Abstract
Most human immunodeficiency virus type 1 (HIV-1)-infected individuals develop an HIV-specific neutralizing antibody (NAb) response that selects for escape variants of the virus. Here, we studied autologous NAb responses in five typical CCR5-using progressors in relation to viral NAb escape and molecular changes in the viral envelope (Env) in the period from seroconversion until after AIDS diagnosis. In sera from three patients, high-titer neutralizing activity was observed against the earliest autologous virus variants, followed by declining humoral immune responses against subsequent viral escape variants. Autologous neutralizing activity was undetectable in sera from two patients. Patients with high-titer neutralizing activity in serum showed the strongest positive selection pressure on Env early in infection. In the initial phase of infection, gp160 length and the number of potential N-linked glycosylation sites (PNGS) increased in viruses from all patients. Over the course of infection, positive selection pressure declined as the NAb response subsided, coinciding with reversions of changes in gp160 length and the number of PNGS. A number of identical amino acid changes were observed over the course of infection in the viral quasispecies of different patients. Our results indicate that although neutralizing autologous humoral immunity may have a limited effect on the disease course, it is an important selection pressure in virus evolution early in infection, while declining HIV-specific humoral immunity in later stages may coincide with reversion of NAb-driven changes in Env.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) has developed multiple mechanisms to evade the host humoral immune response, including oligomeric exclusion, occluded (co)receptor binding sites (12, 14, 17, 28), and heavy glycosylation. Despite its protective structure, Env is a major target of the humoral immune response in HIV-1-infected individuals. Antibodies directed against Env can be detected early in infection and are able to neutralize autologous virus variants with increasing titers over time in most patients (1, 33, 38, 46). However, Env evolves rapidly to escape from these neutralizing antibodies (NAbs). This evokes the development of new NAbs, thus leading to successive cycles of antibody production and viral escape (1, 31, 38, 46).
In response to NAb pressure, the numbers and/or the positions of carbohydrates can evolve to create a continuously changing glycan shield on the surface of Env (46). Large sequence variation in the variable loops, including large insertions and deletions, and changes in the number of potential N-linked glycosylation sites (PNGS) in these regions have also been associated with escape from NAbs. In particular, length and glycosylation characteristics of the V1V2 loop seem to play a role in resistance against NAbs (8, 11, 34, 39, 40, 44), possibly by shielding underlying regions of Env from antibody recognition (20). Finally, single amino acid substitutions may be responsible for the diversification of Env and escape from NAbs (16, 18).
Despite this repertoire of viral escape mechanisms, several studies have indicated that there may be limits to the capacity of Env to escape from NAbs (6, 15, 16). In addition, a sustained HIV-1-specific antibody response with greater breadth and magnitude has been associated with long-term nonprogression (9, 10, 49), although this has not been confirmed by others (2, 22), while loss of neutralizing activity has been associated with disease progression (10, 25). Moreover, rhesus macaques that were prevented from eliciting an anti-simian immunodeficiency virus (SIV) antibody response rapidly progressed to AIDS upon challenge with a pathogenic SIV strain, indicating that antibodies are critical for the control of SIV infection (30).
To date, the development of NAb responses and the subsequent evolution of Env under the influence of NAb pressure have mainly been studied in the initial phase of infection or cross-sectionally in acutely versus chronically infected patients. Here, we present for the first time a longitudinal study of HIV-1 humoral immunity in relation to Env evolution that covers the entire course of infection from seroconversion (SC) to symptomatic disease. We show that, particularly early in infection, NAbs have a large effect on the evolution of Env. Reversion of NAb-induced changes was observed late in infection in the face of declining neutralizing immunity, suggestive of an effect of these changes on viral fitness.
MATERIALS AND METHODS
Patients and viruses.
The patients in our present study were homosexual male participants of the Amsterdam Cohort Studies on HIV/AIDS (ACS) who seroconverted during active follow-up and who progressed to AIDS in the presence of CCR5-using (R5) HIV-1 variants only, as shown by 3-monthly negative MT2 assays. For all virus variants studied here, CCR5 usage was predicted by the V3 loop sequence and confirmed by the inability of these viruses to replicate in the MT2 cell line. For better readability, patient identifiers were recoded as H1 (ACH19999), H2 (ACH19542), H3 (ACH18969), H4 (ACH19768), and H5 (ACH19659). Ranking was based on the abilities of sera to neutralize autologous HIV-1 variants as demonstrated in this study. All patients received antiretroviral (mono)therapy for various periods of time. As this was generally not reflected in changes in viral loads or CD4+ T-cell counts, we considered it ineffective. In those cases where therapy resulted in changes in CD4 counts and/or viral loads, time points of virus isolation were chosen before the start of therapy or after CD4 counts and/or viral loads had returned to the pretreatment levels, with the exception of the fourth time point in patient H3 (Fig. 1). Clonal HIV-1 variants were obtained as previously described (42). For further study, we selected a maximum of 10 virus variants per patient per time point, of which a maximum of five virus variants were studied for neutralization susceptibility. Viruses were selected on the basis of their replication capacities, defined as the first day of detectable p24 production in the microculture after the start of the clonal virus isolation procedure. To prevent a change in neutralization sensitivity of the virus variants during in vitro culture, the number of peripheral blood mononuclear cell (PBMC) passages of viruses was kept to a minimum (4).
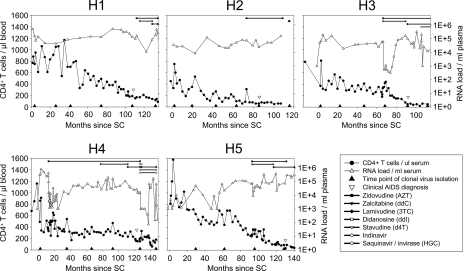
FIG. 1.
CD4+ T-cell numbers, viral loads, and antiretroviral treatments of five typical R5 progressors from the ACS. All patients had an asymptomatic follow-up of 7 to 11 years between SC and clinical AIDS diagnosis. The length and type of antiretroviral therapy are indicated at the top of each diagram.
Primary cells.
PBMCs were isolated from buffy coats obtained from healthy seronegative blood donors by Ficoll-Isopaque density gradient centrifugation. The cells (5 × 106/ml) were stimulated for 3 days in Iscove's modified Dulbecco medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and phytohemagglutinin (PHA) (5 μg/ml). Subsequently, the cells (106/ml) were grown in the absence of PHA in medium supplemented with recombinant interleukin-2 (20 U/ml; Chiron Benelux, Amsterdam, The Netherlands) and Polybrene (5 μg/ml; hexadimethrine bromide; Sigma, Zwijndrecht, The Netherlands).
Autologous neutralization assay.
Viruses were tested for their relative neutralization sensitivities against autologous serum and pooled sera from healthy, uninfected individuals. To prevent possible complement-mediated antibody inhibition of virus infection, complement in human sera and fetal bovine serum was inactivated by 30 min of incubation at 56°C. From each virus isolate, a final inoculum of 20 50% tissue culture infective doses in a volume of 50 μl was incubated for 1 h at 37°C with twofold serial dilutions of serum (range, 1/50 to 1/3,200). Subsequently, the mixtures of virus with serum were added to 105 PHA-stimulated PBMCs derived from healthy blood donors. After 4 h of incubation, the PBMCs were washed once in 100 μl phosphate-buffered saline. On day 7, virus production in culture supernatants was analyzed by an in-house p24 antigen capture enzyme-linked immunosorbent assay (45). Experiments were performed in triplicate. The percent neutralization was calculated by determining the reduction in p24 production in the presence of the agent compared to the cultures with virus only. When possible, 50% inhibitory concentrations (IC50s) were determined by linear regression. For calculations, viruses with IC50s of <50 or >3,200 were assigned a value of 25 or 3,200, respectively.
Sequence analysis.
env was amplified from DNA isolated from infected PBMCs and subsequently sequenced as described previously (5, 7, 37). The nucleotide sequences of all virus clones from an individual were aligned using ClustalW in the software package BioEdit (21) and edited manually. The reference sequence HXB2 was included in the alignment to number each aligned residue according to the corresponding position in this reference sequence. Insertions were given the number of the residue prior to the insertion, followed by a letter to identify each inserted residue. Genetic analyses were performed on gp160 sequences starting at nucleotide position 91, which excludes the Env signal peptide. PNGS were identified using N-Glycosite (48) at the HIV database website (http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html). Net charges of gp160 were calculated by counting all charged amino acid residues per sequence, with R and K counted as +1, H as +0.293, and D and E as −1. Nonsynonymous substitution (dN) and synonymous substitution (dS) rates were calculated using Synonymous Nonsynonymous Analysis Program (SNAP) (27) at the HIV database website (http://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html). To ensure a correct calculation of dN/dS ratios in the variable loops, the codon alignments of these regions were corrected manually. Codons containing indels are excluded in this method. dN/dS ratios between succeeding time points were calculated by averaging the dN/dS ratios between all individual pairs of env sequences from these time points.
Statistical analysis.
Means of gp160 lengths and numbers of PNGS were compared using a t test for independent samples. Correlations between autologous NAb responses and Env characteristics were analyzed using a Spearman's rank test (SPSS software package).
Nucleotide sequence accession numbers.
All sequences included in this study have been deposited in GenBank (accession numbers EU743973 to EU744175).
RESULTS
Neutralizing humoral immunity against autologous HIV-1.
To study the evolution of the HIV-1 envelope gene and changes in neutralization sensitivity over the course of infection, we isolated clonal HIV-1 variants from PBMCs that were obtained at six time points covering the disease course from SC up to 2 to 3 years after clinical AIDS diagnosis from five typical progressors from the ACS who never developed X4 HIV-1 variants (R5 progressors) (Fig. 1). In patient H5, attempts to isolate clonal HIV-1 variants from PBMCs obtained at SC and at time points after AIDS diagnosis were not successful (Fig. 1).
For each patient, autologous neutralizing activities in sera obtained at or close to the time points of virus isolation were measured for a maximum of five randomly selected clonal HIV-1 variants per time point (Fig. 2). The number of HIV-1 variants tested was limited by the amounts of patient sera available. Sequence characteristics of the virus variants used for analyzing the autologous NAb response were similar to those of other virus variants obtained from the same time point (0.2 to 3.6% sequence variation in env between clones from one time point).
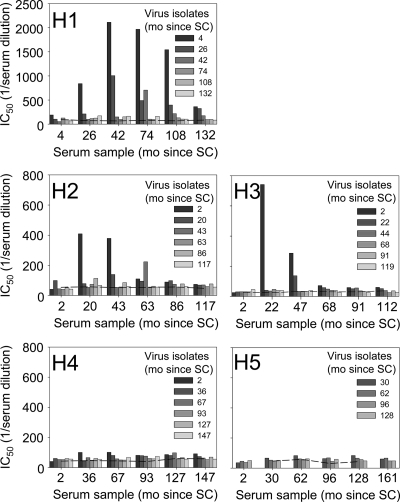
FIG. 2.
Development of autologous humoral immune responses. Average IC50s, determined by linear regression, of ≤5 virus variants per time point are indicated. Bars with identical shading represent inhibition of virus isolates from one time point by sera from different time points (as indicated on the x axis). The dashed lines represent background measurements using pooled sera from uninfected individuals. Note that the maximum value on the y axis in the graph for patient H1 is higher than the others. mo, months.
Large variation was observed between the autologous NAb responses in the five patients. Neutralization of autologous virus variants was observed only in patients H1, H2, and H3. In agreement with findings by others (38, 46), virus variants were not neutralized by contemporaneous serum and sera from earlier time points, suggestive of viral escape. Serum neutralizing activity reached the highest titers against the earliest virus variants and was much less potent against variants from subsequent time points. For viruses from the same time point, serum titers that established neutralization showed only minimal variation. Moreover, this variation was always smaller than the variation in neutralizing serum titers for viruses from different time points. In patient H1, autologous NAb responses persisted until clinical AIDS diagnosis (108 months after SC), whereas the autologous antibody responses in patients H2 and H3 were already lost during the asymptomatic phase of infection. Interestingly, heterologous neutralizing activity, which was analyzed longitudinally in patient H2, continued to increase in breadth and magnitude until 73 months after SC (M. van Gils, unpublished data). Patients H4 and H5 did not develop a detectable autologous NAb response, as has been described for other patients (38). However, as we failed to isolate clonal virus variants from patient H5 at SC, we cannot exclude the presence of NAbs against earlier virus variants from this individual. Interestingly, sera from patients H4 and H5 were both able to neutralize three out of a panel of five heterologous subtype B viruses with low to moderate titers (van Gils, unpublished), indicating that low-level antibody responses had indeed developed in these patients.
Evolution of Env in the course of infection.
To analyze molecular changes in the viral envelope, full-length gp160 genes from a median of 8 virus variants (range, 2 to 10) from each time point were sequenced. From the first time point of H5, we generated a single env sequence from proviral DNA in patient PBMCs. Phylogenetic trees of all sequences were constructed using the neighbor-joining and the maximum-likelihood methods. In both trees, sequences from each individual patient grouped together (data not shown), excluding superinfection and contamination of samples. For each patient, the consensus gp160 sequence of virus variants isolated at SC is shown in Fig. S1 in the supplemental material.
gp160 length and number of PNGS.
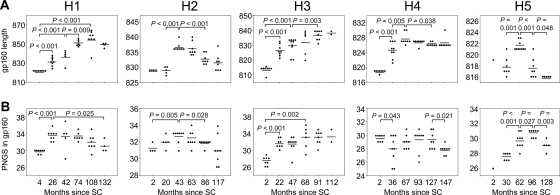
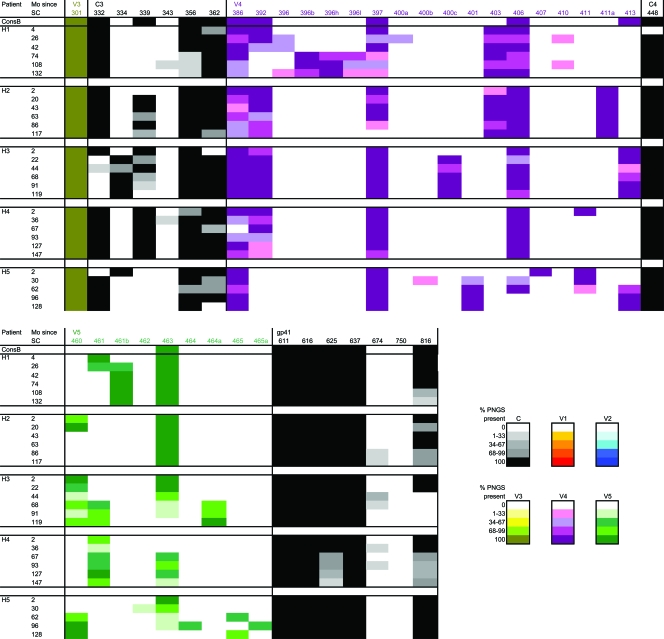
Escape from NAbs of HIV-1 has been associated with increases in both the length and glycosylation of Env (13, 40, 46). Viruses from all of our patients showed an increase in gp160 length early in infection, followed by a decrease toward the end stage of disease in patients H1, H2, H4, and H5 (Fig. 3A). A similar pattern over the course of infection was observed for the number of PNGS, although patient H4 did not show the initial increase but only a significant decrease in the number of PNGS later in the course of infection (Fig. 3B and Table 1). Changes in gp160 length and number of PNGS were not always temporally related. In patient H5, for example, the average gp160 length of coexisting HIV-1 variants peaked at 62 months after SC, whereas the number of PNGS continued to increase until 128 months after SC. In viruses from patient H1, on the other hand, the peak in the number of PNGS preceded the peak in gp160 length. Of note, in this patient, viruses showed an exceptional increase in gp160 length, from an average of 823 amino acids (aa) at 4 months after SC to an average of 855 aa at 108 months after SC (Table 1), as a result of continuous expansion of the V1, V2, and V4 regions (data not shown).
FIG. 3.
Longitudinal analysis of changes in gp160 length and number of PNGS. Each dot represents one clonal virus variant. The horizontal bars indicate average values per time point. P values were calculated using a t test for independent samples. (A) Length of gp160. (B) Number of PNGS in gp160.
TABLE 1.
Correlates of autologous neutralizationa
| Patient | Autologous NAb rank | Length (aa)
|
No. of PNGS in gp160
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gp160
|
gp120 V (peak) | V1V2 (peak) | V4 (peak) | |||||||
| SC | Peak | Δ | SC | Peak | Δ | |||||
| H1 | 1 | 823.1 | 855.4 | 32.3 | 174.4 | 82.6 | 47.3 | 30.9 | 33.2 | 2.3 |
| H2 | 2 | 829.0 | 836.6 | 7.8 | 155.8 | 76.5 | 35.0 | 31.3 | 32.6 | 1.4 |
| H3 | 3 | 813.4 | 836.5 | 23.1 | 154.5 | 75.6 | 34.0 | 27.4 | 33.0 | 5.6 |
| H4 | 4 | 818.9 | 827.6 | 8.8 | 144.9 | 70.0 | 31.0 | 29.6 | 29.3 | -0.4 |
| H5 | 5 | 819.0 | 821.6 | 4.0 | 140.6 | 69.2 | 29.0 | 26.0 | 30.8 | 5.0 |
Correlations between potency of the autologous NAb response and the length of gp160 or the number of PNGS of the virus population early after seroconversion (SC), the maximum average gp160 length or number of PNGS during infection (peak), and the increase in gp160 length or number of PNGS during infection (Δ) are shown. The regions in gp160 where maximum length correlated significantly with the potency of the NAb response are also presented. P values (Spearman) for lengths and PNGS numbers, from left to right, were as follows: 0.391, <0.001, 0.188, <0.001, <0.001, <0.001, 0.104, 0.104, and 0.873.
Length variation in gp160 could be attributed almost completely to changes in the variable loops. Insertions and deletions were observed in V1 for all patients, and additionally, in V2, V4, and/or V5 for some patients (data not shown). There were no associations between the region of the indels and the presence or absence of an autologous NAb response (data not shown).
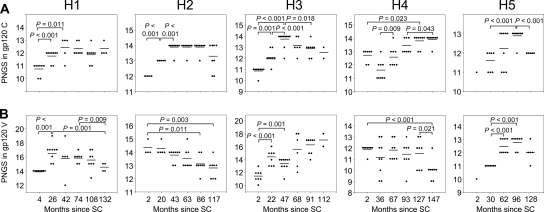
Viruses from four out of five patients showed an increase in PNGS in the constant regions early in infection, followed by stabilization or a decrease at later time points (Fig. 4A). For the variable regions, acquisition and/or loss of PNGS were more variable (Fig. 4B). Viruses from patient H3 showed a continuous increase, viruses from patients H2 and H4 a continuous decrease, and viruses from patients H1 and H5 an increase followed by a decrease in PNGS in the variable regions. These results indicate that PNGS in both the constant and the variable regions of gp120 are involved in Env evolution.
FIG. 4.
Longitudinal analysis of changes in the number of PNGS in the constant and variable regions of gp120. Each dot in the graphs indicates the characteristics of one clonal virus variant. The horizontal bars indicate the averages per time point. P values were calculated using a t test for independent samples. (A) Number of PNGS in the constant regions of gp120. (B) Number of PNGS in the variable regions of gp120.
Locations of PNGS.
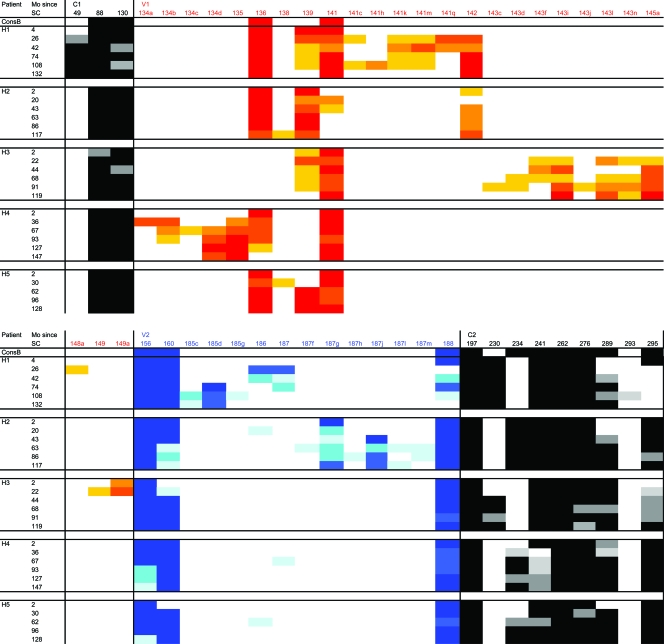
To study changes in the glycan shield in more detail, we analyzed the locations of PNGS in all Env sequences (Fig. 5). Most variation in the locations of the PNGS could be observed in the variable loops V1, V2, V4, and V5 as a result of insertions and deletions in these regions. The PNGS at position 301 in V3 was preserved in the course of infection in all virus variants from all subjects. In the constant regions, the locations of PNGS appeared to be restricted to more specific positions within the molecule. Introductions of glycans at nonconserved sites, such as N49 in patient H1, were not frequently observed. In general, PNGS that were conserved among subtype B sequences were also relatively conserved among virus variants from our patients. A number of PNGS were present in all sequences we analyzed, including N197, N262, N301, N611, N616, and N637. Moreover, we observed that some conserved PNGS present early in infection were absent during later stages of disease, such as N816 in gp41, which was lost in some of the viruses from patients H1, H2, and H4 and completely absent in the late viral quasispecies in patient H3. On the other hand, a number of relatively conserved PNGS that were (partially) absent in the earliest virus population emerged in the course of infection, for example, N160 in V2 in patients H3 and H5 and N289 in C2 in patients H1, H2, H4, and H5. Of note, the absence of N289 in early virus variants predicts the resistance of these viruses against monoclonal antibody 2G12, as we previously observed for early viruses in other patients (37).
FIG. 5.
Changes in the locations of PNGS in gp160 sequences over the course of infection. The percentage of viruses with a PNGS at a given position per patient per time point is color coded using increasingly darker shades of gray for the constant regions of gp120 and gp41 and increasingly darker shades of red, blue, yellow, purple, and green for the V1, V2, V3, V4, and V5 regions, respectively. Mo, months.
Net charge of gp160.
Since changes in the net charge of the V1V2 loop during infection have been reported previously (6), we calculated the net charges of all separate variable regions of gp160. No uniform changes in the Env net charge were observed over the course of infection, nor did we observe a decrease in the net charge in any of the variable regions of Env at the later time points (data not shown).
Single amino acid substitutions.
The consensus sequences of each time point per patient were aligned with the consensus sequence of HIV-1 subtype B Env to analyze changes in single amino acids that were not part of PNGS. Most amino acid substitutions that became dominant in the viral quasispecies were either forward or reverse mutations relative to the consensus sequence, whereas, for only a minority of substitutions, a nonconsensus B amino acid residue mutated to another nonconsensus B amino acid residue. Most mutations became fixed in the viral quasispecies, although some forward mutations reverted at a later time point. A number of forward (n = 34) and reverse (n = 11) mutations occurred in viruses from more than one patient: we observed 25 identical forward mutations and 7 identical reversions in viruses from two patients and 8 identical forward mutations and 4 identical reversions in viruses from three patients, whereas forward mutation D325N occurred in virus variants from four patients (Table 2). Although the majority of mutations that occurred in virus variants from more than one patient were not linked to a specific region in the gp160 env gene, six forward and four reverse mutations were found in relatively close proximity in V3 and C3 (between HXB2 aa 308 and 360). In gp41, 3 identical amino acid reversions and 13 identical forward mutations were observed, 2 of which were located in the epitope for 4E10 (T676S and N677K). Identical mutations in other parts of the env gene (i.e., from C1 to C2 and from V4 to C5) occurred in amino acid residues that to our knowledge have not been described as being involved in NAb escape or (co)receptor usage. Of the 13 mutations that occurred in viruses from three or more patients, 2 mutations were located in close proximity in C1 (V84I and E87K), 3 mutations were located in the V3 region (K315R, E321aD, and D325N), 1 mutation was observed close to the V3 loop (L333I), and 7 mutations were found in gp41 (Table 2).
TABLE 2.
Single amino acid substitutions that became dominant in the viral quasispecies in at least three of five patients
| Gene | Region | Mutationa | Directionb | Presence in patientc
|
||||
|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | ||||
| gp120 | C1 | V84I | F | + | + | + | ||
| E87K | F | + | + | + | ||||
| V3 | K315R | R | + | + | + | |||
| E321aD | F | + | + | + | ||||
| D325N | F | + | + | + | + | |||
| C3 | L333I | R | + | + | + | |||
| gp41 | HR1 | L543Q | R | + | + | + | ||
| K588R | F | + | + | + | ||||
| HR2 | N624D | R | + | + | + | |||
| D624E | F | + | + | + | ||||
| E630Q | F | + | + | + | ||||
| Intracellular domain | P724Q | F | + | + | + | |||
| V829I | F | + | + | + | ||||
Amino acid residues are numbered according to the corresponding positions in the reference sequence HXB2.
Substitutions were categorized as either a forward (F) or a reverse (R) mutation relative to the consensus B sequence.
+, present.
Selection pressure on Env.
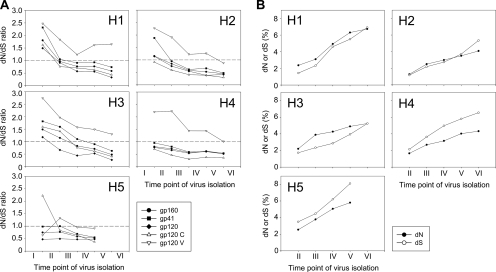
Selection pressure is generally measured as the ratio of the rates of dN and dS, where a dN/dS ratio of >1 is indicative of positive selection. We calculated dN/dS ratios between every pair of successive time points for gp160, gp120, and gp41 and also separately for the constant and variable regions of gp120. Positive selection pressure on gp160, gp120, and gp41, albeit decreasing over time and absent at AIDS diagnosis, was observed in patients H1, H2, and H3, who all had autologous serum neutralizing activity (Fig. 6A). However, selection pressure on the variable domains of gp120 was evident up to or even past the point of AIDS diagnosis in all patients except H5. Interestingly, the dN/dS ratio of gp120 showed a relatively linear decrease, whereas the selection pressure on the variable regions of gp120 in patients H1, H2, and H4 stabilized or increased temporarily toward AIDS (between time points IV and V). For patient H5, selection pressure was observed on the constant regions of gp120 only between the first and second time points, shifting to the variable regions of gp120 between the second and third time points.
FIG. 6.
dN and dS rates in env during the course of infection. (A) Selection pressure on Env, expressed as the ratio between dN and dS. Ratios are shown for full-length gp160, gp41, and gp120 and constant regions and variable regions of gp120 between viruses from successive time points. The dashed lines indicate a dN/dS ratio of 1.0. dN/dS ratios of >1.0 are indicative of positive selection. (B) Divergence of dN and dS over the course of infection relative to the virus population present shortly after SC (time point I in panel A). For all patients, time point V corresponds to the moment of clinical AIDS diagnosis.
In the course of infection, divergence of dN from the earliest virus population was higher than the divergence of dS in the patients who showed autologous neutralizing activity (Fig. 6B). In agreement with previous reports (29, 47), dN divergence stabilized at later time points in these patients, whereas the divergence of dS continued to increase. In patients H4 and H5, divergence of dN was lower than divergence of dS over the entire course of infection, compatible with the absence of detectable autologous neutralizing activity in their sera.
Correlates of autologous neutralization.
To determine if certain Env characteristics are important for eliciting an effective humoral immune response, we analyzed a potential association between neutralizing titers against autologous virus variants and the number of PNGS and lengths of the V1, V2, V4, and V5 regions and of full-length gp160 of early virus variants. Patients were ranked according to the potency of autologous neutralizing activity in their sera. In this ranking, patient H1 performed best, followed by H2, H3, H4, and finally H5 (Table 1). Although the IC50 titers of patient H2 were lower than those of patient H3, patient H2 was given a higher rank, because the NAb response was maintained for a longer time during the chronic phase of infection. For patient H5, we were unable to determine the autologous NAb response against an early virus population due to our inability to isolate virus variants early after SC. However, the absence of a broad heterologous NAb response (data not shown) and high and/or long-term selection pressure on gp160 all suggest that this patient failed to mount a vigorous immune response. As shown in Table 1, the efficiency of the NAb response did not correlate with gp160 length or the number of PNGS of the earliest viruses.
Finally, we analyzed whether changes in gp160 length or the number of PNGS during chronic infection was associated with the potency of the humoral immune response. Although there was no correlation between the efficiency of the NAb response and the increase in the number of PNGS and gp160 length, we did observe a significant association between the NAb response and the maximum average length of virus variants, both in total gp160 length and in V1V2 and V4 separately, later in infection (Table 1). The peak average number of PNGS did not significantly correlate with the efficiency of the NAb response, although viruses from patients H1, H2, and H3, who showed neutralizing activity against their autologous viruses, had a higher maximum number of PNGS (range, 32.6 to 33.2) than patients H4 and H5, who did not develop an effective autologous NAb response (range for peak average number of PNGS, 29.3 to 30.8).
DISCUSSION
HIV-1 evolution in relation to escape from humoral immunity has been studied during the early phase of infection (38, 46). Here, for the first time, we report the neutralizing humoral immune response against HIV-1 and viral escape from this response in relation to molecular changes of the viral envelope during the entire course of infection in five participants in the ACS. We selected individuals with a typical disease course who never developed CXCR4-using variants, as that would have obscured our analysis of Env evolution.
Three patients included in this study developed a NAb response against their autologous viruses. In agreement with previous observations (46), autologous neutralization declined during chronic infection as a result of both viral escape and the inability of the humoral immune system to respond to these newly emerging viral escape variants. Our patients had more or less similar typical disease courses (time between SC and AIDS diagnosis, 7 to 11 years) yet demonstrated a large variation in the development of a potent autologous NAb response. We therefore conclude that, at least in these patients, humoral immunity does not have a major impact on disease progression (35). However, we focused only on the neutralizing capacity of the humoral immune response, whereas antibodies might also be involved in other protective processes, such as antibody-dependent cellular cytotoxicity (3, 24). Moreover, the possibility that humoral immunity in combination with other host factors, such as HIV-1-specific cytotoxic T lymphocytes, and/or innate immune factors, such as APOBEC3F/G or TRIM5α, may protect against HIV-1 disease progression cannot be excluded.
In agreement with a previous report (18), we observed early in infection an increase in the lengths of the variable loops and in the number of PNGS in gp160 irrespective of detectable autologous neutralizing activity in serum. However, HIV-1 variants from patients H1, H2, and H3, who all developed a potent autologous NAb response, showed evidence of positive selection on the envelope gene, as well as a relatively high nonsynonymous divergence over synonymous divergence in the first phase of infection. These findings most likely reflect a higher NAb pressure on Env in these patients than in the patients who did not elicit a detectable autologous NAb response.
In general, positive selection was strongest on the variable regions of gp120, in agreement with the widely accepted view that a large proportion of the autologous antibody response is directed against these regions. Selection pressure was high in the initial phase of infection and decreased over time, possibly reflecting the inability of humoral immunity to respond to newly evolving virus variants and/or the limits to sequence variation compatible with viral replication. Stabilization of the dN late in infection may be a reflection of attenuated humoral immunity (29, 47), which may also allow Env to lower its defenses, leading to a reduction in gp160 length and the number of PNGS. However, the absence of an absolute temporal relationship between the dynamics of the length and level of glycosylation of Env indicate that the influence of selection processes on these Env characteristics may be different.
We do not exclude the possibility that selection pressures other than NAbs may also influence the evolution of Env. For example, the efficiency of (co)receptor usage may be an important selective process in HIV-1 Env evolution during later stages of disease (26, 43), possibly driven by a reduction in the availability of target cells. In this respect, the stabilization and/or increase of the selection pressure on the variable regions of gp120 observed late in infection in three out of five patients may be indicative of the evolution of Env toward more efficient coreceptor use, although a correlation between selection pressure on the variable regions of gp120 and CD4+ T-cell decline was absent in our patients. Furthermore, single amino acid mutations may have been selected by cytotoxic-T-lymphocyte pressure. Indeed, we have recently observed that up to 50% of amino acid changes in Env occur in known or predicted cytotoxic-T-lymphocyte epitopes (32).
The occurrence of identical mutations in HIV-1 variants from three or more patients studied here may indicate that HIV-1 has developed general ways to adapt to changing environments, for example, during or after transmission, to escape from NAbs or in response to decreasing numbers of target cells. This may be true in particular for V3, known as a conserved neutralizing domain (19), as four out of six mutations in gp120 observed in viruses from three patients (or in four patients for mutation D325N) were located in or near the V3 loop. Functional requirements render the V3 loop very conserved (50) with respect to length and glycosylation, which contrasts with the other variable regions, in which large variations, both indels and changes in glycosylation, seem to be allowed without consequences for viral replication fitness (41). The fact that multiple common reverse mutations relative to the consensus B sequence were observed may be indicative of sequence constraints on at least some regions of Env (23). However, common amino acid changes observed in this study may be related to the fact that we studied only a small group of patients infected with relatively closely related virus strains (see Fig. S1 in the supplemental material). A general role for the common mutations in Env, as observed here, in HIV-1 neutralization sensitivity remains to be established. Indeed, functional analyses may reveal for each substitution described here whether it is involved in NAb escape or is, for example, a compensatory mutation.
Analysis of the changes in the glycan shield over the course of infection showed that although virus populations in all patients displayed similar profiles of addition and loss of PNGS, most changes occurred in a patient-specific way. Changes in PNGS were not confined to the variable regions of Env but were also observed in the constant regions, although PNGS in the constant regions appeared to be more restrained with respect to their positions. A number of PNGS, including N88, N197, and N262 in the constant regions of gp120 and N301 in V3, as well as N611, N616, and N637 in gp41, were very conserved, indicating that glycans at these positions are likely to be important for the conformation and/or the functionality of the Env trimer. The acquisition of relatively conserved PNGS early in infection and the loss of such PNGS late in infection may suggest that these glycans are involved in the resistance of Env against NAbs. Moreover, the overall density of glycans on the Env trimer may be another important mechanism of escape from NAbs, as suggested by the trend observed here, where the potency of the NAb response was associated with the peak number of PNGS during infection.
The virus variants that were used in this study were propagated on PBMCs, which may coincide with sequence changes in Env (4, 36). However, these sequence changes seem to be rare, as multiple parallel long-term cultures of clonal virus variants on PBMCs resulted in only very few mutations in the V3-V4 regions (32), pointing to the genetic stability of the virus clones.
In vaccine design, it is important to understand which viral characteristics are important for the elicitation of a broad and high-titer humoral immune response. One could argue that a virus with a compact envelope and a relatively low level of glycosylation would expose many neutralization epitopes and could therefore serve as an effective immunogen. However, when we ranked our patients according to the autologous NAb response, we did not find an association between the NAb response and gp160 length or the number of PNGS of the earliest virus population. This suggests that the presentation of neutralization epitopes depends on more factors than the envelope length and glycosylation characteristics of the transmitted virus variants. Moreover, host factors, such as the B-cell repertoire and humoral immune activation, may also play roles in the development of an efficient antibody response after vaccination.
In conclusion, we studied the HIV-1-specific humoral immune response in relation to viral escape and molecular evolution in Env. Escape from the autologous NAb response coincided with specific amino acid changes and with increases in gp160 length and the number of PNGS, followed by reversions later in infection, when the NAb response subsided. Knowledge of the Env conformation that elicits NAbs and, on the other hand, the Env conformation that resists antibody-mediated neutralization may be applied to the design of immunogens optimally capable of eliciting protective humoral immunity.
Supplementary Material
Acknowledgments
The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, and the University Medical Center Utrecht, are part of The Netherlands HIV Monitoring Foundation and are financially supported by The Netherlands National Institute for Public Health and the Environment. This study was financially supported by the Dutch AIDS Fund (grant 2004064).
We thank Angélique van ′t Wout for critical reading of the manuscript.
Footnotes
Published ahead of print on 4 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyö. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4107-112. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 2031357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum, L. L., K. J. Cassutt, K. Knigge, R. Khattri, J. Margolick, C. Rinaldo, C. A. Kleeberger, P. Nishanian, D. R. Henrard, and J. Phair. 1996. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1572168-2173. [PubMed] [Google Scholar]
- 4.Beaumont, T., E. Quakkelaar, A. van Nuenen, R. Pantophlet, and H. Schuitemaker. 2004. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 785651-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaumont, T., A. van Nuenen, S. Broersen, W. A. Blattner, V. V. Lukashov, and H. Schuitemaker. 2001. Reversal of HIV-1 IIIB towards a neutralization resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 752246-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blay, W. M., S. Gnanakaran, B. Foley, N. A. Doria-Rose, B. T. Korber, and N. L. Haigwood. 2006. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80999-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. Van der Noordaa. 1991. A rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 719808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, Y., L. Qin, L. Zhang, J. T. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332201-208. [DOI] [PubMed] [Google Scholar]
- 10.Cecilia, D., C. A. Kleeberger, A. Munoz, J. V. Giorgi, and S. Zolla-Pazner. 1999. A longitudinal study of neutralizing antibodies and disease progression in HIV-1-infected subjects. J. Infect. Dis. 1761365-1374. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 717719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433834-841. [DOI] [PubMed] [Google Scholar]
- 13.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 735294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks, S. G., B. Schweighardt, T. Wrin, J. Galovich, R. Hoh, E. Sinclair, P. Hunt, J. M. McCune, J. N. Martin, C. J. Petropoulos, and F. M. Hecht. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 806155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draenert, R., T. M. Allen, Y. Liu, T. Wrin, C. Chappey, C. L. Verrill, G. Sirera, R. L. Eldridge, M. P. Lahaie, L. Ruiz, B. Clotet, C. J. Petropoulos, B. D. Walker, and J. Martinez-Picado. 2006. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J. Exp. Med. 203529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. LaBranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 755230-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 10218514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 782394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 22.Harrer, T., E. Harrer, S. A. Kalams, T. Elbeik, S. I. Staprans, M. B. Feinberg, Y. Cao, D. D. Ho, T. Yilma, A. M. Caliendo, R. P. Johnson, S. P. Buchbinder, and B. D. Walker. 1996. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable non-progressing HIV type-1 infection. AIDS Res. Hum. Retrovir. 12585-592. [DOI] [PubMed] [Google Scholar]
- 23.Herbeck, J. T., D. C. Nickle, G. H. Learn, G. S. Gottlieb, M. E. Curlin, L. Heath, and J. I. Mullins. 2006. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J. Virol. 801637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hessell, A. J., L. Hangartner, M. Hunter, C. E. Havenith, F. J. Beurskens, J. M. Bakker, C. M. Lanigan, G. Landucci, D. N. Forthal, P. W. Parren, P. A. Marx, and D. R. Burton. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449101-104. [DOI] [PubMed] [Google Scholar]
- 25.Jolly, P. E., and H. L. Weiss. 2000. Neutralization and enhancement of HIV-1 infection by sera from HIV-1 infected individuals who progress to disease at different rates. Virology 27352-59. [DOI] [PubMed] [Google Scholar]
- 26.Koning, F. A., D. Kwa, B. Boeser-Nunnink, J. Dekker, J. Vingerhoed, H. Hiemstra, and H. Schuitemaker. 2003. Decreasing sensitivity to RANTES neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 188864-872. [DOI] [PubMed] [Google Scholar]
- 27.Korber, B. 2000. Computational analysis of HIV molecular sequences, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), HIV signature and sequence variation analysis. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemey, P., S. L. Kosakovsky Pond, A. J. Drummond, O. G. Pybus, B. Shapiro, H. Barroso, N. Taveira, and A. Rambaut. 2007. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 3e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, C. J., M. Genesca, K. Abel, D. Montefiori, D. Forthal, K. Bost, J. Li, D. Favre, and J. M. McCune. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J. Virol. 815024-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moog, C., H. J. A. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 713734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navis, M., D. Edo Matas, A. Rachinger, F. A. Koning, P. van Swieten, N. A. Kootstra, and H. Schuitemaker. 2008. Molecular evolution of human immunodeficiency virus type 1 upon transmission between human leukocyte antigen disparate donor-recipient pairs. PLoS ONE 3:e2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176924-932. [DOI] [PubMed] [Google Scholar]
- 34.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poignard, P., R. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. H. I. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10431-438. [DOI] [PubMed] [Google Scholar]
- 36.Pugach, P., S. E. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. M. Wolinsky, B. T. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 3218-22. [DOI] [PubMed] [Google Scholar]
- 37.Quakkelaar, E. D., F. P. van Alphen, B. D. Boeser-Nunnink, A. C. van Nuenen, R. Pantophlet, and H. Schuitemaker. 2007. Susceptibility of recently transmitted subtype B human immunodeficiency virus type 1 variants to broadly neutralizing antibodies. J. Virol. 818533-8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 1004144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 811350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 809586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 799069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. Y. De Goede, R. P. Van Steenwijk, J. M. A. Lange, J. K. M. Eeftink Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J. Virol. 661354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stalmeijer, E. H., R. P. van Rij, B. Boeser-Nunnink, J. A. Visser, M. A. Naarding, D. Schols, and H. Schuitemaker. 2004. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J. Virol. 782722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 727840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tersmette, M., I. N. Winkel, M. Groenink, R. A. Gruters, P. Spence, E. Saman, G. van der Groen, F. Miedema, and J. G. Huisman. 1989. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV-p24 gag. Virology 171149-155. [DOI] [PubMed] [Google Scholar]
- 46.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 47.Williamson, S., S. M. Perry, C. D. Bustamante, M. E. Orive, M. N. Stearns, and J. K. Kelly. 2005. A statistical characterization of consistent patterns of human immunodeficiency virus evolution within infected patients. Mol. Biol. Evol. 22456-468. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 141229-1246. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y. J., C. Fracasso, J. R. Fiore, A. Bjorndal, G. Angarano, A. Gringeri, and E. M. Fenyo. 1997. Augmented serum neutralizing activity against primary human immunodeficiency virus type 1 (HIV-1) isolates in two groups of HIV-1-infected long-term nonprogressors. J. Infect. Dis. 1761180-1187. [DOI] [PubMed] [Google Scholar]
- 50.Zolla-Pazner, S. 2005. Improving on nature: focusing the immune response on the V3 loop. Hum. Antibodies 1469-72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.