Abstract
A second cytoplasmic dynein heavy chain (cDhc) has recently been identified in several organisms, and its expression pattern is consistent with a possible role in axoneme assembly. We have used a genetic approach to ask whether cDhc1b is involved in flagellar assembly in Chlamydomonas. Using a modified PCR protocol, we recovered two cDhc sequences distinct from the axonemal Dhc sequences identified previously. cDhc1a is closely related to the major cytoplasmic Dhc, whereas cDhc1b is closely related to the minor cDhc isoform identified in sea urchins, Caenorhabditis elegans, and Tetrahymena. The Chlamydomonas cDhc1b transcript is a low-abundance mRNA whose expression is enhanced by deflagellation. To determine its role in flagellar assembly, we screened a collection of stumpy flagellar (stf) mutants generated by insertional mutagenesis and identified two strains in which portions of the cDhc1b gene have been deleted. The two mutants assemble short flagellar stumps (<1–2 μm) filled with aberrant microtubules, raft-like particles, and other amorphous material. The results indicate that cDhc1b is involved in the transport of components required for flagellar assembly in Chlamydomonas.
INTRODUCTION
The dyneins are a large family of motor proteins that drive microtubule sliding in cilia and flagella and contribute to microtubule-based transport in eucaryotic cells (reviewed in Holzbaur and Vallee, 1994; Porter, 1996; Hirokawa et al., 1998). These enzymes convert the energy derived from ATP binding and hydrolysis into the minus-end–directed movement of cellular cargoes along the surfaces of microtubules (Sale and Satir, 1977; Paschal and Vallee, 1987). The dyneins also play an important role in the spatial organization of microtubule arrays (Verde et al., 1991; Li et al., 1993; Dillman et al., 1996; Koonce and Samso, 1996).
Dyneins have traditionally been separated into two distinct groups, axonemal and cytoplasmic. At least 11 different dynein heavy chain (Dhc)1 subspecies are present in the inner and outer dynein arm structures of the flagellar axoneme, where they play highly specialized roles in the generation of the flagellar waveform (Kagami and Kamiya, 1992; reviewed in Asai and Brokaw, 1993; Gibbons, 1995; Porter, 1996). In contrast, a single cytoplasmic Dhc species has been implicated in a variety of microtubule-based movements, including vesicle transport, mitotic spindle assembly and positioning, nuclear migration, and chromosome movements (reviewed in Holzbaur and Vallee, 1994; Hirokawa et al., 1998). Recently, a second cytoplasmic Dhc sequence, Dyh1b, was discovered in sea urchin embryos as a minor transcript whose expression could be stimulated by deciliation (Gibbons et al., 1994). A homologous cDhc sequence has since been detected in a wide variety of cells and tissues (Tanaka et al., 1995; Criswell et al., 1996; Vaisberg et al., 1996), although it appears to be most abundant in cells involved in some aspect of axoneme assembly (Tanaka et al., 1995; Neesen et al., 1997; Criswell and Asai, 1998). Immunolocalization studies have indicated that the cDhc1b polypeptide is concentrated in the apical cytoplasm of ciliated epithelial cells (Criswell et al., 1996), but it can also be found in close association with the Golgi apparatus in human tissue culture cells (Vaisberg et al., 1996). These studies have suggested that the Dyh1b/cDhc1b isoform might be involved in some aspect of membrane trafficking and/or ciliary and flagellar assembly.
Flagellar assembly has been most thoroughly studied in the biflagellate green alga Chlamydomonas. Both flagellar assembly and flagellar length are precisely regulated (Lefebvre and Rosenbaum, 1986; Tuxhorn et al., 1998), and >33 different genetic loci that affect flagellar assembly have been identified (reviewed in Dutcher, 1989, 1995; Harris, 1989). Experimental deflagellation leads to the rapid induction of flagellar protein synthesis (Lefebvre et al., 1978), and within 90 min, >250 flagellar proteins are assembled into two flagella, each 10–14 μm in length (reviewed in Lefebvre and Rosenbaum, 1986; Johnson and Rosenbaum, 1993). This process requires the rapid delivery of flagellar precursors to the anterior end of the cell, their specific sorting into the flagellar compartment, and their selective transport to the tips of the growing flagella, which is the site of flagellar assembly (Rosenbaum et al., 1969; Witman, 1975; Johnson and Rosenbaum, 1992). Recently, the discovery of a bidirectional transport system within the flagellum (Kozminski et al., 1993) has led to a search for motors that might mediate the process of intraflagellar transport (IFT). The initial studies identified several kinesin-related proteins associated with different classes of flagellar microtubules (Bernstein et al., 1994; Fox et al., 1994; Johnson et al., 1994), including one isoform that is the gene product of the FLA10 locus (Walther et al., 1994). The FLA10 kinesin is required for both the maintenance of IFT and the incorporation of flagellar components onto preexisting flagella (Kozminski et al., 1995; Piperno et al., 1996; Cole et al., 1998). The process of IFT appears to be widespread, because FLA10 kinesin–related proteins have also been implicated in the process of axoneme assembly in Caenorhabditis elegans sensory cilia (Shakir et al., 1993; Tabish et al., 1995), sea urchin blastula cilia (Morris and Scholey, 1997), and mouse embryonic cilia (Nonaka et al., 1998).
Because the FLA10 kinesin–related proteins are plus-end–directed microtubule motors (Yamazaki et al., 1995) and IFT is a bidirectional process, it was proposed that retrograde IFT must be driven by a minus-end–directed motor, such as cytoplasmic dynein, whose delivery to the distal end of the flagellum depended on FLA10 kinesin activity (Kozminski et al., 1995). Indeed, studies in mammalian cells and Drosophila have indicated that cytoplasmic dynein is abundant in the testis and appears to be involved in some aspect of spermatogenesis and male fertility (Collins and Vallee, 1989; Rasmusson et al., 1994; Gepner et al., 1996). Recently, a dynein light chain (LC8) has been found to be essential for retrograde IFT in Chlamydomonas (Pazour et al., 1998). Although LC8 has been associated with a number of different protein complexes (King and Patel-King, 1995; King et al., 1996; Espindola et al., 1996; Harrison et al., 1998), the defect in retrograde IFT observed in the LC8 mutant strongly suggested that a cytoplasmic dynein motor was involved in both IFT and flagellar assembly (Pazour et al., 1998).
In this study, we have asked whether a cytoplasmic Dhc has a role in flagellar assembly in Chlamydomonas. Previous PCR screens have identified nine different Dhc genes distinct from the outer arm Dhc genes described by others (Mitchell and Brown, 1994; Wilkerson et al., 1994), but none of these sequences appeared to encode a cytoplasmic Dhc (Porter et al., 1996). By modifying the PCR reaction conditions, we have now recovered four additional Chlamydomonas Dhc genes, two of which encode cytoplasmic Dhc sequences. One of these sequences, cDhc1b, is closely related to a Dhc gene that is required for the formation of sensory cilia in C. elegans (Grant, personal communication). The Chlamydomonas cDhc1b sequence is a relatively low-abundance transcript whose expression is stimulated in response to deflagellation. Restriction fragment length polymorphism (RFLP)-mapping procedures have indicated that the cDhc1b gene is closely linked to a locus implicated previously in flagellar assembly. To identify null alleles of the cDhc1b gene, we used the cDhc1b clones to screen a new collection of flagellar assembly mutants generated by insertional mutagenesis. Southern blot analysis of >70 flagellar mutants has identified two strains that are associated with significant deletions of the cDhc1b gene. Structural studies have revealed that these mutants typically assemble short flagellar stumps (<1–2 μm) filled with highly aberrant microtubules, raft-like particles, and other amorphous material. These studies indicate that the cDhc1b isoform plays an important role in flagellar assembly in Chlamydomonas. Because of the high degree of sequence conservation observed in cDhc1b sequences, it seems likely that cDhc1b isoforms may serve a similar function in other organisms.
MATERIALS AND METHODS
Cell Culture and Mutant Strains
All cells used in this study were maintained as vegetatively growing cultures at 21°C on rich medium containing sodium acetate (Sager and Granick, 1953) as described previously (Porter et al., 1992). Large-scale liquid cultures were supplemented with additional potassium phosphate as described by Witman (1986).
RNA Purification and Reverse Transcription (RT)–PCR Procedures
Total RNA was isolated from wild-type Chlamydomonas (137c, mt+) cells both before and 45 min after deflagellation induced by pH shock (Witman et al., 1972) as described previously (Wilkerson et al., 1994; Porter et al., 1996). To remove minor amounts of contaminating genomic DNA, we treated the total RNA with RQ1 DNase (Promega, Madison, WI), extracted with phenol and chloroform, and recovered by ethanol precipitation. cDNA was made from 1 μg of total RNA using AMV reverse transcriptase and random primers (Promega). To control for residual genomic DNA contamination, a second set of reactions was performed without reverse transcriptase. The resulting cDNA products were then used as templates in a series of PCR reactions containing a sense primer based on the conserved amino acid sequence KTESVKA [5′-AAG-AC(CGT)-GAG-(AT)(GC)(CGT)-GT(CG)-AAG-GC-3′] and an antisense primer based on the amino acid sequence CFDEFNR [5′-TG(CT)TTCGA(CT)GA(AG)TT(CT)AAC(CA)G-3′]. The PCR reactions contained 2 μl of cDNA, 0.2 mM dNTPs, 2 μM of each primer, 1× reaction buffer, 1.5 mM MgCl2, and 2.5 U of Taq polymerase in a total volume of 50 μl. These reactions were incubated at 95°C for 5 min, followed by 30 cycles of 58°C for 2 min, 72°C for 3 min, and 94°C for 1 min and 1 cycle of 58°C for 2 min and 74°C for 2 min, and then held at 4°C. The PCR products were run on a 1.5% agarose gel and compared against a 100-bp ladder (Life Technologies, Grand Island, NY). Products of the sizes predicted for mature transcripts were gel-purified and subcloned as described previously (Porter et al., 1996). Twenty-one PCR positive clones were sequenced, and four different Dhc sequences were identified among the subclones: cDhc1a (10 copies), cDhc1b (4 copies), Dhc10 (1 copy), and Dhc3 (1 copy).
DNA Isolation and Southern Blot Analysis
Genomic DNA was isolated from wild-type and mutant Chlamydomonas cells as described in Johnson and Dutcher (1991) and modified in Porter et al. (1996). DNA samples (3–4 μg per lane) were digested with a series of restriction enzymes, separated on 0.8–1.0% agarose gels, and transferred overnight to either Zetabind (Cuno, Meriden, CN) or Magnagraph (Micron Separation Systems, Westboro, MA) membranes according to standard procedures (Sambrook et al., 1989) and the manufacturer’s instructions. DNA probes for hybridization were purified in low-melting point agarose and radiolabeled with [32P]dCTP and random primers using either the Prime-it II labeling kit (Stratagene, La Jolla, CA) or the Rediprime labeling kit (Amersham, Uppsala, Sweden). Conditions for prehybridization and hybridization were as described previously (Porter et al., 1996; Myster et al., 1997).
Construction of a SacI Minilibrary
To facilitate the specific recovery of the cDhc1b gene, an ∼7.7-kb SacI genomic fragment spanning the region that encodes the proposed ATP-binding site was isolated from a genomic minilibrary. Twenty-five micrograms of genomic DNA were digested with the restriction enzyme SacI and size-fractionated on a 0.8% agarose gel. The region between 6 and 9 kb was cut into seven 1-mm slices, and the DNA was extracted with phenol and chloroform, ethanol precipitated, and resuspended in TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). An aliquot of each slice (one-tenth volume) was rerun on a second gel, transferred to a Magnagraph membrane, and hybridized overnight with the 150-bp fragment corresponding to the cDhc1b PCR product. The three fractions with the strongest signals were pooled and ligated overnight into SacI-digested pBluescript II. Two microliters of the ligation mixture were transformed into the Escherichia coli strain DH5αF′ using a BTX electroporator (Biotechnologies and Experimental Research, San Diego, CA) and following the manufacturer’s protocols. The transformed cells were plated on LB-Amp, and ampicillin-resistant colonies were transferred to Magnagraph membranes and hybridized overnight with the 150-bp cDhc1b PCR product. A single positive clone containing a 7.7-kb SacI fragment of the cDhc1b gene was identified out of 5000 colonies. The 7.7-kb SacI subclone was purified by CsCl centrifugation and used to screen a large insert genomic library as described below.
Recovery of Large Insert Genomic Clones
Each Dhc sequence was used to screen a λFIX library containing wild-type (21gr) genomic DNA (Schnell and Lefebvre, 1993) as described previously (Porter et al., 1996; Myster et al., 1997). The resulting phage clones were mapped with the restriction enzymes NotI and SacI, and the appropriate fragments were subcloned and sequenced to confirm the identity of the Dhc clones. Initial screens with the cDhc1b PCR product resulted in the recovery of phage clones containing either the cDhc1a gene or a new axonemal Dhc sequence, Dhc11 (see Figure 1). Rescreening the library with the 7.7-kb SacI subclone permitted the specific recovery of seven phage clones that span an ∼23-kb region of genomic DNA containing the cDhc1b gene.
Figure 1.
Identification of additional Dhc genes in Chlamydomonas. The deduced amino acid sequences of four new Dhc sequences in the region surrounding the conserved ATP hydrolytic site (P-loop 1) are shown. cDhc1a, cDhc1b, and Dhc10 were initially recovered in the RT–PCR screen, whereas Dhc11 was identified during subsequent screening of a genomic library (see MATERIALS AND METHODS for details). Dhc3 is an axonemal Dhc sequence (Porter et al., 1996) that was fortuitously isolated in the PCR screen.
Subcloning and Sequencing of Genomic DNA
Restriction fragments from the phage clones were subcloned into pBluescript KSII (Stratagene), and plasmid DNA was purified using either CsCl centrifugation, Wizard Maxi-Preps (Promega), or Quantum Preps (Bio-Rad, Riverside, CA). Selected subclones were sequenced by primer walking using ABI Prism Sequencers (Perkin Elmer, Norwalk, CT) available in the DNA Sequencing Facility (Iowa State University, Ames, IA) or the Microchemical Facility (University of Minnesota, Minneapolis, MN). The sequence data were assembled and analyzed using both the MacVector software, version 3.0, and the Genetics Computer Group (GCG; Madison, WI) sequence analysis programs, version 9.0, available through the Advanced Biosciences Computing Center (University of Minnesota, St. Paul, MN).
Potential open-reading frames were identified using the GCG program Codon Preference and a codon usage table based on Chlamydomonas nuclear sequences (see Myster et al., 1997; Nakamura et al., 1997) (Myster, Knott, and Porter, unpublished results). Potential splice donor and acceptor sites were identified on the basis of the consensus sequences found in Chlamydomonas nuclear genes (Mitchell and Brown, 1994; LeDizet and Piperno, 1995; Zhang, 1996; Myster et al., 1997) (Myster, Knott, and Porter, unpublished results; Schnell, University of Arkansas, personal communication). All splice junctions were confirmed directly by sequence analysis of RT–PCR products derived from the cDhc1b transcript.
cDNA was made from 5 μg of total RNA using Superscript II reverse transcriptase and random hexamers (Life Technologies). PCR reactions were then performed using sequence-specific primers and the Expand PCR kit containing both Taq DNA polymerase and Pwo DNA polymerase (Boehringer-Mannheim, Indianapolis, IN). All reactions were initiated by a single cycle of 94°C for 3 min, 51°C for 1 min, and 74°C for 3 min, followed by 29 cycles of 94°C for 1 min, 51°C for 2 min, and 74°C for 5 min. The PCR products were analyzed on a 1.5% agarose gel, and RT–PCR products of the appropriate size were purified using 0.8% low-melt agarose gels and Wizard PCR Preps (Promega) for direct sequencing with sequence-specific primers.
The proposed translation start site was identified by the recovery of a RT–PCR product using a forward primer located downstream of the TATA box sequence and a reverse primer designed in exon 2. Sequence analysis of the resulting product identified stop codons in all three frames preceding an ATG, which was thereby designated the translation start site.
The predicted amino acid sequence of the cDhc1b gene was compared with other Dhc sequences using the GCG programs Bestfit, Compare, and Pileup. Potential nucleotide-binding sites were identified using the GCG program Motifs, and regions with the potential to form α-helical coiled-coils were identified using the program COILS, version 2.2 (Lupus et al., 1991; Lupus, 1996).
Northern Blot Analysis
Aliquots containing 20 μg of total RNA were size fractionated on 0.75% agarose–formaldehyde denaturing gels and then transferred to either a Zetabind or Magnagraph membrane as described previously (Porter et al., 1996). RNA was immobilized on the membrane by baking at 80°C for 2 h and UV irradiation at 20,000 μJ (Stratalinker II; Stratagene). Prehybridization and hybridization conditions were as described previously (Porter et al., 1996; Myster et al., 1997). To ensure that the signals were gene specific, we obtained probes for hybridization from the 5′ end of the Dhc gene. To control for equal loading of the RNA samples, we also hybridized blots with a probe corresponding to a fragment of the CRY1 gene, which encodes the ribosomal S14 protein (Nelson et al., 1994), as described previously (Porter et al., 1996; Myster et al., 1997; Perrone et al., 1998).
RFLP Mapping
To identify a potential RFLP that might be used as a molecular marker for mapping the cDhc1b gene, we screened selected subclones by hybridization on Southern blots of genomic DNA isolated from two C. reinhardtii strains, 137c and S1-D2, that are polymorphic at the DNA sequence level (Gross et al., 1988). A specific RFLP could be observed using genomic DNA that was double digested with EcoRI–XhoI and a 3.1-kb SacI fragment derived from the 5′ end of the cDhc1b gene. The 3.1-kb SacI fragment was next hybridized to a series of mapping filters containing genomic DNA that had been isolated from tetrad progeny derived from crosses between multiply marked C. reinhardtii strains and S1-D2. The segregation pattern of the cDhc1b gene was then analyzed with respect to 42 genetic and molecular markers covering all of the known Chlamydomonas linkage groups. The mapping filters and the associated genetic and molecular markers are described in detail by Porter et al. (1996).
Isolation of Stumpy Flagella Mutations by Insertional Mutagenesis
The strain A54-e18 (ac17, nit1-1, sr1) was provided by R. Schnell and P. Lefebvre (University of Minnesota, St. Paul, MN). This strain contains an ∼10-kb deletion in the nitrate reductase (NIT1) gene and can be transformed with the plasmid pMN56. Approximately 20,000 nit+ transformants were generated as described by Nelson et al. (1994). After growth on selective medium for 10 d, positive transformants were picked into liquid medium and analyzed for flagellar assembly defects by phase-contrast light microscopy. Approximately 100 transformants were chosen for further study by electron microscopy (Dentler, unpublished results). A similar number of strains with potential flagellar assembly defects was also isolated by transformation of a nit1-305 strain with the pMN24 plasmid. These strains were generously provided by K. Kozminski and J. Rosenbaum (Yale University, New Haven, CT) and were further analyzed by both light and electron microscopy (Dentler, unpublished results).
Electron Microscopy
For structural studies of flagellar mutants, cells were grown under a 12:12 h light/dark cycle in 100-ml liquid cultures of M or R medium with air bubbling (Harris, 1989). Immotile cells were harvested from the bottom of the culture flasks with a large bore pipette and then concentrated in polypropylene tubes using an IEC clinical centrifuge at speed #3 for 3 min. The cells were resuspended in M medium containing 2% glutaraldehyde, fixed for 1 h at room temperature, pelleted again, and then resuspended in 100 mM Na cacodylate, pH 7.2, and 2% glutaraldehyde for fixation overnight at 4°C. The next day, cells were washed three times in fresh 100 mM cacodylate buffer and then post-fixed in cacodylate buffer containing 1% OsO4 for 30–60 min on ice. After three washes with distilled water, the cells were resuspended in 1% aqueous uranyl acetate for 3–12 h at room temperature. The samples were then dehydrated in an acetone series and embedded in BEEM capsules using Embed 812 resin (Electron Microscopy Services, Fort Washington, PA). Thick (∼200 nm) sections were cut on a Dupont (Wilmington, DE) MT6000 microtome, stretched with xylene vapors, and then picked up on naked 300-mesh grids. Sections were stained with 1% uranyl acetate in 50% methanol, followed by lead citrate (Hyatt, 1970), and then imaged with a JEOL 1200EXII microscope operating at 125 kV.
Extraction of Flagellar Stumps
For studies of extracted cells, wild-type and mutant strains were grown and collected as described above, resuspended in buffer (20 mM HEPES, pH 7.5, 3 mM MgSO4, 1 mM EGTA), put on ice, and then diluted with an equal volume of the above buffer containing 1% Nonidet P-40 and 6 mM EGTA. After incubation on ice for 5 min, the extracted cells were pelleted, resuspended in 100 mM Na cacodylate, pH 7.2, containing 2.5% glutaraldehyde, and fixed overnight at 4°C. In some experiments, cells were extracted with 4% Nonidet P-40 for up to 30 min before fixation. All samples were then rinsed, post-fixed, stained, and embedded as described above. Thin sections, ∼30–40 nm, were cut and stained as described above and then observed at 80 kV.
Immunofluorescence Microscopy
Cells were fixed and stained using the methods described by Sanders and Salisbury (1995). Cells were attached to polyethyleneamine-coated coverslips, fixed with cold methanol, air-dried, and rehydrated in phosphate-buffered saline (PBS). Cells were incubated with mouse anti-β-tubulin (diluted 1:500) and mouse anti-kinesin II (K2.4; diluted 1:200) antisera for 1–2 h at 37°C. Coverslips were washed with PBS and incubated with Alexa 594-labeled anti-mouse antibodies (Molecular Probes, Eugene, OR) for 1 h at 37°C. Coverslips were rinsed in PBS and mounted on slides with Gelvatol antibleach solution (Rodriguez and Deinhardt, 1960). Cells were examined with a Zeiss (Thornwood, NY) WL epifluorescence microscope, and images were captured using a DAGE SIT camera, Image Σ frame averaging computer, and Macintosh 6500 computer equipped with a Scion video board (Scion, Frederick, MD). Some cells were viewed and photographed with a Bio-Rad MRC 1000 confocal microscope.
The tubulin antibody was raised against bovine brain β-tubulin and was generously provided by Dr. R. Himes (University of Kansas, Lawrence, KS). The anti-kinesin II antibody (K2.4) was raised against the 85-kDa subunit of sea urchin kinesin II (Cole et al., 1993; Henson et al., 1997) and was generously provided by Dr. J. Scholey (University of California, Davis, CA). The K2.4 antibody specifically cross-reacts with the 90-kDa FLA10 kinesin subunit in Chlamydomonas (Cole et al., 1998).
RESULTS
Recovery of New Dhc Sequences in Chlamydomonas
To recover the cDhc genes from Chlamydomonas, we designed a series of oligonucleotide primers based on regions of sequence conservation surrounding the primary nucleotide-binding site (P-loop 1) in cDhc sequences in other organisms. These primers were then used to amplify the Dhc sequences present in cDNA prepared from vegetatively growing, nondeflagellated cells. A specific PCR product of the expected size (∼150 bp) was observed using a sense primer based on the amino acid sequence KTESVKA and an antisense primer based on the amino acid sequence CFDEFNR. The 150-bp product was subcloned, and 21 different reaction products were sequenced, yielding four distinct Dhc sequences (see MATERIALS AND METHODS). Comparison of the predicted amino acid sequences with that of other Dhc genes revealed that two of the sequences, cDhc1a and cDhc1b, are related to cDhc genes identified in other organisms (Figures 1 and 2). The other two sequences correspond to axonemal Dhc genes, Dhc3 and Dhc10, that were fortuitously amplified along with the cytoplasmic Dhc sequences (Porter et al., 1996) (our unpublished results). To verify and extend the Dhc sequences, we recovered longer clones from a genomic library (see MATERIALS AND METHODS). Because of the high degree of sequence conservation within the P-loop 1 region, the library screen yielded another axonemal Dhc sequence, Dhc11. The predicted amino acid sequences through the hydrolytic domain of the Dhc clones are shown in Figure 1.
Figure 2.
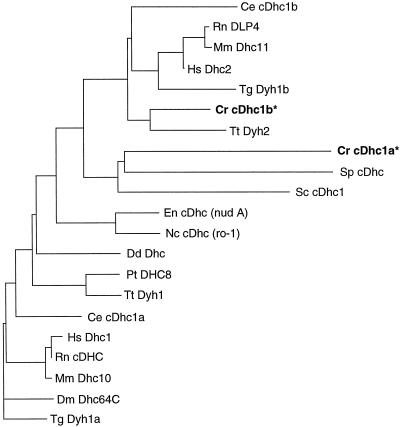
Diagrammatic alignment of cytoplasmic Dhc sequences. The Chlamydomonas reinhardtii (Cr) cytoplasmic Dhc sequences were aligned with cytoplasmic Dhc sequences identified in other organisms using the program CLUSTAL W (Thompson et al., 1994) and were displayed using the program DRAWGRAM from the Phylip package, version 3.57c (Felsenstein, 1998). The abbreviations and GenBank accession numbers for the sequences are as follows: Caenorhabditis elegans (Ce), L33260 and Z75536; Dictyostelium discoideum (Dd), Z15124; Drosophila melanogaster (Dm), L23195; Emericella nidulans (En), U03904; Homo sapiens (Hs), L23958 and U20552; Mus musculus (Mm), Z83808 and Z83809; Neurospora crassa (Nc), L31504; Paramecium tetraurelia (Pt), L17132; Rattus norvegicus (Rn), D13893, L08505, and D26495; Saccharomyces cerevisiae (Sc), Z21877 and L15626; Schizosaccharomyces pombe (Sp), AB006784; Tetrahymena thermophilia (Tt), AF025312 and AF025313; and Tripneustes gratilla (Tg), Z21941 and U03969.
Comparison with Cytoplasmic Dhc Sequences in Other Organisms
Previous studies in other organisms have shown that the cytoplasmic Dhc sequences fall into two distinct groups (Gibbons et al., 1994; Gibbons, 1995; Tanaka et al., 1995). The major cytoplasmic Dhc sequence is ubiquitously expressed in all eucaryotic organisms (reviewed in Gibbons, 1995). The minor cytoplasmic Dhc sequence, known as DYH1b, DLP4, cDHC1b, or Dhc2, has thus far only been detected in those organisms that assemble cilia or flagella at some stage during their life cycle (Gibbons et al., 1994; Tanaka et al., 1995; Vaisberg et al., 1996; Vaughan et al., 1996; Neesen et al., 1997). Comparison of the two Chlamydomonas cytoplasmic Dhc sequences with that of other cytoplasmic Dhc genes confirms that the Chlamydomonas sequences also fall into these two groups (Figure 2). The Chlamydomonas cDhc1a sequence is most similar to the cytoplasmic Dhc sequences identified in the budding yeast Saccharomyces cerevisiae (Eschel et al., 1993; Li et al., 1993) and the fission yeast Schizosaccharomyces pombe (West and McIntosh, personal communication), whereas the Chlamydomonas cDhc1b appears to be most closely related to the cDhc1b isoforms identified in C. elegans, sea urchin, and Tetrahymena (Gibbons et al., 1994; Wilson et al., 1994; Lee et al., 1999). Because both the sea urchin and C. elegans cDhc1b isoforms have been proposed to function in some aspect of flagellar assembly (Gibbons et al., 1994) (Grant, personal communication), we were interested in characterizing the Chlamydomonas cDhc1b gene further.
Recovery and Sequence Analysis of the cDhc1b Gene
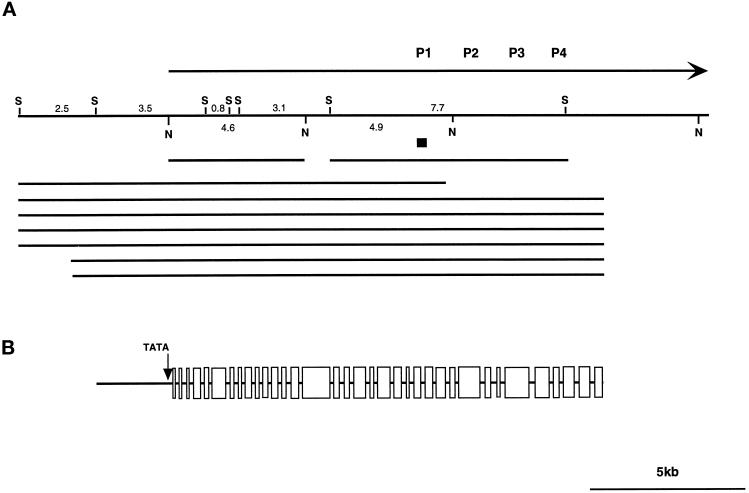
To obtain longer clones of the cDhc1b gene, we screened a series of genomic libraries (see MATERIALS AND METHODS) and eventually recovered seven phage clones spanning >23 kb of genomic DNA (see Figure 3). Restriction mapping and sequence analysis of selected subclones indicated that the 23-kb region contained ∼8 kb of genomic DNA located 5′ of the proposed translation start site and ∼14.5 kb of the cDhc1b transcription unit. A partial restriction map of the region containing the cDhc1b gene and a diagram of the associated subclones used in this study are shown in Figure 3.
Figure 3.
Recovery of the cDhc1b transcription unit. (A) A partial restriction map of the genomic DNA region containing the cDhc1b transcription unit. Also indicated on the diagram are the 150-bp fragment recovered in the PCR screen, the ∼7.7-kb SacI fragment cloned from the size-fractionated minilibrary, the ∼4.6-kb NotI fragment used as a Northern probe, and the approximate positions of the seven phage clones obtained from the large insert genomic library. S, SacI sites; N, NotI sites. (B) Intron–exon structure of the N-terminal and central region of the cDhc1b gene. A diagram of the relative sizes of the introns and exons is shown. All splice sites were confirmed by RT–PCR. Also indicated are the approximate positions of the TATA box sequences and the proposed translation start site.
Sequence analysis of both genomic DNA and RT–PCR products derived from the cDhc1b transcript indicated that the 5′ end of the cDhc1b gene is located within a 3.5-kb SacI subclone (see Figure 3). This region also contains several TATA and tub box sequences (Brunke et al., 1984; Davies and Grossman, 1994) that are presumably required for the regulated transcription of the cDhc1b gene. On the basis of the analysis of the RT–PCR products, the remaining 14.5 kb of the cDhc1b transcription unit contains ∼70% of the coding region located in 35 exons ranging in size from 71 to 905 bp. The predicted amino acid sequence obtained thus far (see Figure 4) corresponds to 3074 amino acids out of an expected ∼4200 residues and extends from the N terminus through to the central region containing the predicted motor domain (Koonce and Samso, 1996; Gee et al., 1997).
Figure 4.
Partial amino acid sequence of cDhc1b. The deduced amino acid sequence of the N-terminal and central region of the cDhc1b gene is shown. The four conserved P-loop motifs are indicated in bold letters. EMBL accession number AJ132478.
A search for potential nucleotide-binding sites in the Chlamydomonas cDhc1b amino acid sequence identified four consensus or near consensus phosphate-binding (P-loop) motifs with the sequence GXXXGKT/S (Walker et al., 1982) in the central region of the polypeptide (Figure 4). These four P-loops (P1–P4) are spaced ∼300 amino acids apart at conserved positions relative to other Dhc sequences. The amino acid sequence around P1 is the most highly conserved among all Dhc sequences, consistent with the proposal that this P-loop corresponds to the primary ATP hydrolytic site (Gibbons, 1995). Comparison with cDhc1b-related sequences in other organisms (Gibbons et al., 1994; Wilson et al., 1994; Lee et al., 1999) indicates that the region around P2 is more conserved than that around either P3 or P4; this differs from previous observations with cDhc1a-related sequences, in which P3 is more highly conserved, or with axonemal Dhc sequences, in which P4 is more highly conserved (reviewed in Gibbons, 1995).
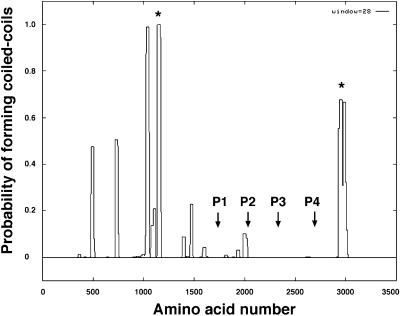
The predicted amino acid sequence of the cDhc1b gene was also analyzed using programs that predict secondary structure to identify regions with the potential to form α-helical coiled-coil domains (Lupus et al., 1991; Lupus, 1996). As indicated in Figure 5, one region before the first P-loop (residues 1023–1056 and 1133–1172) and another region after P4 (residues 2926–3000) show a high probability of forming coiled-coil domains. The presence of predicted coiled-coil domains separating the central region containing the four P-loop sequences from both the N-terminal and C-terminal regions has also been observed in many other Dhc sequences (Mitchell and Brown, 1994, 1997).
Figure 5.
Structural domains within the cDhc1b polypeptide. The probability of forming regions of α-helical coiled-coil structure was determined using the program COILS (Lupus et al., 1991; Lupus, 1996). Peaks of high probability that are also encoded by homologous regions in other Dhc sequences are indicated by the asterisks (see Mitchell and Brown, 1994, 1997).
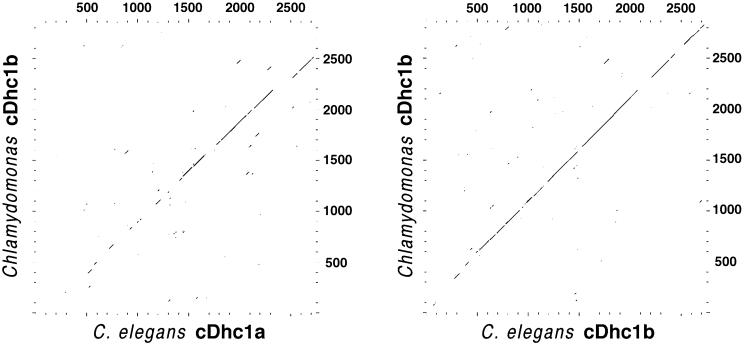
The predicted amino acid sequence of the cDhc1b gene was compared with several other full-length or near full-length Dhc sequences, including the three Dhc sequences (α, β, and γ) that form the outer dynein arm in Chlamydomonas (Mitchell and Brown, 1994, 1997; Wilkerson et al., 1994), the 1α and 1β Dhcs of the I1 inner dynein arm (Myster, Knott, Bower, Perrone, and Porter, unpublished results), and cytoplasmic Dhc sequences from several organisms (Koonce et al., 1992; Eschel et al., 1993; Li et al., 1993; Mikami et al., 1993; Vaisberg et al., 1993, 1996; Zhang et al., 1993; Gibbons et al., 1994; Plamann et al., 1994; Wilson et al., 1994; Xiang et al., 1994; Lye et al., 1995; Lee et al., 1999). In each case, a high degree of sequence similarity was evident over long stretches of the central region of the Dhc (Figure 6) (our unpublished results). However, the Chlamydomonas cDhc1b also shares significant sequence homology in its N-terminal region with the cDhc1b sequence identified in C. elegans (Figure 6). Because the N-terminal region is thought to be important in the association of a Dhc with its specific intermediate and light chain subunits (Mocz and Gibbons, 1993; Sakakibara et al., 1993), these observations suggest that the Chlamydomonas and C. elegans cDhc1b sequences could assemble into motor complexes containing related accessory subunits and perform similar functions.
Figure 6.
Pairwise comparison of Dhc sequences. The Chlamydomonas cDhc1b polypeptide was compared with other full-length Dhc sequences using the GCG program Compare with a window size of 50 and a stringency of 22. Shown are the plots against the two C. elegans cytoplasmic dynein sequences, cDhc1a (Lye et al., 1995) and cDhc1b (Wilson et al., 1994). The GenBank accession numbers for the C. elegans sequences are L33260 and Z75536, respectively.
Expression of the cDhc1b Transcript in Chlamydomonas
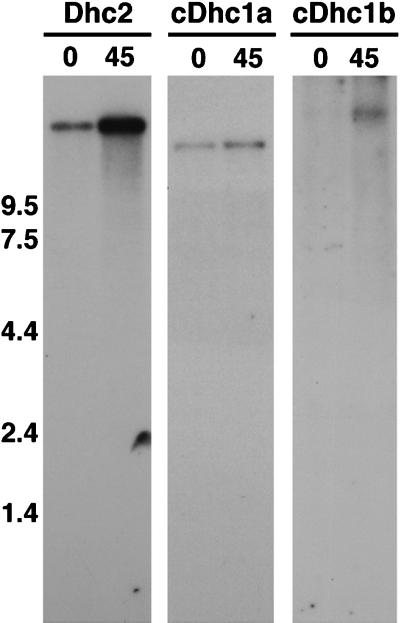
Although the Chlamydomonas cDhc1b gene was recovered by RT–PCR using RNA isolated from nondeflagellated cells, previous work has indicated that the expression of the sea urchin cDhc1b gene can be upregulated in response to deciliation (Gibbons et al., 1994). We therefore isolated RNA both before and after deflagellation of Chlamydomonas cells and analyzed the expression of the cDhc1b transcript on Northern blots. Because we were concerned about possible cross-hybridization between the cDhc1b sequence and the abundant axonemal Dhc transcripts present in deflagellated cells (Mitchell, 1989; Wilkerson et al., 1994; Porter et al., 1996), we used a restriction fragment derived from the 5′ end of the gene as the hybridization probe. This fragment encodes the divergent N-terminal region (see Figure 3). As shown in Figure 7, the expression of the Chlamydomonas cDhc1b transcript (>13 kb) is stimulated by deflagellation. The signal both before and after deflagellation is significantly weaker than that observed with a control probe for an axonemal Dhc transcript (Dhc2), as indicated by the difference in exposure times (see Figure 7 legend), but the cDhc1b transcript is consistently more abundant after deflagellation than is the cDhc1a transcript (see Figure 7) (our unpublished results). These results indicated a potential role for cDhc1b in either flagellar motility or assembly.
Figure 7.
Expression of Chlamydomonas Dhc genes in response to deflagellation. Shown are autoradiograms of Northern blots loaded with total RNA isolated from wild-type cells before (0) and 45 min after (45) deflagellation. Left, the blot was hybridized with a control probe for the axonemal sequence Dhc2 and was exposed overnight. Middle, the blot was hybridized with an ∼6.0-kb SacI fragment from the 5′ end of the cDhc1a transcription unit and was exposed for 20 d, although faint signals could be observed on shorter exposures. Right, the blot was hybridized with a 4.6-kb NotI fragment isolated from the 5′ end of the cDhc1b transcription unit (see Figure 3) and was exposed for 10 d. All probes were labeled to the same specific activity, and all of the Dhc transcripts are estimated to be >13 kb in length (see also Porter et al., 1996).
Identification of cDhc1b Mutations
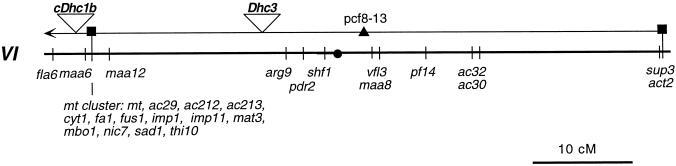
To determine whether the cDhc1b gene might be linked to a previously identified flagellar mutation, we used RFLP-mapping procedures to place the sequence on the genetic map of Chlamydomonas (see MATERIALS AND METHODS). As shown in Figure 8, the cDhc1b gene is closely linked to the mating type (mt) locus on linkage group VI. This location places the cDhc1b gene in close proximity to the reported map position of a temperature sensitive, flagellar assembly mutation, fla6 (Adams et al., 1982). We have been unable to analyze this linkage further in a direct cross with fla6 because the original mutation has apparently reverted (Bower and Porter, unpublished results), but because of these observations, we decided to screen a new collection of flagellar assembly mutants generated by insertional mutagenesis with the goal of identifying other potential cDhc1b mutations.
Figure 8.
Genetic map position of the cDhc1b gene. The genetic map of linkage group VI (redrawn from Harris, 1989; Porter et al., 1996) is shown on the bottom line. The approximate map locations of the Dhc clones (open triangles) and another molecular marker (black triangle) are shown on the top line. The black squares indicate the genetic markers in the C. reinhardtii strain that were used to anchor the two maps relative to one another. The black circle marks the position of the centromere. The parental ditype:nonparental ditype:tetratype ratios and estimated map distances in centiMorgans (cM) are as follows: cDhc1b versus mt (22:0:1; 2.2 cM), cDhc1b versus Dhc3 (11:0:6; 17.6 cM), cDhc1b versus pcf8–13 (8:0:22; 36.7 cM), and cDhc1b versus act2 (3:0:11; 39.3 cM). The distance from the centromere was estimated using the centromere-linked markers ac17 (4:5:21; 35 cM) and y1 (4:4:20; 35.7 cM). Additional mapping data for the linkage group VI markers are provided in Porter et al. (1996).
Transformation of Chlamydomonas cells with exogenous DNA containing a selectable marker is a highly efficient method for the recovery of new mutations that affect either flagellar assembly or flagellar motility (Tam and Lefebvre, 1993). The selectable marker integrates nonhomologously into genomic DNA, and as a result, the site of the new mutation is often marked by plasmid sequences that can be used as a molecular tag for the recovery of flanking genomic DNA. In addition, plasmid insertion is often accompanied by deletion or rearrangement of the host cell DNA, and the resulting mutant phenotype is very stable. Finally, if cloned genes corresponding to a potential mutant locus are available, it is relatively straightforward to identify mutations in the gene of interest simply by screening genomic DNA from the mutants on Southern blots and looking for changes in the restriction pattern of the gene. We have used this approach previously to identify mutations in several genes that encode subunits of the axonemal dyneins (Myster et al., 1997; Perrone et al., 1998) (Perrone and Porter, unpublished results).
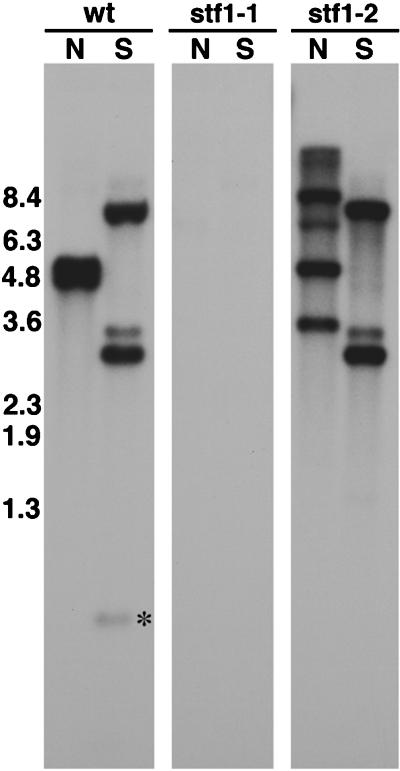
To identify a potential cDhc1b mutation, we screened DNA samples isolated from >70 different flagellar assembly mutants by hybridization with different fragments of the cDhc1b gene. As shown in Figure 9, we have thus far recovered two independently isolated, stumpy flagellar (stf) mutants with different defects in the cDhc1b gene. The stf1-1 strain is associated with a deletion of >15 kb of genomic DNA, which includes at least two-thirds of the cDhc1b coding region. The stf1-2 strain is missing ∼1 kb of genomic DNA located in the 5′ end of the coding region (see Figure 9 legend). Defects of this magnitude in the N-terminal region of a Dhc are likely to be null mutations. Hybridization of the blots with control probes for other Dhc sequences has confirmed that the RFLPs observed are specific to the cDhc1b gene and are not caused by problems with the loading or digestion of the DNA samples (Wysocki and Porter, unpublished results). Hybridization with probes for the NIT1 gene used as the selectable marker has also demonstrated that both mutants contain only a single plasmid insert (Bower and Porter, unpublished results) (see Figure 9). The stf1-1 and stf1-2 strains therefore contain bona fide cDhc1b mutations that are associated with plasmid insertions.
Figure 9.
Identification of cDhc1b mutations. Shown are autoradiograms of three duplicate Southern blots containing genomic DNA isolated from wild-type (wt) and two stumpy flagellar mutants, stf1-1 and stf1-2. Four micrograms of genomic DNA were digested with the restriction enzymes NotI (N) and SacI (S), separated on a 0.8% agarose gel, transferred to a Magnagraph membrane, and hybridized overnight with the 4.6- and 4.9-kb NotI fragments of the cDhc1b gene (see Figure 3). No cross-hybridizing sequence is detected in the stf1-1 genomic DNA, whereas an RFLP in one of the NotI fragments is detected in stf1-2 genomic DNA. Longer exposures indicated that the stf1-2 genomic DNA is missing the two SacI fragments of 0.8 and 0.3 kb located within the wild-type 4.6-kb NotI fragment (see Figure 3). Hybridization with probes for the NIT1 gene indicated the presence of a single plasmid insert in each strain and the hybridization of this sequence to the polymorphic cDhc1b restriction fragments in stf1-2 (Bower and Porter, unpublished results).
cDhc1b Mutants Assemble Short, Defective Flagella
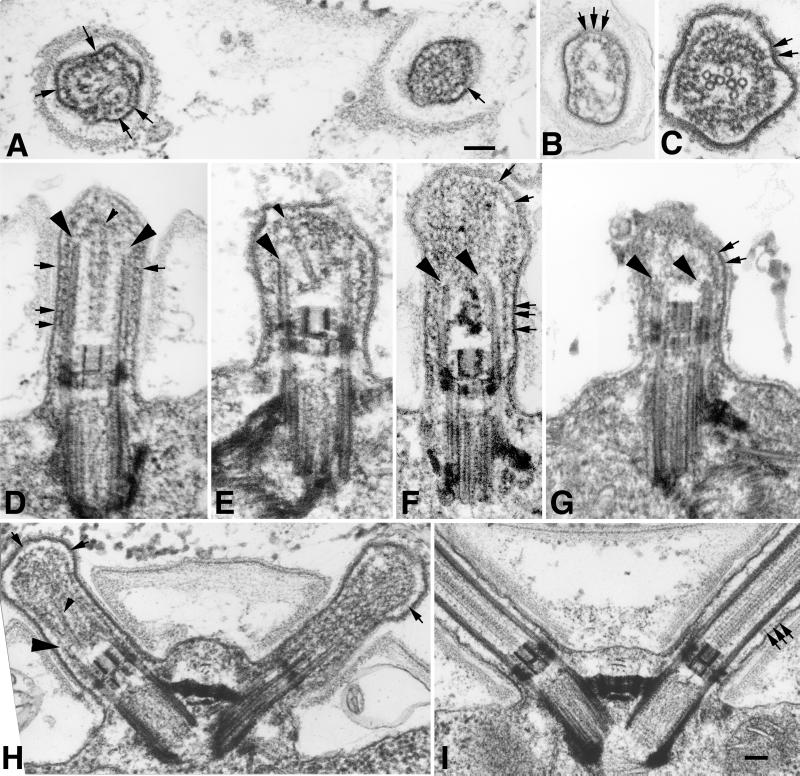
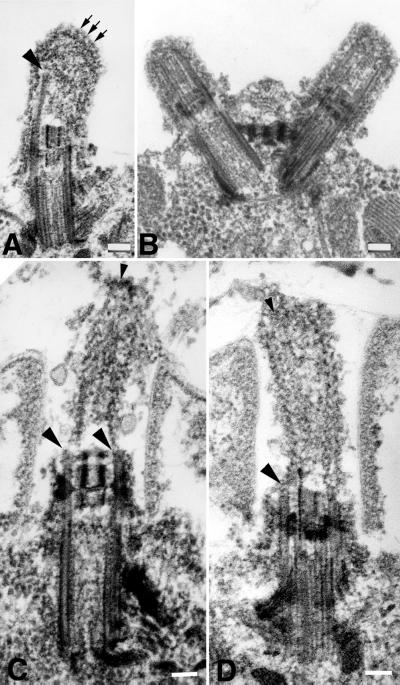
Short flagellar stumps were observed on both stf1 mutants using differential interference contrast microscopy. Examination of thin-sectioned cells by transmission electron microscopy revealed that both stf1 strains fall into a class of stumpy mutants whose flagella are shorter than 1–2 μm in length (Figure 10). The basal body and transition zone structures appeared identical to those found in wild-type cells (Figure 10, D–I), but the microtubules within the flagellar stumps were extremely short (0.5–1.0 μm) and aberrantly organized. When viewed in cross section (Figure 10, A–C), it was evident that most stf1 flagella did not contain the typical “9+2” array of microtubules, although a few examples could be found. In most flagella, one or more singlet microtubules could be seen, usually collapsed into the center of the flagellum (Figure 10, B and C). When viewed in longitudinal section (Figure 10, D–H), the flagellar microtubules extended from the basal body microtubules, but many appeared to end in open, slightly frayed sheets of protofilaments (Figure 10, F and G). Few convincing examples of a normal central pair apparatus were found, although some flagella contained microtubules with free proximal ends, similar to central pair microtubules (Figures 10E and 11C). Others contained a core of amorphous material similar in appearance to that found in central pair mutants (Figure 10D). No clearly defined capping structures were observed, but the distal ends of the central microtubules were embedded in an amorphous substance (Figures 10E and 11C), similar to the cap material seen in growing, wild-type flagella (Dentler, unpublished results).
Figure 10.
Electron microscopic analysis of stumpy flagellar mutants (A–H) and wild-type Chlamydomonas cells (I). Cross sections of flagella on a single stf1 cell (A) and on individual cells (B and C) typically reveal few complete microtubules and no outer doublet microtubules. Compared with the wild-type flagella (I), the flagella on the stf1 mutants rarely extend beyond the cell wall (D and H). The transition zones and basal bodies appear normal in the mutants, but the flagellar microtubules are short and often terminate as open-ended or filamentous structures (see large arrowheads). Occasionally, microtubules extend from a distal cap-like structure and terminate with the free ends just distal to the basal cup (D, E, and H, small arrowheads). Flagella are filled with electron dense material, and stalked bead structures line the flagellar membrane (small arrows). Bars, 0.1 μm.
Figure 11.
Electron microscopic analysis of detergent-extracted stf1 mutants. Whole cells were extracted with 0.5% Nonidet P-40 for 5 min (A and B) or 2% Nonidet P-40 for 30 min (C and D) before fixation. Both the cell and flagellar membranes have been removed, but amorphous granular and filamentous material remain associated with the flagellar stumps. Filamentous structures extend from the ends of the flagellar or basal body microtubules (A, C, and D, large arrowheads). In some flagella, the filaments associated with the microtubules coalesce at the distal tip (A, small arrows). In others, microtubules with free proximal ends are occasionally found linked to a distal cap-like structure (C, small arrowhead). Bars, 0.1 μm.
The flagella of the stf1 mutants were also filled with an electron dense, amorphous matrix that surrounded the microtubules (Figures 10 and 11). Numerous spherical particles with short stalks were found adjacent to flagellar membrane, and free particles of a similar size also filled the flagellar matrix (Figure 10, A–H, small arrows). In longitudinal sections, the particles often appeared in rows below the flagellar membrane (Figure 10, D–H), similar to the raft particles associated with IFT in wild-type flagella (see Figure 10I) (see Kozminski et al., 1995).
To determine whether the amorphous granular material was directly associated with the microtubules or whether it was simply excess flagellar raft material that filled the matrix, we extracted cells with detergent before fixation (Figure 11). Initial extractions for 2–10 min with 0.5% Nonidet P-40 completely removed the flagellar membrane and most of the stalked bead structures but left a large amount of material still firmly attached to the flagellar stumps (Figure 11, A and B). Microtubules and amorphous material filled the matrix, and the flagella appeared nearly identical to that in control stf1 cells whose membrane was intact. Organized arrays of raft particles were not observed, although some raft-like structures were visible (Figure 11A, arrows). Thus, the amorphous material seen in the stf1 flagellar stumps is not simply soluble matrix protein.
Additional extraction of the cells with 2% Nonidet P-40 for 30 min removed more granular material and clearly revealed the filamentous material extending from the flagellar and basal body microtubules (Figure 11, C and D, large arrowheads). In some flagella, the filaments coalesced at the distal tips (see Figure 11C, small arrowhead), in association with cap-like material. Additional extraction of the cells with 2 mM Mg-ATP did not release either the matrix material or the filaments associated with microtubules (Dentler, unpublished results).
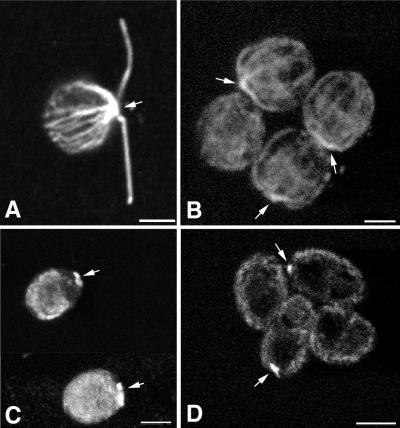
In addition to the flagella, the stf1 mutants were analyzed for other microtubule-related structural defects. Examination of wild-type and mutant cells by thin-section transmission electron microscopy indicated no morphological differences in the organization of the Golgi apparatus (Dentler, unpublished results). Examination of cytoplasmic microtubule arrays using both conventional and confocal immunofluorescence microscopy revealed that the stf1 mutants contained apparently normal arrays of basal body rootlet microtubules. However, the number of cytoplasmic microtubules was lower in the stf1 mutants than in wild type, and in many mutant cells, the cytoplasmic microtubules were shorter than those in wild type (see Figure 12, A and B). In wild-type cells, microtubule arrays extending from the basal bodies were easily observed in all focal planes (Figure 12A), but in the stf1 mutants, the microtubule arrays were only evident around the edges of the cells (Figure 12B). Staining wild-type cells (Figure 12C) with an antibody specific for the FLA10 subunit indicated that the kinesin II complex was present throughout the cell body but concentrated in the basal body region, consistent with previous reports (Vashishtha et al., 1996; Cole et al., 1998). Staining of stf1 mutant cells with the same antibody revealed that the FLA10 kinesin II complex was more concentrated in the anterior region of the cell (Figure 12D).
Figure 12.
Immunofluorescence microscopy of wild-type (A and C) and stf1 mutant (B and D) cells stained with monoclonal antibodies to β-tubulin (A and B) or one of the kinesin II subunits (C and D). Bars, 2 μm.
DISCUSSION
Recovery of Additional Dhc Sequences
In this study, we report the recovery of four Dhc sequences in Chlamydomonas that are distinct from the three outer arm Dhc genes (Mitchell and Brown, 1994, 1997; Wilkerson et al., 1994) and the nine putative inner arm genes (Dhc1-Dhc9) identified previously (Porter et al., 1996). Sequence comparisons indicate that two genes (Dhc10 and Dhc11) are closely related to the axonemal Dhc sequences, and consistent with this hypothesis, the expression of both genes is enhanced by deflagellation (Knott and Porter, unpublished results). More recent work has demonstrated that Dhc10 encodes an inner arm Dhc (Perrone and Porter, unpublished results). The two remaining sequences (cDhc1a and cDhc1b) are more similar to the cytoplasmic Dhc sequences identified in other organisms (Figures 1 and 2). Sequence data beyond the region represented by the PCR primers have shown that the cDhc1a gene also encodes a dynein sequence identified previously as pcr4 (Wilkerson et al., 1994) (Bower and Porter, unpublished results; Witman, personal communication). Together with previous estimates based on Southern blot analyses (Porter et al., 1996), these observations indicate that the Chlamydomonas genome contains ∼16 different Dhc genes. The size of the Dhc gene family in Chlamydomonas is comparable with that found in other species such as sea urchin (14), Paramecium (12), Drosophila (>7), rat (13–15), mouse (11), and humans (>8) (Asai et al., 1994; Gibbons et al., 1994; Rasmusson et al., 1994; Tanaka et al., 1995; Andrews et al., 1996; Vaisberg et al., 1996; Vaughan et al., 1996; Neesen et al., 1997). The remarkable conservation of the Dhc gene family between such diverse organisms is consistent with the proposal that the Dhc gene family diverged into a small number of groups relatively early in the evolution of eucaryotes, but after these groups were established, they remained largely unchanged (Gibbons, 1995).
Alignment of the region encoding the ATP hydrolytic domain suggests that the cDhc1a sequence is the Chlamydomonas homologue of the major cytoplasmic dynein isoform (Figure 1). This isoform is the only Dhc sequence that has been identified thus far in both budding and fission yeast, the slime mold Dictyostelium, and filamentous fungi, where it plays important roles in the assembly and positioning of the mitotic spindle, nuclear migration, and vesicle transport (Koonce et al., 1992; Koonce and Samso, 1996; Koonce and Knecht, 1998; Eschel et al., 1993; Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994; Inoue et al., 1998; Pollock et al., 1998) (West and McIntosh, personal communication). Because of these observations, we would predict that the cDhc1a sequence might be involved in cell division and/or the positioning of basal body structures in Chlamydomonas. Although cDhc1a-related sequences are abundant in the testes of both Drosophila and vertebrates (Collins and Vallee, 1989; Rasmusson et al., 1994; Criswell and Asai, 1998), our Northern blot analyses indicate that the Chlamydomonas cDhc1a sequence is a relatively low-abundance transcript whose expression is not dramatically altered by deflagellation (Figure 8), consistent with what has been observed during ciliogenesis in other organisms (Asai et al., 1994; Gibbons, et al., 1994; Kandl et al., 1995; Andrews et al., 1996). Whether the cDhc1a sequence plays a role in flagellar assembly remains to be determined. Recent studies on the associated 8-kDa dynein LC have demonstrated that this polypeptide is required for flagellar assembly and retrograde IFT (Pazour et al., 1998), but no cDhc1a defects have thus far been detected in the present collection of the flagellar assembly mutants (Wysocki, Porter, and Dentler, unpublished results). Further insight into the functions of the cDhc1a sequence in Chlamydomonas will require both more information about its subcellular location and the identification and characterization of a specific cDhc1a mutation. We are obtaining N-terminal sequence for production of an isoform-specific antibody and screening additional insertional mutants with our cDhc1a clones to address these questions.
Alignment of the Chlamydomonas cDhc1b sequence with that of other cytoplasmic Dhc genes has identified homologues in sea urchin (Dyh1b), rat (DLP4), humans (Dhc2), mouse (Dhc11), and the worm C. elegans (Dhc1b) (Gibbons et al., 1994; Wilson et al., 1994; Tanaka et al., 1995; Vaisberg et al., 1996; Neesen et al., 1997). These sequence similarities extend into the N-terminal region of the polypeptide (Figure 6). Because the N-terminal region is thought to be involved in the association of the Dhc with isoform-specific intermediate and light chains (Sakakibara et al., 1993), these observations suggest that the cDhc1b-related sequences are likely to be assembled into similar multisubunit complexes, but nothing is yet known about the cDhc1b-associated subunits in any organism. Sucrose density gradient centrifugation of the dynein isoforms in rat testis indicates that the cDhc1b heavy chain does not cosediment with any of the intermediate chain or LC subunits typically found in association with the cDhc1a isoform, including the 8-kDa LC (Criswell and Asai, 1998) (Vaisberg, Grissom, and McIntosh, personal communication). Thus it is not clear how the flagellar assembly defects observed in the 8-kDa dynein LC mutants (Pazour et al., 1998) may be related to those observed with the cDhc1b mutants. Additional work is clearly needed to characterize the components of the cDhc1b motor complex.
The cDhc1b Mutant Phenotype in Chlamydomonas
To test the possible role of cDhc1b in flagellar function in Chlamydomonas, we used gene-specific probes to place the cDhc1b gene on the genetic map and to screen collections of flagellar mutants generated by insertional mutagenesis. The cDhc1b gene maps near the reported position of the FLA6 locus (Adams et al., 1982) (Figure 8), but because the original fla6 strain is no longer available, we were unable to determine directly whether fla6 is a temperature-sensitive mutation in the cDhc1b gene. However, using Southern blot analyses, we have identified two flagellar assembly mutants associated with significant deletions in the cDhc1b gene (Figure 9). Although the size of the deletion varies between the two strains, in both cases, the region encoding the N-terminal portion of the Dhc has been disrupted, and the resulting mutant phenotype is the same. The basal body and transition zone structures are wild-type in appearance, but most of the microtubules distal to the basal bodies within the flagellar stumps are highly aberrant (Figure 10). Doublet microtubules were rarely found, and most flagella contained fewer than seven singlet microtubules. The singlet microtubules present were abnormally short and often ended in open sheets resembling the protofilaments seen at the ends of microtubules assembled in vitro. Detergent extraction of stf1 mutants revealed filamentous structures, possibly incomplete microtubules, continuous with some of the basal body microtubules (Figure 11). The microtubule-capping structures normally observed in wild-type flagella (Dentler, 1980; Dentler and LeCluyse, 1982) were not found at the ends of the microtubules in the stf1 flagella. However, amorphous material was observed at the distal ends of stf1 microtubules (Figures 10 and 11), and this material is similar in appearance to that seen at the distal ends of microtubules in growing cilia that are shorter than 2 μm in length (Portman et al., 1987).
The stf1 mutant phenotype is quite distinct from the short flagellar (shf) mutants, which assemble outer doublet and central pair microtubules but fail to reach wild-type lengths (Jarvik and Chojnacki, 1985; Kuchka and Jarvik, 1987; Pazour et al., 1998) (Dentler, unpublished results), but very similar to other stumpy flagellar mutants, which lack normal microtubule arrays (McVittie, 1972; Jarvik and Chojnacki, 1985). Whether the stf1 mutants represent new alleles of the other stumpy flagellar mutant strains remains to be determined.
Another striking feature of the stf1 mutant phenotype in Chlamydomonas is that the flagellar matrix is filled with an amorphous, electron dense material. Similar material has been described in other stumpy mutants (McVittie, 1972; Jarvik and Chojnacki, 1985), where it was presumed to represent unassembled flagellar protein. Small particles resembling the raft structures associated with IFT (Kozminski, et al., 1993, 1995; Cole et al., 1998) are also found in the stf1 mutants (Figure 10). Although the biochemical composition of the matrix is largely unknown, its appearance resembles the material found in the LC8 mutant fla14, which includes some unassembled flagellar precursors and raft particle polypeptides (Cole et al., 1998; Pazour et al., 1998). However, on the basis of our morphological analysis of detergent-extracted cells, a significant amount of matrix material does remain associated with the extracted flagellar stumps (Figure 11). These observations suggest that the stf1 flagella lack some component(s) critical for flagellar microtubule assembly or stability.
Comparison with cDhc1b Defects in Other Organisms
The phenotype of the Chlamydomonas cDhc1b mutants is also similar to the phenotype of several sensory cilia mutants in C. elegans (reviewed in Bargmann, 1993; Mori and Ohshima, 1997). These mutants fail to assemble the nonmotile cilia located at the distal end of their sensory neurons. Such structural defects alter the ability of the sensory neurons to monitor the local environment, leading to defects in such behaviors as chemotaxis (che) and osmotic avoidance (osm) (Bargmann, 1993; Starich et al., 1995). One of these genes, osm-3, encodes a FLA10-related kinesin homologue (Shakir et al., 1993; Tabish et al., 1995), whereas two others, osm-1 and osm-6, encode homologues of the raft particle polypeptides (Collet et al., 1998; Cole et al., 1998) (Stone and Shaw, personal communication). A fourth sensory cilium mutant, che-3, encodes the C. elegans homologue of the cDhc1b gene (Grant, personal communication), and its phenotype is particularly striking. In che-3, the sensory cilia are shortened, the microtubule structures are highly aberrant, and the distal tips of the neurons become filled with an amorphous matrix material (Lewis and Hodgkin, 1977; Albert et al., 1981; Perkins et al., 1986). Analysis of several green fluorescent protein–labeled proteins in the che-3 mutant neurons has revealed that the matrix material includes homologues of the raft particle polypeptides (OSM-6) as well as ciliary membrane receptors (ODR-10) that accumulate in the distal tips at levels significantly above those observed in wild-type neurons (Collet et al., 1998; Dwyer et al., 1998). The accumulation of material in the che-3 mutant neurons differs from what has been observed in other sensory cilia mutants such as osm-3 (Collet et al., 1998). These results demonstrate that most of the components of the sensory cilia are transported from the cell body to the site of assembly in che-3 neurons, but once there, they fail to be incorporated into a functional cilium.
The observation that cDhc1b mutations disrupt axoneme assembly in both Chlamydomonas flagella and C. elegans sensory cilia suggests that the cDhc1b isoform may participate in the formation of axoneme structures in a variety of tissues. These include the motile axonemes in sperm flagella and ciliated epithelia, as well as the nonmotile axonemes in vertebrate photoreceptors and inner ear kinocilia. An involvement of cDhc1b in axoneme assembly would be consistent with its high level of expression in respiratory epithelia and the testis (Tanaka et al., 1995; Criswell et al., 1996; Criswell and Asai, 1998; Neesen et al., 1997). A minus-end–directed dynein motor would also complement the plus-end–directed activity of the FLA10-related kinesin II motors (Cole et al., 1993; Kondo et al., 1994; Walther et al., 1994) that have been found in association with these structures (Kondo et al., 1994; Yamazaki et al., 1995; Beech et al., 1996; Henson et al., 1997).
Observations on Dyh2 knockouts in Tetrahymena indicate that the cDhc1b homologue is not required for axoneme assembly in all cells (Lee et al., 1999). Tetrahymena Dyh2 mutant cells can regenerate motile cilia with apparently normal kinetics and without any visible defects in ciliary ultrastructure. However, if the cortical microtubule cytoskeleton is disrupted in the Dyh2 mutants during deciliation, the basal body rows become disorganized, and cilia regenerate randomly over the cell surface (Lee et al., 1999). These apparent discrepancies in the mutant phenotypes may reflect a difference in the dynamic behavior of cilia and flagella in the two organisms (see discussion in Lee et al. [1999]). Tetrahymena cilia are relatively stable organelles that do not shorten or elongate during the life cycle, whereas Chlamydomonas flagella assemble and disassemble with each cell cycle, as well as adjust their lengths in response to a variety of environmental conditions (Johnson and Porter, 1968; Lefebvre and Rosenbaum, 1986; Tuxhorn et al., 1998). These differences indicate that it will be important to evaluate the function of the cDhc1b isoform in the context of several different cell types.
The phenotype of the cDhc1b mutations in Chlamydomonas is not strictly limited to a defect in flagellar assembly. Although we have observed no gross defects in cell size or shape, as might have been predicted from the phenotype of the Dyh2 knockouts in Tetrahymena (Lee et al., 1999), the cytoplasmic microtubule array does appear to be altered in the stf1 mutants (Figure 12). As analyzed by immunofluorescence, both the number and length of the cytoplasmic microtubules appear to be reduced, although further work will be needed to quantify these defects. In addition, the FLA10 kinesin II complex appears to be more concentrated in the anterior region of the mutant cells as compared with wild type (Vashishtha et al., 1996; Cole et al., 1998), which may reflect an accumulation of the kinesin II in the flagellar stumps (Figure 12D). However, no significant changes in the appearance of the Golgi apparatus were observed in the stf1 mutants, as might have been expected from studies in cultured mammalian cells in which Dhc2 colocalizes with markers for the Golgi apparatus and microinjection of a Dhc2 antibody leads to fragmentation of the Golgi (Vaisberg, et al., 1996). Whether these discrepancies reflect a bona fide difference in the function of the Dhc2/cDhc1b homologues in these two cell types or simply the presence of multiple cDhc1b-like isoforms in vertebrates that perform specialized functions (Criswell and Asai, 1998) remains to be determined.
The Role of cDhc1b in Flagellar Assembly
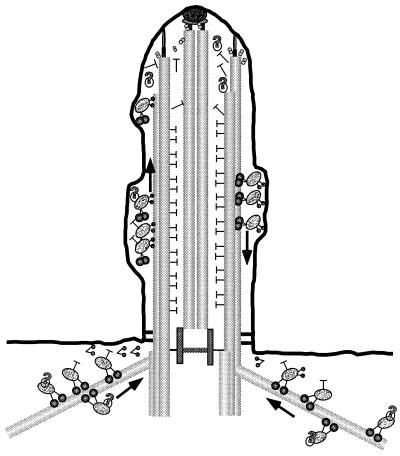
Within the past few years, it has become clear that motor activities are essential for flagellar assembly. Flagellar assembly requires the delivery of flagellar precursors to the distal ends of the flagellar microtubules (Johnson and Rosenbaum, 1992; Piperno et al., 1996), and plus-end–directed, kinesin II complexes seem to be essential for axonemal growth (Walther et al., 1994; Kozminski et al., 1995; Morris and Scholey, 1997; Nonaka et al., 1998). What then would be the role of a minus-end–directed motor such as cytoplasmic dynein? We suggest three possibilities: 1) to transport flagellar precursors to the basal body region or flagellar base, 2) to recycle cargoes (rafts) carried to the flagellar tips by the FLA10 kinesin II, and/or 3) to carry signals from the flagellar tip so the cell can monitor flagellar length (see Figure 13).
Figure 13.
Model for cDhc1b activity in flagellar assembly and maintenance. Transport of IFT particles toward the flagellar tips (the microtubule plus ends) is mediated by FLA10 kinesin II motors (small, paired spheres). Cytoplasmic dynein (large, paired spheres) may be required for the minus-end–directed transport of flagellar components along cytoplasmic microtubules to the base of the flagellum. Alternatively, cytoplasmic dynein may be required to transport IFT particles from the flagellar tip to the flagellar base or for the recycling of IFT particles in the cytoplasm to pick up new components. Although the nature of material transported up the flagellum is unknown, possible components include radial spokes (T-shaped structures), unidentified complexes (large question marks), and/or signaling components involved in regulating flagellar length.
In most ciliated cells, the basal bodies are associated with the minus ends of a cytoplasmic microtubule array that extends its plus ends into the cell body. Because microtubule-based transport appears to be essential for moving axonemal precursors within the flagellum (Kozminski et al., 1995; Piperno et al., 1996; Morris and Scholey, 1997), it seems reasonable to propose that some of these components are also transported along cytoplasmic microtubules to the basal body region by a cytoplasmic dynein. The accumulation of material in the stf1 mutant flagella indicates that not all flagellar components require dynein 1b–mediated transport to the basal body region, but the absence of normal flagellar microtubules also suggests that one or more components essential for flagellar assembly are not present in the mutant flagella. Cytoplasmic dynein 1b could therefore be involved in the delivery of some essential but as yet unidentified components from the cell body to the basal body region. Because microtubule caps are always present on doublet and central pair microtubules in full-length cilia and flagella (Dentler, 1990) and because the caps form as cilia and flagella grow beyond 2 μm, it is reasonable to suggest that part of the stf1 defect might be the inability to assemble the microtubule caps. Whether this is due to a failure in the transport of cap components remains to be determined.
Cytoplasmic dynein 1b could also complement the activity of a plus-end–directed FLA10 kinesin II and serve as the retrograde motor for IFT. Disrupting the retrograde motor would lead to an excess of both the FLA10 kinesin II motor and associated raft components in the flagellar compartment, as was observed in fla14 (Pazour et al., 1998). The failure to recycle these components back to the cell body could eventually result in a “traffic jam” that blocks flagellar assembly. However, a raft-recycling defect may not completely explain the stf1 mutant phenotype, because so few intact microtubules are observed in the stf1 flagella. One might predict that a recycling defect would produce short flagella with normal axonemes, similar to that seen in the dynein LC mutant fla14 (Pazour et al., 1998). Still, the loss of a dynein heavy chain subunit may have a more drastic effect on retrograde IFT that results in the excess or deficiency of some component that affects microtubule stability.
If flagellar assembly requires the retrograde movement of some component or signal that allows the cell to monitor flagellar assembly or length, a defect in this signaling pathway might also led to a block in flagellar assembly (reviewed in Lefebvre and Rosenbaum, 1986; Johnson and Rosenbaum, 1993). Studies of the long flagella (lf) mutants in Chlamydomonas have revealed that the flagellar length control is a dynamic process requiring the interaction of several components (McVittie, 1972; Barsel et al., 1987). Single lf1, lf2, or lf3 mutants assemble flagella nearly twice the wild-type length, but double lf mutants or null mutants of the LF loci fail to assemble flagella (Barsel et al., 1987) (Tam and Lefebvre, personal communication). Other experiments have shown that Chlamydomonas can adjust the lengths of its flagella in response to changes in its environment (Dentler and Adams, 1992; Tuxhorn et al., 1998). Interestingly, the sequence analysis of raft polypeptide homologues has indicated that some of these proteins contain PxxP motifs that might interact with SH3 domains and could potentially be involved in signal transduction (Wick et al., 1995; Collet et al., 1998; Cole et al., 1998). In addition, recent work has demonstrated that a G protein α subunit is involved in the specification of sensory cilia morphology in C. elegans (Roayaie et al., 1998), and G protein subunits have also been detected in flagellar membrane preparations from the green alga Gonium pectorale (Haller and Fabry, 1998). How such signaling pathways may be related to the process of intraflagellar transport and dynein motor activity remains to be determined. To better understand the specific role of the cDhc1b motor, we need additional information on its subcellular location in the cell body and/or the flagellum. The identification of the specific cargoes of the cDhc1b motor may also provide new insights into the mechanism by which the cDhc1b isoform contributes to the process of flagellar assembly.
ACKNOWLEDGMENTS
We acknowledge the support and encouragement from several of our colleagues at the University of Minnesota and the University of Kansas, including Pete Lefebvre, Carolyn Silflow, Dick Linck, Jocelyn Shaw, Bob Herman, Tom Hays, and Kathy Suprenant. We also thank several members of the Porter and Dentler laboratories for both technical support and helpful discussion. W.D. especially thanks Pete Lefebvre, Carolyn Silflow, and the members of their laboratories for their help and encouragement during a sabbatical leave in their laboratories. We also thank Keith Kozminski and Joel Rosenbaum of Yale University for sharing the flagellar assembly mutants that were recovered in their screen for gliding mutants. Dick Himes (University of Kansas, Lawrence, KS) and Jon Scholey (University of California, Davis, CA) generously provided antisera for tubulin and kinesin II, respectively. This work was supported by grants from the National Science Foundation (MCB-9305217) and the National Institutes of Health (GM-55667 to M.E.P. and GM-32556 to W.D.).
Abbreviations used:
- cDhc
cytoplasmic dynein heavy chain
- che
chemotaxis
- cM
centiMorgan
- GCG
Genetics Computer Group
- IFT
intraflagellar transport
- LC
light chain
- lf
long flagella
- mt
mating type
- osm
osmotic avoidance
- PBS
phosphate-buffered saline
- RFLP
restriction fragment length polymorphism
- RT
reverse transcription
- shf
short flagellar
- stf
stumpy flagellar
REFERENCES
- Adams GMW, Huang B, Luck DJL. Temperature-sensitive, assembly defective flagella mutants of Chlamydomonas reinhardtii. Genetics. 1982;100:579–586. doi: 10.1093/genetics/100.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert PS, Brown SJ, Riddle DL. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- Andrews KL, Nettesheim P, Asai DJ, Ostrowski LE. Identification of seven rat axonemal dynein heavy chain genes: expression during ciliated cell differentiation. Mol Biol Cell. 1996;7:71–79. doi: 10.1091/mbc.7.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai DJ, Beckwith SM, Kandl KA, Keating HH, Tjandra H, Forney JD. The dynein genes of Paramecium tetraurelia. Sequences adjacent to the catalytic P-loop identify cytoplasmic and axonemal heavy chain isoforms. J Cell Sci. 1994;107:839–847. doi: 10.1242/jcs.107.4.839. [DOI] [PubMed] [Google Scholar]
- Asai DJ, Brokaw CJ. Dynein heavy chain isoforms and axonemal motility. Trends Cell Biol. 1993;3:398–402. doi: 10.1016/0962-8924(93)90090-n. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Genetic and cellular analysis of behavior in C. elegans. Annu Rev Neurosci. 1993;16:47–71. doi: 10.1146/annurev.ne.16.030193.000403. [DOI] [PubMed] [Google Scholar]
- Barsel S-E, Wexler DE, Lefebvre PA. Genetic analysis of Long-flagellar mutants of Chlamydomonas reinhardtii. Genetics. 1987;118:637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech PL, Pagh-Roehl K, Noda Y, Hirokawa N, Burnside B, Rosenbaum JL. Localization of kinesin superfamily proteins to the connecting cilium of fish photoreceptors. J Cell Sci. 1996;109:889–897. doi: 10.1242/jcs.109.4.889. [DOI] [PubMed] [Google Scholar]
- Bernstein M, Beech PL, Katz SG, Rosenbaum JL. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J Cell Biol. 1994;125:1313–1326. doi: 10.1083/jcb.125.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunke K, Anthony J, Sterberg E, Weeks D. Repeated consensus sequence and pseudopromoters in the four coordinately regulated tubulin genes of Chlamydomonas reinhardtii. Mol Cell Biol. 1984;4:1115–1124. doi: 10.1128/mcb.4.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Vallee RB. Preparation of microtubules from rat liver and testis: cytoplasmic dynein is a major microtubule-associated protein. Cell Motil Cytoskeleton. 1989;14:491–500. doi: 10.1002/cm.970140407. [DOI] [PubMed] [Google Scholar]
- Criswell PS, Asai DJ. Evidence for four cytoplasmic dynein heavy chain isoforms in rat testis. Mol Biol Cell. 1998;9:237–247. doi: 10.1091/mbc.9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell PS, Ostrowski LE, Asai DJ. A novel cytoplasmic dynein heavy chain: expression of DHC1b in mammalian ciliated epithelial cells. J Cell Sci. 1996;109:1891–1898. doi: 10.1242/jcs.109.7.1891. [DOI] [PubMed] [Google Scholar]
- Davies JP, Grossman AR. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol. 1994;14:5165–5174. doi: 10.1128/mcb.14.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler WL. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- Dentler WL. Ciliary and Flagellar Membranes. R.A. Bloodgood, New York: Plenum Press; 1990. Linkages between microtubules and membranes in cilia and flagella; pp. 31–64. [Google Scholar]
- Dentler WL, Adams C. Flagellar microtubule dynamics in Chlamydomonas: cytochalasin D induces periods of microtubule shortening and elongation; colchicine induces disassembly of the distal, but not proximal, half of the flagellum. J Cell Biol. 1992;117:1289–1298. doi: 10.1083/jcb.117.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler WL, LeCluyse EL. The effects of structures attached to the tips of tracheal ciliary microtubules on the nucleation of microtubule assembly in vitro. Cell Motil Suppl. 1982;1:13–18. doi: 10.1002/cm.970020705. [DOI] [PubMed] [Google Scholar]
- Dillman JF, III, Dabney LP, Pfister KK. Cytoplasmic dynein is associated with slow axonal transport. Proc Natl Acad Sci USA. 1996;93:141–144. doi: 10.1073/pnas.93.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Algae as Experimental Systems. New York: Alan R. Liss; 1989. Linkage group XIX in Chlamydomonas reinhardtii (Chlorophyceae): genetic analysis of basal body function and assembly; pp. 39–53. [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- Eschel D, Urrestarazu LA, Vissers S, Jauniaux J-C, van Vleit-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;1993:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espindola FS, Cheney RE, King SM, Suter DM, Mooseker MS. Myosin-V and dynein share a similar light chain. Mol Biol Cell. 1996;7:372a. [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inferences and reliability. Annu Rev Genet. 1998;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fox LA, Sawin KE, Sale WS. Kinesin-related proteins in eukaryotic flagella. J Cell Sci. 1994;107:1545–1550. doi: 10.1242/jcs.107.6.1545. [DOI] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gepner J, Li M-G, Ludmann S, Kortas C, Boylan K, Iyadurai SJP, McGrail M, Hays TS. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996;142:865–878. doi: 10.1093/genetics/142.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Asai DJ, Tang WJY, Hays TS, Gibbons IR. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. Dynein family of motor proteins: present status and future questions. Cell Motil Cytoskeleton. 1995;32:136–144. doi: 10.1002/cm.970320214. [DOI] [PubMed] [Google Scholar]
- Gross CH, Ranum LPW, Lefebvre PA. Extensive restriction length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr Genet. 1988;13:503–508. doi: 10.1007/BF02427756. [DOI] [PubMed] [Google Scholar]
- Haller K, Fabry S. Brefeldin A affects synthesis and integrity of a eukaryotic flagellum. Biochem Biophys Res Commun. 1998;243:597–601. doi: 10.1006/bbrc.1997.8015. [DOI] [PubMed] [Google Scholar]
- Harris E. The Chlamydomonas Sourcebook. San Diego, CA: Academic Press; 1989. [Google Scholar]
- Harrison A, Olds-Clarke P, King S. Identification of the t complex-encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol. 1998;140:1137–1147. doi: 10.1083/jcb.140.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Cole DG, Roesener CD, Capuanon S, Mendola RJ, Scholey JM. The heterotrimeric motor protein kinesin-II localizes to the midpiece and flagellum of sea urchin and sand dollar sperm. Cell Motil Cytoskeleton. 1997;38:29–37. doi: 10.1002/(SICI)1097-0169(1997)38:1<29::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- Holzbaur ELF, Vallee RB. Dyneins: molecular structure and cellular function. Annu Rev Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- Hyatt MA. Biological Applications. New York: VanNostrand Reinhold; 1970. Principles and techniques of electron microscopy. , 263. [Google Scholar]
- Inoue S, Turgeon BG, Yoder OC, Aist JR. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J Cell Sci. 1998;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Chojnacki B. Flagellar morphology in stumpy-flagella mutants of Chlamydomonas reinhardtii. J Protozool. 1985;32:649–656. doi: 10.1111/j.1550-7408.1985.tb03095.x. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Dutcher SK. Molecular studies of linkage group XIX of Chlamydomonas reinhardtii: evidence against a basal body location. J Cell Biol. 1991;113:339–346. doi: 10.1083/jcb.113.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Haas MA, Rosenbaum JL. Localization of kinesin-related protein to the central pair apparatus of the Chlamydomonas reinhardtii flagellum. J Cell Sci. 1994;107:1551–1556. doi: 10.1242/jcs.107.6.1551. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Flagellar regeneration in Chlamydomonas: a model system for studying organelle assembly. Trends Cell Biol. 1993;3:156–161. doi: 10.1016/0962-8924(93)90136-o. [DOI] [PubMed] [Google Scholar]
- Johnson UK, Porter KR. Fine structure of cell division in Chlamydomonas reinhardi: basal bodies and microtubules. J Cell Biol. 1968;38:403–425. doi: 10.1083/jcb.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandl KA, Forney JD, Asai DJ. The dynein genes of Paramecium tetraurelia: the structure and expression of the ciliary β and cytoplasmic heavy chains. Mol Biol Cell. 1995;6:1549–1562. doi: 10.1091/mbc.6.11.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci. 1992;103:653–664. [Google Scholar]
- King SM, Barbarese E, Dillman JFI, Patel-King RS, Carson JH, Pfister KK. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271:19356–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS. The Mr = 8000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem. 1995;265:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Grissom PM, McIntosh JR. Dynein from Dictyostelium: primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J Cell Biol. 1992;119:1597–1604. doi: 10.1083/jcb.119.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Knecht DA. Cytoplasmic dynein heavy chain is an essential gene product in Dictyostelium. Cell Motil Cytoskeleton. 1998;39:63–72. doi: 10.1002/(SICI)1097-0169(1998)39:1<63::AID-CM6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Samso M. Overexpression of dynein’s globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol Biol Cell. 1996;7:935–948. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka MR, Jarvik JW. Short-flagella mutant of Chlamydomonas reinhardtii. Genetics. 1987;115:695–691. doi: 10.1093/genetics/115.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. ida4–1, ida4–2, and ida4–3 are intron splicing mutations affecting the locus encoding p28, a light chain of Chlamydomonas axonemal inner dynein arms. Mol Biol Cell. 1995;6:713–723. doi: 10.1091/mbc.6.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wisniewski JC, Dentler WL, Asai DJ. Gene knockouts reveal separate functions for two cytoplasmic dyneins in Tetrahymena thermophila. Mol Biol Cell. 1999;10:771–784. doi: 10.1091/mbc.10.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA, Nordstrom SA, Molder JE, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J Cell Biol. 1978;78:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA, Rosenbaum JL. Regulation of the synthesis and assembly of ciliary and flagella proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Hodgkin JA. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J Comp Neurol. 1977;172:489–510. doi: 10.1002/cne.901720306. [DOI] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupus A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Lupus A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lye RJ, Wilson RK, Waterston RH. Genomic structure of a cytoplasmic dynein heavy chain gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 1995;32:26–36. doi: 10.1002/cm.970320104. [DOI] [PubMed] [Google Scholar]
- McVittie A. Genetic studies on flagellum mutants of Chlamydomonas reinhardtii. Genet Res Camb. 1972;19:157–164. [Google Scholar]
- Mikami A, Paschal BM, Mazumdar M, Vallee RB. Molecular cloning of the retrograde transport motor cytoplasmic dynein. Neuron. 1993;10:787–796. doi: 10.1016/0896-6273(93)90195-w. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. Molecular analysis of the alpha and beta dynein genes of Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 1989;14:435–445. doi: 10.1002/(SICI)1097-0169(1997)37:2<120::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Brown KS. Sequence analysis of the Chlamydomonas α and β dynein heavy chain genes. J Cell Sci. 1994;107:635–644. doi: 10.1242/jcs.107.3.635. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Brown KS. Sequence analysis of the Chlamydomonas reinhardtii flagellar α dynein gene. Cell Motil Cytoskeleton. 1997;37:120–126. doi: 10.1002/(SICI)1097-0169(1997)37:2<120::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Mocz G, Gibbons IR. ATP-insensitive interaction of the amino-terminal region of the β heavy chain of dynein with microtubules. Biochemistry. 1993;32:3456–3460. doi: 10.1021/bi00064a032. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Molecular neurogenetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. BioEssays. 1997;19:1055–1064. doi: 10.1002/bies.950191204. [DOI] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9 + 2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster S, Knott J, O’Toole E, Porter M. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8:607–620. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1997;25:244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neesen J, Koehler M, Kirschner R, Steinlein C, Kreutzberger J, Engel W, Schmid M. Identification of dynein heavy chain genes expressed in human and mouse testis: chromosomal localization of an axonemal dynein gene. Gene. 1997;200:193–202. doi: 10.1016/s0378-1119(97)00417-4. [DOI] [PubMed] [Google Scholar]
- Nelson JAE, Savereide PB, Lefebvre PA. The Cry1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol Cell Biol. 1994;14:4011–4019. doi: 10.1128/mcb.14.6.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF-3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule associated protein MAP 1C. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, Porter ME. The Chlamydomonas IDA7 locus encodes a 140 kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell. 1998;9:3351–3365. doi: 10.1091/mbc.9.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1FLA10 to reach the distal part of flagella in Chlamydomonas. J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp-1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N, Koonce MP, de Hostos EL, Vale RD. In vitro microtubule-based organelle transport in wild-type Dictyostelium and cells overexpressing a truncated dynein heavy chain. Cell Motil Cytoskeleton. 1998;40:304–314. doi: 10.1002/(SICI)1097-0169(1998)40:3<304::AID-CM8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Porter ME. Axonemal dyneins: assembly, organization, and regulation. Curr Opin Cell Biol. 1996;8:10–17. doi: 10.1016/s0955-0674(96)80042-1. [DOI] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, Farlow SJ. The dynein gene family in Chlamydomonas reinhardtii. Genetics. 1996;144:569–585. doi: 10.1093/genetics/144.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman RW, LeCluyse EL, Dentler WL. Development of microtubule capping structures in ciliated epithelial cells. J Cell Sci. 1987;87:85–94. doi: 10.1242/jcs.87.1.85. [DOI] [PubMed] [Google Scholar]
- Rasmusson K, Serr M, Gepner J, Gibbons I, Hays TS. A family of dynein genes in Drosophila melanogaster. Mol Biol Cell. 1994;5:45–55. doi: 10.1091/mbc.5.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Deinhardt F. Preparation of a semipermanent mounting medium for fluorescent antibody studies. Virology. 1960;12:316–317. doi: 10.1016/0042-6822(60)90205-1. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Molder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas: the use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:601–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies in Chlamydomonas reinhardtii. Ann NY Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takada S, King SM, Witman GB, Kamiya R. A Chlamydomonas outer arm dynein mutant with a truncated β heavy chain. J Cell Biol. 1993;122:653–661. doi: 10.1083/jcb.122.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. The direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci USA. 1977;74:2045–2049. doi: 10.1073/pnas.74.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanders MA, Salisbury JL. Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 1995;47:163–169. doi: 10.1016/s0091-679x(08)60805-5. [DOI] [PubMed] [Google Scholar]
- Schnell RA, Lefebvre PA. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics. 1993;134:737–747. doi: 10.1093/genetics/134.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir MA, Fukushige T, Yasuda H, Miwa J, Siddiqui SS. C. elegans osm-3 gene mediating osmotic avoidance behavior encodes a kinesin-like protein. NeuroReport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh W-H, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabish M, Siddiqui ZK, Nishikawa K, Siddiqui SS. Exclusive expression of C. elegans osm-3 kinesin gene in chemosensory neurons open to the external environment. J Mol Biol. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Zhang Z, Hirokawa N. Identification and molecular evolution of new dynein-like protein sequences in rat brain. J Cell Sci. 1995;108:1883–1893. doi: 10.1242/jcs.108.5.1883. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive, multiple sequence alignment through sequence weighting, position-sensitive gap penalties, and weight-matrix choice. Nucleic Acids Res. 1994;22:4673–4689. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxhorn J, Daise T, Dentler WL. Regulation of flagellar length in Chlamydomonas. Cell Motil Cytoskeleton. 1998;40:133–146. doi: 10.1002/(SICI)1097-0169(1998)40:2<133::AID-CM3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Grissom PM, McIntosh JR. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J Cell Biol. 1996;133:831–842. doi: 10.1083/jcb.133.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, Mcintosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:840–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha M, Walther Z, Hall JL. The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J Cell Sci. 1996;109:541–549. doi: 10.1242/jcs.109.3.541. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, et al. Multiple mouse chromosomal loci for dynein-based motility. Genomics. 1996;36:29–38. doi: 10.1006/geno.1996.0422. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez J-M, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α-and β-subunits of ATP synthase myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther Z, Vashishtha M, Hall JH. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick MJ, Ann DK, Loh HH. Molecular cloning of a novel protein regulated by opioid treatment of NG108–15 cells. Mol Brain Res. 1995;32:171–175. doi: 10.1016/0169-328x(95)00090-f. [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Witman GB. Molecular analysis of the γ heavy chain of Chlamydomonas flagellar outer arm dynein. J Cell Sci. 1994;107:497–506. doi: 10.1242/jcs.107.3.497. [DOI] [PubMed] [Google Scholar]
- Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Witman GB. The site of in vivo assembly of flagellar microtubules. Ann NY Acad Sci. 1975;253:178–191. doi: 10.1111/j.1749-6632.1975.tb19199.x. [DOI] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonas flagella I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/3B: a heterotrimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Regulation of Nitrate Assimilation in Chlamydomonas reinhardtii. Ph.D. Thesis. Minneapolis, MN: University of Minnesota.; 1996. [Google Scholar]
- Zhang Z, Tanaka Y, Nonaka S, Aizawa H, Kawasaki H, Nakata T, Hirokawa N. The primary structure of rat brain (cytoplasmic) dynein heavy chain, a cytoplasmic motor enzyme. Proc Natl Acad Sci USA. 1993;90:7928–7932. doi: 10.1073/pnas.90.17.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]