear or PML NB-associated proteins (Fig. 2A).
FIG. 1.
γHV68 infection induces the loss of PML NBs. Different mouse fibroblast cell lines were mock infected or infected with γHV68 at an MOI of 5 and fixed at the indicated time points after infection. The cells were stained with a monoclonal antibody against mPML and a secondary antibody conjugated with Alexa Fluor 594. The cells were costained with DAPI.
FIG. 2.
γHV68 induces proteasome-dependent degradation of PML. (A) Immunoblot of infected 3T12 cell extracts harvested at the indicated time points postinfection. The blots were probed with antibodies to various cellular proteins, indicated on the right-hand end of the panels. Molecular weights are indicated to the left of each panel. (B) RNA extracted from γHV68-infected cells was analyzed by quantitative RT-PCR (qRT-PCR). mRNA levels for PML and γHV68 ORF57 were normalized to β-actin. The data are representative of three independent experiments. The panel on the right shows a small sample of the infected cells used to obtain RNA, which was analyzed by immunoblotting for PML as a control. (C) Immunoblot of mock-infected and γHV68-infected 3T12 cells treated with either nothing (−), solvent (dimethyl sulfoxide [DMSO]), MG132, or lactocystin. The cell extracts were probed for expression levels of the proteins indicated to the left of each panel.
To determine whether γHV68 was inducing PML protein loss through reduction of PML RNA levels, we analyzed PML mRNA from mock- and γHV68-infected cells by quantitative RT-PCR. The results indicate that PML RNA levels remain constant following γHV68 infection, while RNA for the immediate-early/early transcript encoding the gene for ORF57 was induced (Fig. 2B).
The quantitative RT-PCR results suggested that γHV68 induces the reduction of PML protein by a posttranscriptional mechanism. To further test this notion, we asked whether PML degradation was mediated by a proteasome-dependent mechanism. Results shown in Fig. 2C indicate that both MG132 and lactacystin significantly inhibited γHV68-induced PML degradation, suggesting that PML turnover in γHV68-infected cells is regulated by a proteasome-dependent mechanism.
γHV68-induced PML degradation is mediated by a component of the virion.
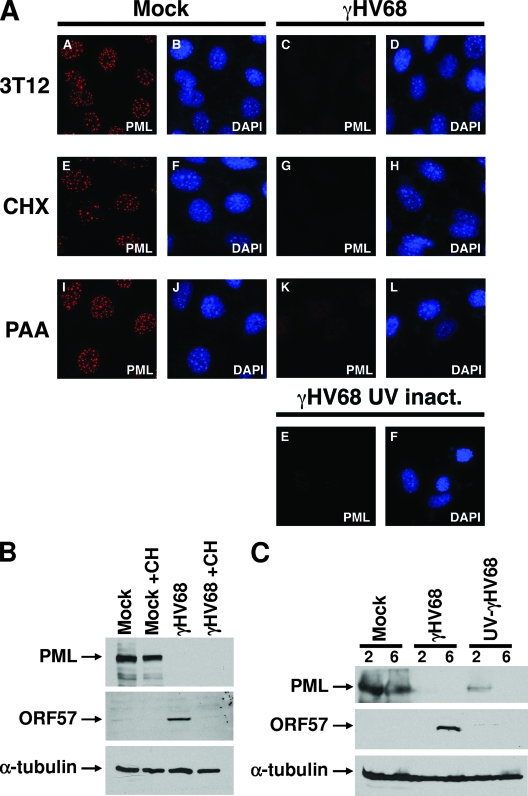
To determine whether γHV68 induced PML degradation was dependent on synthesis of viral proteins, we treated 3T12 cells with cycloheximide or PAA to block protein synthesis and viral DNA replication, respectively. Neither treatment inhibited γHV68-induced destruction of PML (Fig. 3A and B). UV-inactivated γHV68 also induced PML destruction (Fig. 3A and C), consistent with the notion that this function is mediated by a virion component. In both experiments expression of the viral ORF57 protein was inhibited, indicating that the treatments were effective (Fig. 3B and C).
FIG. 3.
γHV68-induced PML degradation is associated with a virion component. (A) 3T12 cells were mock infected or infected with γHV68 at an MOI of 5 and fixed at the indicated time points after infection. The cells were stained with a monoclonal antibody against mPML and a secondary antibody conjugated with Alexa Fluor 594. The cells were costained with DAPI. Some cells were treated prior to and during infection with cycloheximide (CHX) or PAA, as indicated to the left of appropriate panels. UV-inactivated (inact.) virus was also used to infect cells, which was done under conditions similar to those used for wild-type virus infections. (B) Immunoblots of 3T12 cells mock infected or γHV68 infected for 5 h that were either untreated or treated with cycloheximide (CH). The blots were probed with antibodies indicated to the left of each panel. (C) Immunoblots of mock-infected, γHV68-infected, or UV-inactivated γHV68-infected 3T12 cells. The cells were harvested at 2 or 6 h postinfection as indicated above the panels. The blots were probed with antibodies indicated to the left of each panel.
The ORF75c tegument protein induces PML degradation.
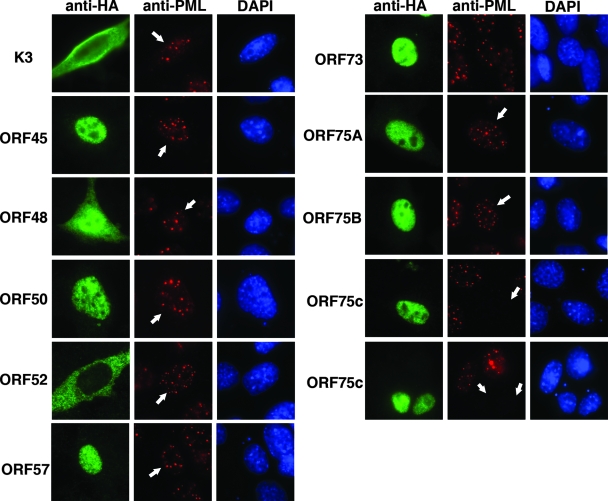
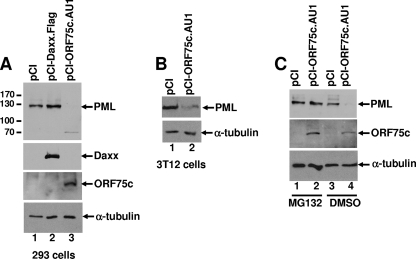
The data presented in Fig. 3 indicated that a virion component was responsible for inducing PML degradation upon infection with γHV68. An earlier study identified viral polypeptides that were present in purified γHV68 virions (12). Therefore, we initially focused on previously identified virion tegument proteins (i.e., ORFs 45, 48, 52, and 75c) as candidates for possessing PML-degrading function. In addition, because PML degradation is mediated by immediate-early proteins encoded by other herpesviruses (26) and K3 has been reported to have ubiquitin-ligase activity (10), we tested γHV68 proteins ORF50, ORF57, ORF73 (LANA), and K3 to determine if they could induce PML degradation. NIH 3T12 cells were transfected with expression plasmids for several HA-tagged γHV68-encoded proteins, and the cells were stained with anti-HA to detect the expressed protein and anti-PML to monitor PML expression. Only ORF75c was able induce PML destruction (Fig. 4). Two paralogs of ORF75c, ORF75a and ORF75b, are encoded by γHV68. Neither ORF75a nor ORF75b was able to induce PML destruction (Fig. 4). Cotransfection of ORF75c and mPML expression plasmids in either 293 or 3T12 cells also demonstrated that ORF75c was sufficient to induce PML degradation (Fig. 5A and B). Interestingly, γHV68 infection of human cells does not result in PML degradation (i.e., endogenous human PML), and ORF75c is unable to induce degradation of human PML in cotransfection experiments with human PML cDNAs (data not shown). The reason for this is unclear but is likely to involve specific unique sequences or structures in mPML rather than differences in cellular factors since ORF75c was able to induce mPML degradation in both human and mouse cells. To investigate whether ORF75c-mediated degradation of PML was proteasome dependent, we included the proteasome inhibitor MG132 in the cotransfection experiments. The results suggest that ORF75c is sufficient to induce PML degradation in a proteasome-dependent manner (Fig. 5C).
FIG. 4.
ORF75c is sufficient to induce the loss of PML NBs. cDNAs encoding several γHV68 proteins were transfected into 3T12 cells as indicated to the left of each series of panels. At 24 h posttransfection, the cells were fixed and stained with anti-HA (green), anti-PML (red), and DAPI (blue). White arrows in the PML-stained cells indicate the cells that are expressing the HA epitope-tagged γHV68 protein.
FIG. 5.
ORF75c is sufficient to induce the proteasome-dependent degradation of PML. (A) An mPML-HA (mPML) expression plasmid was cotransfected with empty expression vector (pCI), pCI-Daxx.Flag, and pCI-ORF75.AU1 into 293 cells. Cell extracts were harvested 24 h posttransfection and analyzed by immunoblotting. Expression of mPML, Daxx, ORF75c, and α-tubulin was detected using anti-HA, anti-Flag, anti-AU1, and anti-α-tubulin antibodies, respectively. Molecular weights are indicated to the left of the PML immunoblot. (B) Immunoblot analysis of 3T12 cells cotransfected with mPML-HA and either pCI or pCI-ORF75c.AU1. mPML was detected using an anti-HA antibody and a loading control with anti-α-tubulin antibodies as indicated to the right of each panel. (C) 293 cells were cotransfected as described for panel A, but the cells were also treated with dimethyl sulfoxide (DMSO) or 500 nM MG132 at 24 h posttransfection for an additional 24 h before the cells were harvested for immunoblot analysis.
γHV68 infection is less robust in cells expressing PML.
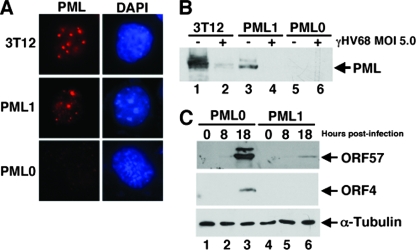
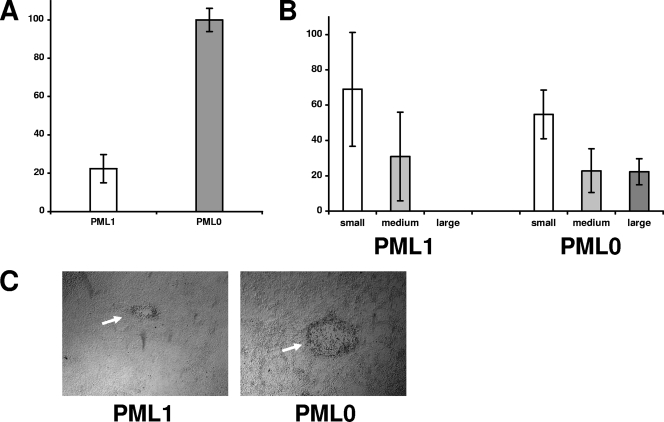
CMV replicates with higher efficiency in cells depleted for PML expression (61), consistent with the idea that PML carries out an intrinsic cellular defense against viral infection. To investigate whether PML exerts a similar effect on γHV68, we tested whether it could replicate better in MEF cells derived from PML knockout mice than in PML-expressing fibroblasts derived from the parental wild-type mice. Consistent with previous observations of more robust CMV infection in PML-depleted human fibroblasts (61), γHV68 replicated better in the PML-null fibroblasts (Table 1). Complementary experiments were done comparing virus replication in PML-null fibroblasts transduced with a control retrovirus vector (PML0) to PML-null fibroblasts converted by retrovirus transduction to express mPML (PML1 cells). PML localized in punctate nuclear dots as expected in PML1 cells (Fig. 6A), although the frequency of PML dots was slightly lower than in the wild-type fibroblasts, consistent with lower overall PML expression in these cells relative to wild-type cells as determined by Western blot analysis (Fig. 6B). One hundred percent of the transduced PML1 cells were PML positive by immunofluorescence. Following infection with γHV68 at high MOIs, the expressed PML protein was degraded similar to wild-type PML in 3T12 cells (Fig. 6B). Collectively, the data suggest that the PML1 cell line expresses PML with characteristics consistent with wild-type PML. Consistent with results shown in Table 1, γHV68 infection of PML0 cells at an MOI of 0.1 yielded 4.6-, 5-, and 7.8-fold more infectious virus in three separate experiments than similarly infected PML1 cells. In addition, virus gene expression (Fig. 6C) and plaque-forming efficiency were more robust in PML0 cells than in PML1 cells (Fig. 7).
TABLE 1.
γHV68 grows efficiently in PML-negative fibroblasts
| MOI | Titer (PFU/ml) in 3T12 cellsa
|
Relative increase (n-fold)b | |
|---|---|---|---|
| PML+/+ | PML−/− | ||
| 0.1 | 4 × 102 | 1 × 104 | 25 |
| 1.0 | 1.7 × 104 | 5 × 105 | 29 |
| 3.0 | 5 × 105 | 8.5 × 106 | 17 |
Titers of virus were determined in from PML−/− and PML+/+ cells infected for 24 h with γHV68.
The relative increase in viral titers between PML−/− and PML+/+ cells was obtained by dividing the number of PFU/ml of PML−/− cells by the number of PFU/ml from PML+/+ cells. Because the PML−/− cells grew slightly faster, titers were also calculated as the number of PFU/cell. However, the relative increases were similar to the values shown in the table (data not shown). Results are representative of three separate experiments.
FIG. 6.
Characteristics of PML knockout MEFs engineered to express mPML. (A) PML−/− cells transduced with a retrovirus containing an mPML cDNA (PML1), and PML−/− cells transduced with a retrovirus lacking the PML cDNA (PML0) were stained with anti-PML (red) or DAPI (blue). (B) 3T12, PML1, and PML0 cells were mock infected (−) or infected with γHV68 (+) at an MOI of 5.0 for 4 h. The cell extracts were analyzed by Western blotting using an antibody against mPML. (C) Western blot of viral protein expression using anti-ORF57 and anti-ORF4 antibodies from PML0 and PML1 cells that were infected at an MOI of 0.1 for the indicated time points.
FIG. 7.
Plaque-forming efficiency of PML-negative MEFs relative to PML-positive MEFs. (A) The relative number of plaques detected on PML0 versus PML1 cells infected with similar dilutions of γHV68 wild-type virus. The results are from three experiments, and the standard deviations are indicated. (B) In addition to fewer plaques in PML1 cells relative to PML0 cells, the average size of the plaques in PML1 cells was smaller. The percentages of small, medium, and large plaques in infected PML0 and PML1 cells are shown. The results are from three experiments, and the standard deviations are indicated. (C) Phase contrast of an average plaque commonly found in PML0 infected cells compared to the average small plaque detected in PML1 cells. Both photographs were taken at ×40 magnification.
γHV68 ORF75c is required for production of infectious virus.
Using transposon mutagenesis, a previous study indicated that ORF75c might be important for γHV68 growth in culture (57). However, it is unclear whether the transposon insertion altered expression of neighboring genes, whether the insertion caused the synthesis of a dominant negative form of ORF75c, or if the mutant virus contained other inactivating mutations. To investigate the importance of the PML-degrading activity of ORF75c more succinctly, we generated γHV68 mutant viruses lacking the ability to produce the protein. Using allelic exchange, we introduced a stop codon following the initiation codon for ORF75c into the γHV68 BAC (Fig. 8A and B). In addition, the stop codon was followed by a BamHI restriction site and a second frameshift mutation. Two clones derived from independent experiments were isolated (75c.Stop1 and 75c.Stop2), and one of these, 75c.Stop1, was converted back to the wild-type sequence by allelic exchange to generate 75c.MR (marker rescue). By restriction enzyme analysis, the mutant viral BACs contained the expected additional BamHI restriction site compared to the wild-type γHV68 BAC, while the 75c.MR had lost the site (Fig. 8C). No additional changes were observed between the viral DNAs when digested with Hind III (Fig. 8C). To confirm that faithful recombination had occurred in each of the clones, we PCR amplified and sequenced genomic DNA, which included the boundaries of the targeting sequence used for allelic exchange (Fig. 8A). Aside from the introduced mutations in the 75c.Stop clones, no other changes were observed (data not shown). PCR amplification of the same region in the 75c.MR genome indicated that it was identical to the wild-type γHV68 sequence (data not shown). Wild-type, marker-rescued, and 75c.Stop viral DNAs were purified and transfected into either NIH 3T12 or Vero-Cre cells. Virus was easily recovered from wild-type- and 75c.MR-transfected cells. These viruses grew with similar kinetics (Fig. 8D). No virus was ever recovered from 75c.Stop1 or 75c.Stop2 clones. Attempts to recover virus by transfecting PML knockout cells with the 75c.Stop virus BACs were also not successful (data not shown). Therefore, ORF75c appears to be required for production of infectious virus, possibly due to factors other than or in addition to its PML degrading activity.
DISCUSSION
In this study we found that γHV68 ORF75c induces the rapid degradation of PML by a proteasome-dependent mechanism and that loss of PML correlates with more robust viral replication. This conclusion is supported by data indicating that PML mRNA levels remain unaltered in γHV68-infected cells relative to mock-infected cells whereas PML protein levels are dramatically decreased in γHV68-infected cells (Fig. 2A and B). Furthermore, PML protein levels are substantially maintained following γHV68 infection when cells are treated with proteasome inhibitors (Fig. 2C). γHV68-mediated PML degradation appears to be modulated primarily by the tegument protein known as ORF75c (Fig. 3 to 5). γHV68 replicated more efficiently in PML-null fibroblasts (Table 1; Fig. 6 and 7). Surprisingly, ORF75c-null viruses were unable to produce infectious virus, suggesting that the protein may provide essential viral functions in addition to inducing PML degradation (Fig. 8). To our knowledge, these data are the first to implicate the importance of PML modulation for rhadinovirus biology. The data are novel because rhadinovirus ORF75 genes, which have homology to FGARAT enzymes, previously had no known function(s).
Degradation of PML following γHV68 infection is reminiscent of observations that HSV induces a proteasome-dependent degradation of PML, which is mediated by the immediate-early gene ICP0 (26). Several studies have implicated PML as having a repressive effect on HSV gene expression following infection, which is counteracted by ICP0 (25, 28). While HSV must synthesize ICP0 after infection, ORF75c is brought into the cell with the virion particle, suggesting that γHV68 might be particularly sensitive to the repressive effects of PML so that the virus must neutralize PML rather quickly following infection. The fact that the PML degradation function is carried out or regulated by ORF75c was somewhat unexpected: first, because no known function has been ascribed for ORF75 proteins and, second, because the protein is associated with the virion. Most herpesvirus gene products implicated in modulating PML are encoded by immediate-early gene products (except CMV pp71, a tegument protein that induces the degradation of Daxx), while ORF75c is a viral tegument protein (12).
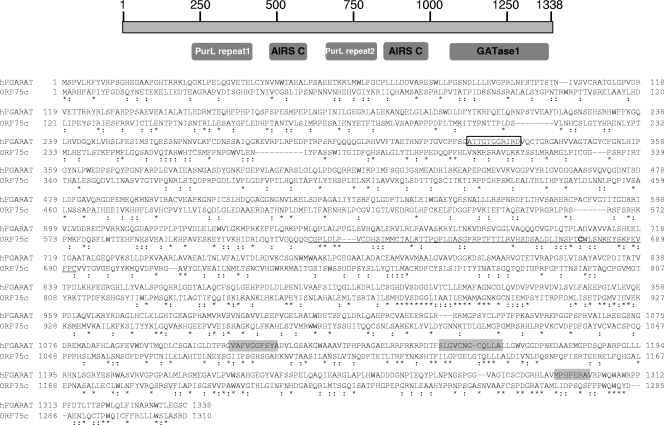
The mechanism by which ORF75c mediates proteasome-dependent PML degradation remains unclear. Unlike ICP0, ORF75c does not contain a recognizable RING finger domain typical of some proteins possessing intrinsic E3 ligase activity. There are two distinct groups of E3 ligase proteins that are characterized by defining motifs. The HECT domain E3s and the zinc finger E3s, which can be further subdivided into two classes of RING finger domains, U-box, and PHD domain E3s. Sequence comparisons using a variety of bioinformatics programs (e.g., ELM, Interproscan, and BLAST) failed to identify any of these motifs in ORF75c. Clustal W sequence comparisons between ORF75a, ORF75b, and ORF75c indicate that they have diverged significantly from each other. ORF75a is 26% identical and 43% similar to ORF75c, and ORF75b is 24% identical and 43% similar to ORF75c. ORF75a and ORF75b are 23% identical and 40% similar relative to each other. Because of the general divergence of the proteins relative to each other, primary sequence motifs unique to ORF75c that might mediate PML degradation function were not readily identified, save for a nondescript Cys-His region between residues 624 to 693. Recently, however, the human herpesvirus 8 (HHV8) Rta protein was found to have intrinsic E3 ligase activity specifically targeting IRF7 for proteasome-dependent degradation (69). Although the HHV8 Rta protein lacks a recognizable RING finger domain, the proposed E3 ligase catalytic activity required a Cys-rich region, which has no significant similarity to any previously described E3 ligase (69). The ORF75c Cys rich-region (i.e., 5 Cys) between residues 624 and 693 may function similarly (Fig. 9). Rubulavirus V proteins induce the proteasome-dependent degradation of cellular STATs through recruitment of cellular STAT-specific ubiquitin ligase complexes (50, 63, 64). Likewise, rather than acting as a direct E3 ligase, ORF75c may act as a functional E3 ligase by recruiting the assembly of PML-specific ubiquitin ligase complexes. Future studies to determine specific domains within ORF75c that mediate PML degradation and possible ORF75c interactions with PML and confirmation that ORF75c induces formation of polyubiquitin should begin to clarify its mode of action.
FIG. 9.
Features of the human FGARAT protein and comparison to ORF75c. A schematic of the human FGARAT protein is shown at the top with amino acid numbers indicated. Below, the approximate locations of conserved regions are shown. PurL repeat, protein domain of eukaryotic and prokaryotic proteins known as PurL, which possesses FGARAT activity; AIRS C, AIR synthase-related protein; and GATase1, type 1 glutamine amidotransferase-like domain found in FGARAT. Sequence comparison between the human FGARAT and ORF75c is shown below the diagram. Asterisks indicate conserved residues and colons indicate similarity. The ATP binding sequence is shown in an open box, and putative glutamine binding regions are boxed in light gray. The underlined sequence between residues 624 and 693 contains a Cys-rich region.
Earlier studies showed that all gammaherpesviruses, but not alpha- or betaherpesviruses, encode proteins with homology to the cellular FGARAT enzyme (Table 2). HHV8 encodes a single FGARAT homolog (ORF75) (53), while herpesvirus saimiri (HVS) encodes two FGARAT homologs (5), one near the right-hand end of the genome analogous to HHV8 and γHV68 and the other near the left-hand end of the genome, called ORF3. γHV68 has amplified the ORF75 gene, encoding three FGARAT homologs, ORFs 75a, 75b, and 75c (66). EBV also encodes an FGARAT homolog, referred to as BNRF1, which is positionally conserved with HVS ORF3. FGARAT enzymes catalyze the fourth step in de novo purine biosynthesis, and it remains unclear why herpesviruses would require such an activity immediately following infection or during other stages of virus infection. One common theme is that purified virion particles from several gammaherpesviruses contain ORF75/BNRF1 in their tegument (12, 34, 46, 70). To our knowledge, however, there is no available data that address whether HVS ORF3 or γHV68 ORFs 75a or 75b proteins are made during infection and if these gene products also associate with the tegument. Aside from being present in the tegument, the role of viral FGARAT homologs in gammaherpesvirus biology remains unclear. Recent data suggest that EBV BNRF1 may contribute important functions for virus entry (29). However, BNRF1 localizes primarily in the cytoplasm and does not affect PML protein levels or localization (unpublished observations), while γHV68 ORF75 localizes predominantly in the nucleus (Fig. 4). HHV8 ORF75 localizes in both compartments (unpublished observations). It is likely that the sequence divergence within the family of gammaherpesvirus-encoded FGARAT genes may enable these proteins to contribute multiple and diverse functions within the gammaherpesvirus subfamily.
TABLE 2.
Similarity of cellular FGARAT gammaherpesvirus ORF75 and BNRF1 proteinsa
| Virusb | Gammaherpesvirus subfamilyc | Protein | Total size (aa)d | Conserved region (nt)e | E value | % Identity |
|---|---|---|---|---|---|---|
| HHV4 | LCV | BNRF1 | 1,318 | 607-1311 | 2e-67 | 32 |
| CeHV15 | LCV | BNRF1 | 1,314 | 618-1310 | 6e-65 | 31 |
| HHV8 | RV | ORF75 | 1,296 | 567-1290 | 3e-61 | 30 |
| OHV2 | RV | ORF75 | 1,316 | 574-1315 | 8e-60 | 28 |
| CeHV17 | RV | ORF75 | 1,298 | 581-1294 | 1e-64 | 28 |
| CalHV3 | LCV | ORF75 | 1,321 | 609-1319 | 6e-64 | 28 |
| BoHV4 | RV | ORF75 | 1,305 | 493-1302 | 7e-54 | 26 |
| AlHV1 | RV | ORF75 | 1,315 | 603-1312 | 3e-53 | 26 |
| SaHV2 | RV | ORF75 | 1,299 | 606-1289 | 1e-47 | 27 |
| MuHV4 | RV | ORF75a | 1,291 | 675-1261 | 1e-23 | 25 |
| MuHV4 | RV | ORF75b | 1,275 | 562-1270 | 1e-29 | 24 |
| MuHV4 | RV | ORF75c | 1,310 | 560-1306 | 1e-38 | 23 |
Results were generated using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi?CMD=Web&PAGE_TYPE=BlastHome).
Abbreviations with HV are as follows: Ce, cercopithecine; O, ovine; Cal, callitrichine; Bo, bovine; Al, alcelaphine; Sa, saimiriine; Mu, murid.
LCV, lymphocryptovirus; RV, rhadinovirus.
aa, amino acids.
nt, nucleotides.
While the overall size of ORF75 genes is similar to the cellular FGARAT (i.e., approximately 1,300 amino acid residues), the homology is biased within the carboxy-terminal half of these proteins (Table 2). Although the sequence identity ranges from 23 to 32%, the E values are quite low, indicating that the homology is significant (Table 2). Among cellular FGARATs encoded by different organisms, including prokaryotes, there are two well-conserved glutamine binding sites as well as an ATP binding site (18, 20, 47). None of the viral FGARAT homologs appears to have retained these motifs, at least at the primary sequence level (Fig. 9; also unpublished observations). This suggests the viral FGARAT genes may have evolved different functions from their cellular counterparts although the possibility remains that they retain FGARAT activity mediated through a noncanonical sequence and/or structure. γHV68 will be an ideal system to analyze whether FGARAT activity is retained and whether it is important for gammaherpesvirus biology. It is clear, however, that herpesvirus genes with homology to cellular enzymes do not always retain the activity of the related cellular gene product. For example, murine CMV and HHV7 encode homologs of the ribonucleotide reductase large subunit. The proteins, however, lack enzymatic activity and appear to have evolved novel functions, including the ability to block infected cells from premature apoptosis (15, 36, 60). Likewise, the CMV UL72 and UL82 (also known as pp71) proteins have sequence homology to the dUTPase family of proteins but lack dUTPase activity. However, UL72 is moderately required for replication in vitro, while pp71 binds to and degrades hDaxx (16, 17).
It is unclear why we were unable to recover infectious virus from ORF75c-null genomes although this is consistent with an earlier study (57). We envision three possibilities for this observation. First, the inability to counteract PML may preclude the virus from establishing productive infection. We consider this possibility somewhat unlikely because we were unable to recover virus when genomes were transfected into PML knockout cells. In addition, previous results indicated that ORF75c might be expressed during the later phases of infection, consistent with its being a structural protein (4, 21, 39). Therefore, it may not play a significant role in modulating viral gene expression from infectious DNA. However, the reduced replication efficiency of HSV ICP0 mutant viruses can be overcome with infection at high MOIs (24, 30). It is possible that transfection of γHV68 BACs represents infection at a low MOI. Development of a system to complement the ORF75c-null mutants and generate virus stocks with high titers would allow testing of this hypothesis. A second possibility is that ORF75c mediates important activities related to viral assembly, egress, or entry, in addition to the PML degradation function. Finally, the stop codon mutation introduced into the ORF75c gene may disrupt the transcription or translation of viral genes adjacent to ORF75c, which may confer a null phenotype. Generation of specific ORF75c mutants within the context of the virus will help to further delineate the role of ORF75c during the virus replication cycle and address these issues. Such mutants will help to clarify whether ORF75c is the only γHV68 viral protein that induces PML degradation or if ORF75c induces degradation of cellular proteins other than PML that facilitate viral replication.
The importance of the findings reported here are that we have identified a rhadinovirus gene product that has potent activity against a cellular intrinsic host response mediated by PML. Furthermore, the viral gene product that mediates this activity, ORF75c, had no assigned functions other than being a tegument protein. Our work has implications for identification of potential ORF75 functions in human rhadinoviruses, exemplified by HHV8, as well as potentially identifying novel functions for cellular FGARAT proteins. Because of the species specificity of many human herpesviruses, it has been difficult to investigate how PML might modulate herpesvirus infections in vivo. γHV68 allows an integrated approach to investigating herpesvirus biology in vitro and in vivo in a facile experimental model system. Evidence that counteracting PML activity is important for γHV68 replication suggests that analysis of γHV68 infection in PML knockout animals may be a rewarding strategy to understand how PML may regulate the amplitude and kinetics of acute and chronic herpesvirus infection in vivo.
Acknowledgments
We thank Samuel Speck for his generous gifts of γHV68 virus, protocols, and advice and Philip Stevenson for his generous gift of monoclonal antibody to γHV68 ORF4. We also thank Rosemary Rochford for protocols and advice and Frank Ramig and Rick Lloyd for critical readings of the manuscript.
This study was supported by NIH RO1 grant CA124309 (P.D.L.).
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 752388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13111-121. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 184899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 766244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, and B. Fleckenstein. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 665047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. (Erratum, 185:946.) [DOI] [PubMed] [Google Scholar]
- 7.Allday, M. J., D. H. Crawford, and B. E. Griffin. 1989. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J. Gen. Virol. 701755-1764. [DOI] [PubMed] [Google Scholar]
- 8.Bernardi, R., and P. P. Pandolfi. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 229048-9057. [DOI] [PubMed] [Google Scholar]
- 9.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 1521099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15627-636. [DOI] [PubMed] [Google Scholar]
- 11.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B 356437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bortz, E., J. P. Whitelegge, Q. Jia, Z. H. Zhou, J. P. Stewart, T. T. Wu, and R. Sun. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 7713425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowling, B. L., and A. L. Adamson. 2006. Functional interactions between the Epstein-Barr virus BZLF1 protein and the promyelocytic leukemia protein. Virus Res. 117244-253. [DOI] [PubMed] [Google Scholar]
- 15.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291303-305. [DOI] [PubMed] [Google Scholar]
- 16.Caposio, P., L. Riera, G. Hahn, S. Landolfo, and G. Gribaudo. 2004. Evidence that the human cytomegalovirus 46-kDa UL72 protein is not an active dUTPase but a late protein dispensable for replication in fibroblasts. Virology 325264-276. [DOI] [PubMed] [Google Scholar]
- 17.Davison, A. J., and N. D. Stow. 2005. New genes from old: redeployment of dUTPase by herpesviruses. J. Virol. 7912880-12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawid, I. B., T. C. French, and J. M. Buchanan. 1963. Azaserine-reactive sulfhydryl group of 2-formamido-N-ribosylacetamide 5′-phosphate: L-glutamine amido-ligase (adenosine diphosphate). II. Degradation of azaserine-C14-labeled enzyme. J. Biol. Chem. 2382178-2185. [PubMed] [Google Scholar]
- 19.Dellaire, G., and D. P. Bazett-Jones. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26963-977. [DOI] [PubMed] [Google Scholar]
- 20.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 2628274-8287. [PubMed] [Google Scholar]
- 21.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 8499-109. [DOI] [PubMed] [Google Scholar]
- 22.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 207266-7273. [DOI] [PubMed] [Google Scholar]
- 23.Everett, R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol. 8365-374. [DOI] [PubMed] [Google Scholar]
- 24.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 781763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89819-830. [DOI] [PubMed] [Google Scholar]
- 26.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 726581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 795078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 807995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feederle, R., B. Neuhierl, G. Baldwin, H. Bannert, B. Hub, J. Mautner, U. Behrends, and H. J. Delecluse. 2006. Epstein-Barr virus BNRF1 protein allows efficient transfer from the endosomal compartment to the nucleus of primary B lymphocytes. J. Virol. 809435-9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 782169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 716611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 786527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lembo, D., M. Donalisio, A. Hofer, M. Cornaglia, W. Brune, U. Koszinowski, L. Thelander, and S. Landolfo. 2004. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J. Virol. 784278-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling, P. D., R. S. Peng, A. Nakajima, J. H. Yu, J. Tan, S. M. Moses, W. H. Yang, B. Zhao, E. Kieff, K. D. Bloch, and D. B. Bloch. 2005. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 243565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, S., I. V. Pavlova, H. W. t. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 742029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 7710488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 21767-75. [DOI] [PubMed] [Google Scholar]
- 41.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129278-287. [DOI] [PubMed] [Google Scholar]
- 42.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 7710295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B 356569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 207234-7242. [DOI] [PubMed] [Google Scholar]
- 45.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 716619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 801574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson, D., J. Bleskan, K. Gardiner, and J. Bowersox. 1999. Human phosphoribosylformylglycineamide amidotransferase (FGARAT): regional mapping, complete coding sequence, isolation of a functional genomic clone, and DNA sequence analysis. Gene 239381-391. [DOI] [PubMed] [Google Scholar]
- 48.Peng, R., A. V. Gordadze, E. M. Fuentes Panana, F. Wang, J. Zong, G. S. Hayward, J. Tan, and P. D. Ling. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng, R., S. C. Moses, J. Tan, E. Kremmer, and P. D. Ling. 2005. The Epstein-Barr virus EBNA-LP protein preferentially coactivates EBNA2-mediated stimulation of latent membrane proteins expressed from the viral divergent promoter. J. Virol. 794492-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Precious, B., K. Childs, V. Fitzpatrick-Swallow, S. Goodbourn, and R. E. Randall. 2005. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 7913434-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 811-19. [DOI] [PubMed] [Google Scholar]
- 52.Preston, C. M., and M. J. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 717807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 9314862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 803863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 736405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, M. J., S. Hwang, W. H. Wong, T. T. Wu, S. Lee, H. I. Liao, and R. Sun. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc. Natl. Acad. Sci. USA 1023805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2403-409. [DOI] [PubMed] [Google Scholar]
- 59.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 672571-2585. [DOI] [PubMed] [Google Scholar]
- 60.Sun, Y., and J. Conner. 1999. The U28 ORF of human herpesvirus-7 does not encode a functional ribonucleotide reductase R1 subunit. J. Gen. Virol. 802713-2718. [DOI] [PubMed] [Google Scholar]
- 61.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 808006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304160-166. [DOI] [PubMed] [Google Scholar]
- 64.Ulane, C. M., A. Kentsis, C. D. Cruz, J. P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 7910180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11371-379. [DOI] [PubMed] [Google Scholar]
- 66.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 715894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 2791547-1551. [DOI] [PubMed] [Google Scholar]
- 68.Wiesmeijer, K., C. Molenaar, I. M. Bekeer, H. J. Tanke, and R. W. Dirks. 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140180-188. [DOI] [PubMed] [Google Scholar]
- 69.Yu, Y., S. E. Wang, and G. S. Hayward. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 2259-70. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]