Abstract
It is known that organic forms of Selenium (Se) inhibit chemically-induced rat mammary carcinogenesis, although the molecular basis remains to be elucidated. To identify signaling pathways involved in carcinogenesis that are also modulated by methylselenocysteine (MSC), we compared the global gene expression profiles in mammary tissues from pubescent female rats maintained on a selenium supplemented (3 ppm) diet with those on a standardized diet after N-nitroso-N-methylurea (NMU). While the Se enriched diet altered the steady state levels of genes involved in various cellular functions, the most dramatic effect was the coordinated changes in the expression of multiple genes that regulate circadian rhythm. Normal mammary tissue of rats fed a standardized diet showed little circadian oscillation relative to liver tissue. By contrast, mammary tissue of rats maintained on the Se enriched diet showed a progressive, time dependent increase in the expression of circadian gene Per 2 and circadian regulated transcription factor DBP. Our results further demonstrated that the expression of Per2 and DBP mRNAs were significantly decreased in mammary tumors arising in rats on the selenium enriched diet, but not in tumors of rats on the control diet, suggesting that Se-induced elevation in the expression of circadian genes was incompatible with mammary carcinogenesis. Given the previously reported role of Per2 as a tumor suppressor, these observations suggest that Per2 is an important target of MSC during chemoprevention in NMU-induced rat mammary carcinogenesis, and for the first time provide a mechanistic link between chemoprevention and circadian rhythm.
Keywords: selenium, chemoprevention, mammary carcinogenesis, microarray, circadian
INTRODUCTION
Selenium (Se), a dietary trace element, has shown to reduce the incidence of several cancers in epidemiological studies (1). The role of Se in chemoprevention of mammary carcinomas was first suggested by animal studies performed by Ip and coworkers. Whereas exposure of virgin female rats to a variety of carcinogens during puberty results in close to 100 % incidence of mammary carcinomas in many susceptible strains, animals fed with garlic grown in selenium enriched soil showed dramatic reductions in tumor incidence (2-5). Moreover, the inhibition of mammary carcinogenesis by organic forms of Se was more effective at early stages of carcinogenesis than later stages (5-8), suggesting that Se mediated its effects at the post-initiation stage of carcinogenesis. Subsequent studies determined that metabolite methylselenol was the active form of chemopreventive Se (reviewed by Ip, (9, 10)
The mechanisms of Se mediated chemoprevention in vivo are still poorly understood at the molecular level. Early hypotheses focused on selenoproteins such as glutathione peroxidase, an enzyme involved in reducing reactive oxygen species (reviewed by Ganther,(11). However, since maximum glutathione peroxidase enzyme activity is attained in animal tissues without selenium supplementation, it is improbable that the latter mechanism plays a significant role (12-16). More recent studies reviewed by Whanger (17) have focused on the ability of methylselenocysteine (MSC) to suppress cell proliferation and induce apoptosis in vitro and in vivo. Nonetheless the exact pathways involved in these processes in vivo are still unclear, and alternative mechanisms of chemoprevention have not been ruled out.
In an attempt to further define the molecular bases for Se chemoprevention, several studies used DNA microarray technology to compare Se-induced changes in gene expression profiles in different types of mammary samples including whole mammary gland, pre-malignant tumor cell lines, and mammary tumors or in vivo system (for review see (18)). The results of these studies demonstrated that chemopreventive Se altered the steady state levels of a wide spectrum of genes that are involved in cell cycle, tumor growth inhibition, metabolism, and apoptosis. Moreover, many of the Se mediated changes in gene expression were dependent on the experimental system, suggesting that selenium probably exerted its chemopreventive effects via multiple mechanisms. In the present study we first investigated the chemopreventive potential of MSC in the Fischer 344 (F344) rat. Our results showed that as in other strains, dietary supplementation with Se (3 ppm) in the form of MSC for 30 days following N-nitroso-N-methylurea (NMU) exposure reduced the incidence of mammary carcinogenesis by ~60% in the F344 strain. Consistent with the results of previous studies, we found that the chemopreventive levels of MSC regulated the expression of genes involved in numerous cellular processes. However, unlike previous studies, our results demonstrated that during the course of Se-induced chemoprevention, there were significant effects on both the steady state levels and temporal oscillations of genes involved in circadian rhythm in mammary tissue. Moreover, the Se-induced increases in circadian genes were suppressed in all tumors that arose in all NMU-treated animals maintained on the Se enriched diet. Given previous observations that circadian gene Per2 is a tumor suppressor (19), and accumulating epidemiological evidence that disruption of normal circadian rhythm may increase the risk of cancer development (20-22), our study provides a new insight into the mechanism of selenium-induced chemoprevention.
Materials and Methods
Animal Maintenance and Diet Preparation
Before conducting any animal experiments, all protocols were reviewed and received the approval of the Institutional Animal Care and Use Committee. All experiments were performed in FHCRC vivarium, which is fully accredited by AAALAC. Pathogen-free female Fisher 344 rats were purchased from Harlan Sprague-Dawley (Indianapolis, IN). Upon arrival, animals were maintained on the standard powdered AIN-76A diet (Harlan Tekland, WI) for a week, for acclimatization to a powdered ration. The AIN-76 mineral mix provides 0.1 ppm Se in the form of sodium selenite. The selenium supplemented diet was made by admixing L-Se-methylselenocysteine (MSC) (Selenium Technologies, Lubbock, TX) with standard, powdered AIN-76A diet to a final concentration of 3 ppm Se. All diets were prepared by Harlan Tekland (Madison, WI) and were provided to the rats fresh every week. All animals were housed under controlled climate conditions and a 12 h light/12 h dark cycle (7am on / 7pm off).
Chemoprevention Study
To induce mammary tumors, sixty female F344 rats between the the ages of 50-55 days were injected i.p. with a single 50mg/kg body weight dose of NMU (Ash Stevens Inc., Detroit, MI). NMU was dissolved in acidified saline at a final concentration of 10mg/ml just prior to injection. After chemical treatment, animals were randomly divided into two groups and maintained on either the basal AIN-76A diet or the selenium supplemented diet (3 ppm Se) until the termination of the experiment. All animals were examined weekly to determine the appearance, size, and location of mammary tumors. All rats were euthanized by CO2 thirty-two weeks after carcinogen administration. At necropsy, the mammary gland was carefully examined for the palpable and non-palpable mammary tumors. All tumors as well as normal mammary gland were excised and fixed in normal buffered formalin for later histopathological diagnosis. Only confirmed adenocarcinomas were included in the data analysis. Tumor incidences of two dietary regimens at the final time point were compared by Chi-square analysis, and the inhibition of tumorigenesis was calculated based on the total tumor yields from both groups.
RNA Isolation
For microarray experiments, total RNA were isolated from mammary glands on the left side of each animal using Qiagen RNeasy Maxi Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. The isolated RNA was then precipitated twice by 4 M LiCl, resuspended in 3M NaOAc (pH5.2) and precipitated from ethanol, followed by two washes with 80% ethanol. Alternatively, total RNA from mammary gland was isolated using Trizol (Invitrogen, Carlsbad, CA) and purified on Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). Liver total RNA was isolated by Qiagen RNeasy Mini Kit. Purified RNA was resuspended in RNase-free water (Sigma, St. Louis, MO) and stored at -80°C.
Microarray Analysis
Gene expression profiles of mammary tissue were determined using Affymetrix GeneChip® oligoarray (Affymetrix, Santa Clara, CA) at Icoria, Inc. (former Paradigm Genetics, Research Triangle Park, NC). Biotinalyted cRNA samples were prepared according to the standard Affymetrix GeneChip protocol (Affymetrix, Santa Clara, CA). In brief, 1 μg of total RNA from each sample spiked with poly A as a labeling control were converted to double-stranded cDNA with GeneChip One-Cycle cDNA Synthesis Kit. After second-strand synthesis, the cDNA is purified with the GeneChip Sample Cleanup Module. The resulting double-stranded DNA is then used to generate multiple copies of biotinylated cRNA by in vitro transcription with the GeneChip 3’-Amplification Reagent Kit for IVT Labeling. The amount and quality of the fluorescently labeled cRNA was assessed using a Nanodrop ND-100 spectrophometer and an Agilent Bioanalyzer. For each sample, 15 μg of biotinylated cRNA spiked with bioB, bioC, bioD and cre (microarray hybridization controls) was hybridized to a GeneChip Rat Genome 230 2.0 Array (Affymetrix, Sanata Clara, CA) for 16 hours at 45°C. Following hybridization, all arrays were washed and stained in an Affymetrix GeneChip Fluidics Station. Stained arrays were scanned with an Affymetrix GeneChip® Scanner 3000. Quality check (visual inspection of image, % Present calls, background, scaling factor, 3’/5’ GAPDH and Beta-actin ratios, detection call of biotin spike-in) and data analysis were carried out using Affymetrix GeneChip Operating Software (GCOS) and Quality Reporter.
Statistical Analysis
To identify the differentially expressed genes between control diet and selenium supplemented diet, data from the selenium samples and control samples were combined using an error-weighted average using Rosetta Resolver version 5.1.0.1.0 (Rosetta Biosoftware, Seattle, WA). The combined data were compared to create a ratio in which the selenium samples were in the numerator and the control samples were in the denominator. Transcripts demonstrating an absolute fold change greater than 1.3, a log ratio p-value less than 0.001, and a combined log10 intensity of greater than -1, were considered significant.
Gene Enrichment Analysis
Genes that were differentially expressed between control and selenium diet were imported into GoMiner (http://discover.nci.nih.gov/gominer), a gene ontology enrichment tool for identifying gene symbols. Transcripts with no associated gene symbol were omitted from these analyses as they have not been attributed by the Gene Ontology consortium to a biological process. The input list was used as a means to identify the enrichment of biological processes using High-Throughput GoMiner (23).
Quantitative Real-Time PCR
To validate the data obtained from microarrays, a fluorogenic 5’ nuclease-based Real-Time PCR assay was used to quantitate the mRNA levels of specific genes in the same samples analyzed on microarrays. Briefly, reverse transcription was performed according to the manufacturer’s established protocol using 1μg of total RNA and iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA.) For gene expression measurements,4 μl of cDNA were included in a PCR reaction (16 μl final volume) that included the appropriate forward (FP) and reverse (RP) primers at 438 nM each, 188 nM TaqMan probe and 1X TaqMan® Fast Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems Inc., Foster City, CA). The PCR primers and the dual-labeled probes (6-carboxy-fluorescein (FAM) and 6-carboxy-tetramethyl-rhodamine (TAMRA)) used were listed in Supplemental Table 1. Beta-actin gene expression levels were utilized as an internal control to normalize the data for all individual genes.
RESULTS
Chemopreventive Effects of Selenium on Mammary Carcinogenesis in F344 Rats
The chemopreventive efficacy of organic Se was not previously examined in the F344 rat strain. We therefore compared the incidence and latency of mammary tumor formation in NMU-treated F344 females maintained on a standardized diet (0.1 ppm Se) with those of animals fed a MSC supplemented diet (3 ppm Se). At a dose of 3 ppm Se, MSC has repeatedly shown to reduce tumor incidence with no reported toxicity in Sprague-Dawley rats using chemical-induced carcinogenesis models (24-27)}. All previous studies examining the effects of Se on mammary carcinogenesis were terminated at 24 weeks after carcinogen treatment, and typically reported reductions of ~50-70% in tumor incidence. As shown in Figure 1, at 24 weeks, we also observed a dramatic reduction in the incidence of mammary carcinomas in F344 female rats maintained on the enriched selenium diet. For rats on the control diet, the first mammary tumor appeared approximately 10 weeks after NMU administration, whereas on the selenium diet no tumors were detected until 23 weeks after NMU exposure.
Figure 1.
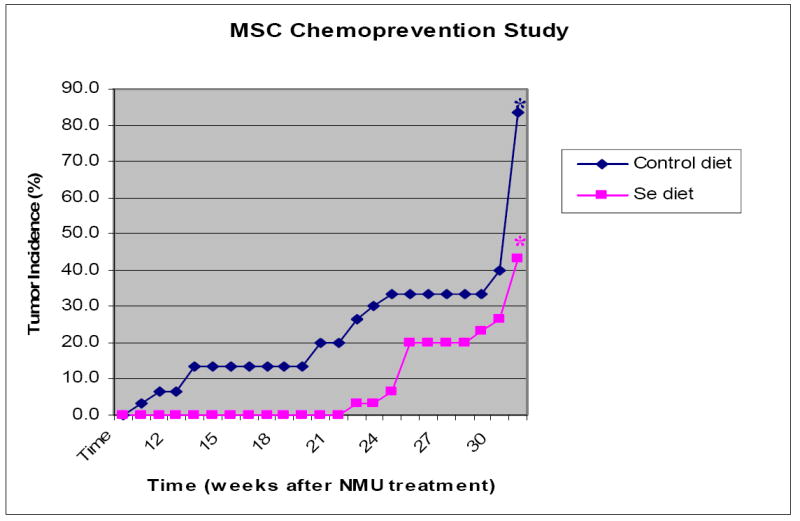
Effects of methylselenocysteine on mammary tumorigenesis in female F344 rats given one single i.p. injection of NMU (50mg/kg body weight). Y: tumor incidence: number of rats with palpable tumors/total number of rats within the group. X: time after NMU treatment in weeks. (* indicates tumor incidence based on visible tumors at necropsy.)
Unlike previous studies, we extended the study beyond 24 weeks. Significantly, the number of animals with palpable mammary tumors were comparable for animals on the control and selenium diets (11/30 and 11/29, respectively). However, at necropsy, we found that many rats maintained on the control diet had small mammary tumors that were not easily detected by palpation. All the tumors were subject to histopathological diagnosis, and only confirmed carcinomas were used in comparisons (Table 1). Including those detected at necropsy, 13 rats on the selenium enriched diet and 25 of the 30 animals on the control diet developed mammary carcinomas. Statistical analyses of these data showed that the tumor incidence was significantly reduced by selenium supplement in the diet (p<0.001). Although multiplicity of tumors was only slightly higher on control diet than selenium diet, the number of mammary carcinomas from selenium diet was 62% less than that of control diet.
Table 1.
Chemopreventive effects of methylselenocysteine on NMU-induced mammary carcinogenesis in female F344 rats.
| Tumor Incidence†,†† | Total # of Tumors†† | Multiplicity‡ | % of Inhibition§ | |
|---|---|---|---|---|
| AIN-76A | 22/30 | 35 | 1.59 | --- |
| AIN-76A + Se (3 ppm) | 9/29* | 13 | 1.44 | 63% |
Tumor incidence was defined by number of tumor-bearing rats divided by total number of rats.
Both tumor incidence and total number of tumors presented in this table are histologically confirmed adenocarcinomas.
Multiplicity was defined by the total number of tumors divided by total number of tumor-bearing rats.
Tumor inhibition was calculated based on total number of tumors.
P<0.05
Histopathologically, the majority of mammary tumors from both dietary groups were adenocarcinomas, some of which were admixed with fibroadenoma. There were also some adenomas in both groups, and one leiomyosarcoma was found among animals on the control diet. Notably, we observed two adenocarcinomas with squamous cell metaplasia in rats on the selenium diet. This rare, invasive form of mammary carcinoma, however, was not observed in rats maintained on the standard diet. The latter finding suggested the possibility that Se was selecting against the predominant cancer phenotype detected in this experimental tumor model.
Global Gene Expression Profiles of Mammary Gland at Day 30 after NMU Exposure
To identify the genes and pathways that are affected during chemoprevention, we generated global gene expression profiles of rat mammary cells in the presence and absence of MSC supplementation. Seventy-two, 50-day old female F344 rats were randomly divided into two groups receiving an i.p. injection of either NMU (50mg/kg body weight) or the vehicle. Rats under each treatment were further divided into two groups - those fed the standard AIN-76A diet and those fed AIN-76A diet supplemented with MSC (3 ppm Se). Six rats from each group were sacrificed on days 1, day 7 and day 30 after chemical treatment. Mammary glands were harvested and quickly transferred to the RNAlater™ to preserve the integrity of RNA. To minimize the variation between animals, total RNA was isolated from mammary glands from the left side of all individual rats.
Previous studies demonstrated that the chemoprevention by organic Se required that animals be fed the enriched diet for a period of one month, after which incidence of mammary tumor in rats was essentially the same as that of rats fed on chemopreventive Se diet for six months (8). This observation suggests that the crucial selenium-induced effects on mammary gland occur during the first month after chemical exposure. Therefore, we first compared gene expression profiles of total RNA samples from the NMU treated group at day 30 after exposure. Using the Affymetrix Rat Genome 230 2.0 oligoarray, which analyzes over 30,000 transcripts and variants from over 28,000 well-substantiated rat genes, we generated profiles of whole mammary gland from rats on control diet (n=6) and selenium diet (n=6). Expression profiles were then analyzed for differentially expressed genes using several analytical software packages, including the Rosetta Resolver, the TIGR Multi Experiment Viewer (MEV) (The Institute for Genomic Research, MD), and ArrayAssist software (Stratagene, La Jolla, CA). The results obtained with all of these analysis tools were similar (data not shown), and identified 79 transcripts that were differentially expressed between control diet and selenium diet using the following criteria: ratio p<0.001, absolute fold change> 1.3, combined log10 intensity > -1. Among these differentially expressed transcripts, 54 transcripts were up-regulated and 25 transcripts were down-regulated by the selenium enriched diet. These genes are involved in various biological processes, such as metabolism, cell proliferation and signal transduction. Data from the 40 differentially expressed genes with good annotation were further evaluated for gene ontology using GoMiner. The gene enrichment analysis (Table 2) showed that several clock genes involved in circadian rhythm, including Per2, Per3, DBP, Cry2, Arntl (Bmal1), were among the set of genes that showed the greatest response to dietary selenium supplementation.
Table 2.
Results of GoMiner Gene enrichment analysis of differentially expressed genes by selenium.
| GO CATEGORY | GENE | P-value |
|---|---|---|
| GO:0007623_circadian_rhythm | Arntl (↓2. 7) | 4.61E-09 |
| Cry2 (↑2.1) | ||
| DBP (↑2.3) | ||
| Per2 (↑3.3) | ||
| Per3 (↑1.7) | ||
| GO:0048511_rhythmic_process | Arntl | 5.90E-08 |
| Cry2 | ||
| DBP | ||
| Per2 | ||
| Per3 | ||
| GO:0006775_fat-soluble_vitamin_metabolism | DBP | 0.001 |
| Ttpa | ||
| GO:0000723_telomere_maintenance | Hspa1a | 0.045 |
| GO:0008207_C21-steroid_hormone_metabolism | Star | 0.045 |
| GO:0009123_nucleoside_monophosphate_metabolism | Prps1 | 0.045 |
| GO:0009636_response_to_toxin | Ephx1 | 0.045 |
| GO:0019439_aromatic_compound_catabolism | Ephx1 | 0.045 |
| GO:0006692_prostanoid_metabolism | Ptgis | 0.050 |
| GO:0006693_prostaglandin_metabolism | Ptgis | 0.050 |
| GO:0006700_C21-steroid_hormone_biosynthesis | Star | 0.040 |
| GO:0006805_xenobiotic_metabolism | Ephx1 | 0.040 |
| GO:0007565_pregnancy | Ttpa | 0.040 |
| GO:0009112_nucleobase_metabolism | Prps1 | 0.040 |
| GO:0009124_nucleoside_monophosphate_biosynthesis | Prps1 | 0.040 |
| GO:0009161_ribonucleoside_monophosphate_metabolism | Prps1 | 0.040 |
| GO:0009410_response_to_xenobiotic_stimulus | Ephx1 | 0.040 |
| GO:0009894_regulation_of_catabolism | Arntl | 0.040 |
To validate the microarray results, we performed Real-Time PCR on the Per2 and DBP genes in mammary tissue of animals from both NMU and vehicle treated groups in the analysis. We also examined three timepoints (day 1, day 7 and day 30) for each group to test whether the effects on circadian genes were treatment- and/or time- dependent. The steady state expression levels of Per2 and DBP transcripts normalized to β-actin are presented in Figure 2A and Figure 2B, respectively. The results confirmed that dietary selenium upregulated Per2 expression in both NMU treated group and saline-treated group. Analysis of the data using Student’s t-test showed that upregulation of Per2 by selenium was statistically significant at day 7 and day 30, but not at day 1, possibly due to greater inter-individual variability. Similarly, DBP gene expression level was higher on the selenium diet relative to control diet at day 7 and day 30, but based on the t-test the upregulation of DBP was only significant in the NMU treated group. Quantitative PCR data not only confirmed the microarray results, but also suggested that the upregulation of circadian regulated clock genes occurred as early as 7 days after being on the selenium-enriched diet.
Figure 2.
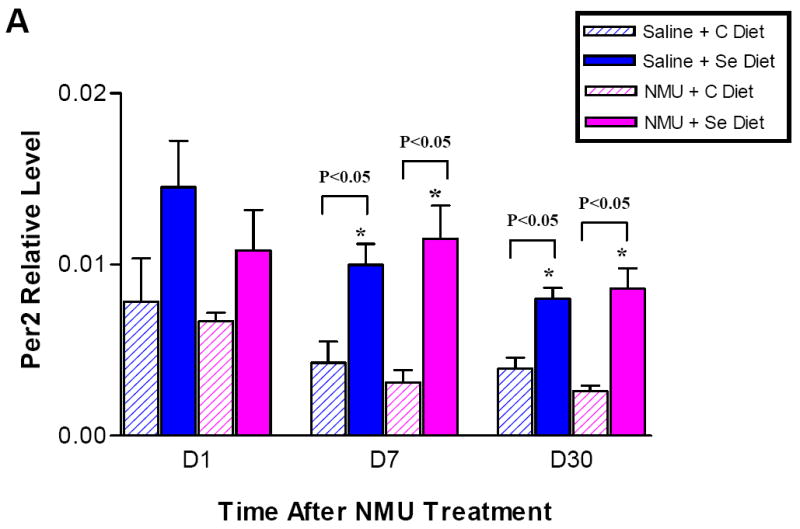
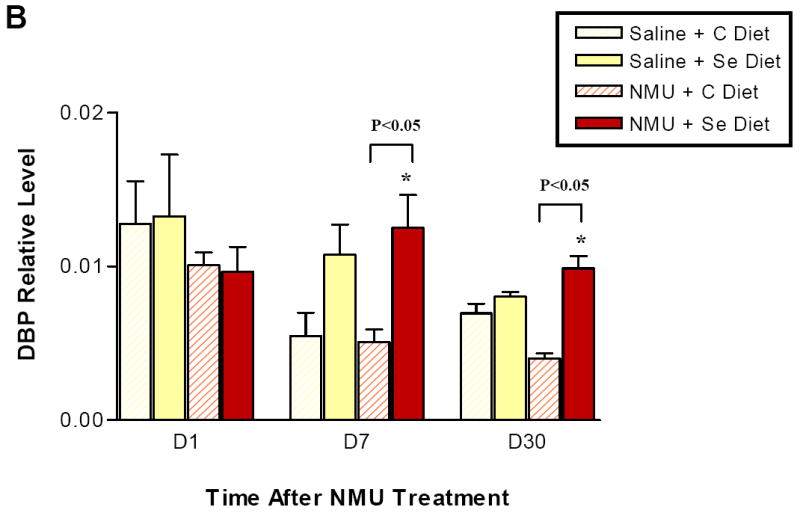
Quantitative analysis of Per2 (A) and DBP (B) in mammary glands. Real-Time PCR with TaqMan probe was performed on mammary gland total RNA samples isolated from rats exposed to NMU or vehicle, and fed on the control diet and the selenium diet. Y-axis: mRNA level of Per2 normalized to β–actin. X: days after NMU treatment. The Per2 and DBP expression levels in mammary glands from animals exposed to the same chemical but fed on either the selenium diet or the control diet was compared by a Student t-test. Asterisk (*) indicates a statistical significance (p< 0.05).
Circadian Gene Per2 and DBP Expression Levels in Mammary Carcinomas
Previous studies showed that Per2 can function as a potent tumor suppressor (19, 28). We therefore compared the level of Per2 in mammary tumors induced in animals on the two diets. We performed Real-Time PCR for Per 2 expression on 8 carcinomas from control diet and 6 carcinomas from selenium diet. For comparison, we isolated total RNA from the adjacent normal mammary gland of each carcinoma. A pair-wise t-test was performed on the normalized gene expression levels between carcinomas and their own neighboring normal tissues. The results suggested that in all cases, both Per2 and DBP were down-regulated in carcinomas compared to the adjacent normal tissue (Table 3). These decreases are likely to be underestimated given that the rat mammary carcinomas typically comprise up to 50% normal stromal cells. Nonetheless, down-regulation of circadian genes was statistically significant in carcinomas arising in animals on the selenium enriched diet, and there was also a trend (p= 0.07) of down-regulation in carcinomas from animals on the control diet. These observations were in sharp contrast to the elevated expression of circadian rhythm observed in normal mammary tissues of animals on the selenium-enriched diet for 30 days. Together, these findings suggested that there was a selection against the expression of Per2 during the process of carcinogenesis, with the most pronounced effect in tumors arising in animals on the selenium-enriched diet.
Table 3.
Pair-wised comparison of Per2 and DBP mRNA levels between mammary carcinomas and their adjacent normal tissues.
| Per2 | DBP | |
|---|---|---|
| Se Carcinoma (n=6) | ↓ 1.83 (p=0.042) | ↓ 2.83 (p=0.002) |
| Control Carcinoma (n=8) | ↓ 2.22 (p=0.072) | ↓ 1.48 (p=0.22) |
Effects of Selenium on the diurnal expression of of Per2 and DBP
As we did not anticipate the involvement of circadian rhythm genes, and levels of circadian genes fluctuate continuously over a 24-hour period, we wanted to eliminate the possibility that minor variations in the time of day when samples were collected, contributed to the differences detected among the experimental groups. More importantly, we were keenly interested in investigating how selenium modulates the expression of circadian genes during the day. We therefore generated 24-hour diurnal expression profiles for the Per2 and DBP genes in both liver and mammary glands from animals on the two diets. Liver was used as a control because of the abundant expression of circadian genes and its obvious display of circadian rhythm (29). In this experiment, we administered NMU to 42 individual, age-matched 50-day-old female F344 rats, which were then randomized to either a control diet or Se diet for 30 days. We collected livers and mammary glands from three animals in each group, every 4 hours during a 24-hour period. In addition, rats from both diets were euthanized alternately at each time point to minimize differences caused by minor difference in the exact time of tissue harvest. The mRNA levels of Per2 and DBP were measured in all the samples using Real-Time PCR. The normalized data are shown in Figure 3, where x-axis indicates the time of harvest in Zeitgeber Time (ZT), with ZT0 indicating when the lights were turned on. Y-axis shows the gene expression levels after normalizing to β-actin. In liver tissues, there clearly was circadian regulation of both genes, indicated by cyclical fluctuation over 24-hour period, and there was essentially no difference between control diet and selenium diet. By contrast, Per2 expression levels in animals on the control diet were maintained at relatively constant and low levels in the mammary gland during the same 24-hour period. For most of the day there was no significant difference in Per2 level between the two diets. However, when the light was turned off at ZT12, there was a dramatic increase in Per2 mRNA levels in the mammary glands of animals on the selenium diet, which dropped sharply from its peak around ZT16. A similar effect was also observed in DBP the regulation gene in the mammary gland, albeit to a lesser degree. These results suggested that in the F344 rats, there is little diurnal variation on the expression of clock gene Per2, and the Per2 regulated DBP in the mammary gland, and that a chemopreventive diet enriched in selenium induced a rhythmical pattern in the expression of these genes.
Figure 3.
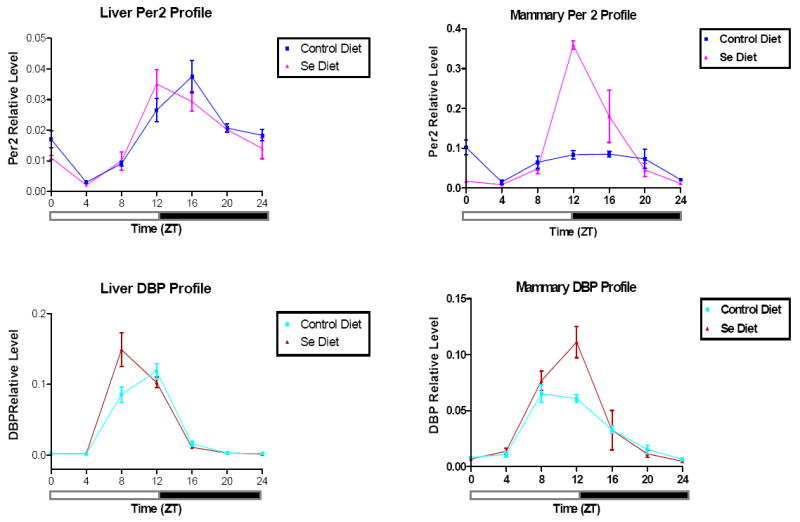
Quantitative analysis of Per2 and DBP mRNA levels over a 24 hour period. Real-Time PCR was performed on total RNA samples isolated from livers and mammary glands of age-matched, 50-day-old rats fed on either the control diet (n=3) or the selenium diet (n=3) for 30 days after NMU exposure (50mg/kg body weight). X-axis: Zeitgeber time; Y-axis: mRNA level normalized to β-actin, shown in mean±SEM.
Discussion
Organic Selenium Inhibits NMU-induced Mammary Carcinogenesis in F344 Rats
Previous studies documented that MSC inhibits chemical-induced mammary carcinogenesis in Sprague-Dawley rats (3, 5), an outbred strain that is highly susceptible to chemical-induced mammary carcinogenesis. However, different rat strains exhibit significant differences in their sensitivities toward chemical-induced mammary carcinogenesis due to yet to be defined genetic variations. The extent to which the response to chemopreventive agents varies across strains has to our knowledge not been investigated in the rat. Since most of our studies have made use of the inbred F344 strain (30-33), we wanted to determine if the latter strain was also responsive to the chemopreventive effects of MSC. Our study showed that dietary supplementation with MSC significantly inhibited NMU-induced mammary carcinogenesis in female inbred F344 rats. The tumor incidence of rats on the selenium diet was reduced to half of what was observed in rats on the control diet, and there was a 62% reduction in total number of tumors. These results are very similar to the data obtained from similar chemoprevention studies using Sprague-Dawley rats suggesting the chemopreventive effects of selenium are not strain dependent (2, 3, 5, 8, 13). The latter observation suggests that Selenium targets common pathways in mammary carcinogenesis that are shared by these two, and perhaps all, susceptible strains of rats.
Gene Expression Profiling of Mammary Glands Identifies Circadian Genes as Novel Targets of Selenium
Previous studies have suggested that selenium might exert its chemopreventive effects at early stage of mammary carcinogenesis (5-7). We hypothesized that by studying the differences in gene expression profiles of mammary cells with NMU exposure, in the presence and absence of supplemental dietary selenium, we could identify key regulatory elements modulated by Se that are essential to the promotion of mammary carcinogenesis. In designing these studies, we first had to decide what cells to use for expression profiling. Although rat mammary carcinomas are considered to originate from ductal epithelium, the importance of the interaction between epithelial cells and stromal cells has been widely acknowledged (34-36). In addition, the percentage of mammary epithelial cells dramatically decreases as mammary gland develops past puberty, making it difficult to isolate and enrich the epithelial cell population. Moreover, techniques used for isolating mammary epithelial cells, such as Laser Capture Microdissection (LCM) or cell surface marker based cell sorting, entail the risk of altering the in vivo gene expression patterns in the mammary cells. For these reasons, we chose to focus our analyses on expression profiles in whole mammary tissue rather than isolated mammary epithelial cells.
Several previous studies examining the chemopreventive effects of selenium using either cDNA or oligonucleotide arrays (37-40) implicated genes involved in multiple biological processes, including cell cycles regulation (e.g. CDK1, CHK2, cyclin A), apoptosis (e.g. bcl-2, bax) and signal transduction. However, all these previous data were derived from cancer cell lines or mammary tumors, and therefore may not have detected Se-induced changes in gene expression at the early stage of carcinogenesis, that are essential to chemoprevention. We therefore examined the effects of selenium on the entire genome in the whole mammary gland at various times early in the process of NMU-induced carcinogenesis. Using a relatively stringent p-value combined with signal intensity and fold change, we identified a number of differentially expressed genes that were associated with organic selenium supplementation. The genes and biochemical pathways involved in regulating circadian rhythm were among most significantly and consistently altered by the selenium enriched diet. Our observation that Per2 expression is reduced in tumors relative to adjacent normal mammary confirms the possibility that important Se-modulated genes are selected against during the process of carcinogenesis.
Circadian Genes are Implicated in Tumorigenesis
The circadian gene family consists of eight core members: Clock, casein kinase Iε (CKI ε), cryptochrome1, 2 (Cry1, Cry2), Period 1, 2, 3 (Per1, Per2, Per3) and Bmal1. The circadian clock sustains rhythms of about 24 hours in the absence of external cues via feedback loops of the circadian genes in the suprachiasmatic nuclei (SCN), as well as in most peripheral tissues (41).
Since circadian clock genes play a role in normal development and differentiation of mouse mammary gland (42), the disruption of circadian genes has led to the speculation of its involvement in mammary tumorigenesis. The possibility that disruption of circadian rhythm in women led to increased mammary tumor development was first reported in the 1960s, and ongoing studies showed that disruption of circadian endocrine rhythm accelerates breast epithelial stem-cell proliferation, induces mammary gland development, and increases the formation of spontaneous mammary tumors in rodents (reviewed by (41)). A breakthrough in the study of circadian rhythm and its connection with cancer development came when Per2, a core circadian gene, was shown to have tumor suppressor activity. Lee and colleagues showed that mice 20 deficient in the Per2 gene not only lost circadian rhythm(43), but were also cancer prone (19, 44). Given the role of Per2 in tumor suppression, we speculated that selenium may exert its chemopreventive effects by upregulating Per2, which may subsequently regulate DNA-damage responses, cell proliferation and apoptosis; thus acting as a tumor suppressor. Our hypothesis is supported by our observation that the up-regulation of Per2 was consistently abolished in mammary carcinomas that arose in NMU treated rats on the organic Se enriched diet, while, Per2 gene expression levels were unaffected in adjacent normal tissues. These findings suggested biological selection against elevated Per2 during the process of carcinogenesis in this setting. Moreover, at least two other studies have reported abnormal expression of Per1, Per2 and Per3 in endometrial carcinomas and human breast cancer tissues (45, 46). A more recent study demonstrated that Per2 binds to the estrogen receptor alpha (ERα), and that binding enhances degradation of the ERα protein (47). Since estrogen is a promoter of mammary carcinogenesis in cells expressing ERα, increased levels of Per2 provides a mechanistic link between a selenium enriched diet and inhibition of mammary carcinogenesis at the post initiation stage.
If circadian gene expression were regulated by organic selenium, then the question is, how does selenium modulate the level of these genes? Our initial studies only compared steady state levels of genes in mammary tissue during a narrow window within 24 hours. To gain a better insight into how dietary selenium affected the regulation on circadian genes, we monitored the expression level of Per2 and DBP every four hours over a 24-hour period. Liver cells showed the anticipated circadian regulation and selenium did not have any impact on the level of Per2 and DBP gene expression. By contrast, Per2 levels in mammary glands from animals on the control diet failed to show the expected circadian oscillation over the 24-hour period. Unlike liver, whose major functions are metabolism and detoxification, the mammary gland is highly sensitive to hormonal regulation. Since autonomic nervous system (ANS) and neuroendocrine systems are regulated by the SCN clock, maintaining a steady level of Per2 in normal mammary gland may have its significance in balancing the hormonal input and output signals essential for normal function of the organ. A similar observation was made in mouse where eight circadian genes, including Per2, showed robust circadian expressions of mRNA in all peripheral tissues examined (heart, lung, liver, stomach, spleen, and kidney) except testis (48).
Unexpectedly, our analysis demonstrated that circadian oscillation over 24 hours was dramatically upregulated in mammary glands by dietary selenium supplementation, beginning few hours following the light to dark transition. DBP, an output gene of the circadian clock, showed a similar expression profile to that of Per2 suggesting that selenium also affects the circadian output signals. Several epidemiological studies have shown the disruption of circadian cycles, particularly among people working predominantly at night, leads to an increased risk of developing breast cancer (20, 21, 49). The proposed mechanism for the observed effects of circadian disruption on mammary carcinogenesis is that exposure to light at night rapidly suppresses melatonin production, which is a rhythmic neuronal activity regulated by SCN (50). Therefore, comparisons of serum melatonin levels between animals on the control and the Se enriched should help establish a putative link between Per2 and melatonin in the future.
Although previous studies reported that circadian gene expression can be induced by serum shock or down-regulated by glucose in cultured Rat-1 fibroblasts (51, 52), we are the first to show that the dietary supplement selenium can regulate circadian genes expression in mammary tissue in vivo. This finding was unexpected, but not biologically implausible. Interestingly, recent studies have shown that transcription feedback loops of circadian genes can also be regulated by intracellular redox pathways. Reduced forms of the redox cofactors, NAD(H) and NADP(H), strongly enhance DNA binding of the Clock:Bmal1 and NPAS2:Bmal1, heterodimeric transcription factors that regulate circadian gene expression, whereas oxidized forms inhibit (53). The targets that the Clock:Bmal1 complex regulates include Per and Cry genes (41). As an antioxidant, selenium-mediated redox reactions involve the cycling of GSH and GSSG which is coupled by NADP/NADPH. The latter may further regulate circadian gene expression.
In summary, the present study showed that dietary selenium in the form of MSC inhibits the NMU-induced mammary carcinogenesis in female F344 rats with the same efficacy as in Sprague-Dawley rats. By combining in vivo chemoprevention studies with genomic microarray analysis of the target tissue, we identified clock genes as novel targets of selenium chemoprevention during the early stage of mammary carcinogenesis. These studies are the first to provide a mechanistic link between regulation of circadian genes and selenium-induced chemoprevention of mammary carcinogenesis.
Supplementary Material
Acknowledgments
This work was supported by Public Health Services grant numbers P30ES007033, P30ES005022 and U19ES011387. We thank Edward K. Lobenhofer and the staff at Icoria (now Cogenics, a Division of Clinical Data) for performing microarrays experiments and data analyses. We thank Denny Liggit for performing histopathological analyses on rat mammary tumors. We thank Isaac Edery and C.S. Yang for critical review of the manuscript.
References
- 1.Clark LC, Combs GF, Jr, Turnbull BW, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. Jama. 1996;276(24):1957–63. [PubMed] [Google Scholar]
- 2.Ip C, Lisk DJ. Efficacy of cancer prevention by high-selenium garlic is primarily dependent on the action of selenium. Carcinogenesis. 1995;16(11):2649–52. doi: 10.1093/carcin/16.11.2649. [DOI] [PubMed] [Google Scholar]
- 3.Ip C, Zhu Z, Thompson HJ, Lisk D, Ganther HE. Chemoprevention of mammary cancer with Se-allylselenocysteine and other selenoamino acids in the rat. Anticancer Res. 1999;19(4B):2875–80. [PubMed] [Google Scholar]
- 4.Ip C, Thompson HJ, Zhu Z, Ganther HE. In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000;60(11):2882–6. [PubMed] [Google Scholar]
- 5.Ip C, Dong Y. Methylselenocysteine modulates proliferation and apoptosis biomarkers in premalignant lesions of the rat mammary gland. Anticancer Res. 2001;21(2A):863–7. [PubMed] [Google Scholar]
- 6.Ip C. Prophylaxis of mammary neoplasia by selenium supplementation in the initiation and promotion phases of chemical carcinogenesis. Cancer Res. 1981;41(11 Pt 1):4386–90. [PubMed] [Google Scholar]
- 7.Ip C, Ganther HE. Activity of methylated forms of selenium in cancer prevention. Cancer Research. 1990;50(4):1206–11. [PubMed] [Google Scholar]
- 8.Lu J, Pei H, Ip C, Lisk DJ, Ganther H, Thompson HJ. Effect on an aqueous extract of selenium-enriched garlic on in vitro markers and in vivo efficacy in cancer prevention. Carcinogenesis. 1996;17(9):1903–7. doi: 10.1093/carcin/17.9.1903. [DOI] [PubMed] [Google Scholar]
- 9.Ip C. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128(11):1845–54. doi: 10.1093/jn/128.11.1845. [DOI] [PubMed] [Google Scholar]
- 10.Ip C, Hayes C, Budnick RM, Ganther HE. Chemical form of selenium, critical metabolites, and cancer prevention. Cancer Research. 1991;51(2):595–600. [PubMed] [Google Scholar]
- 11.Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis. 1999;20(9):1657–66. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 12.Ip C, Lisk DJ. Modulation of phase I and phase II xenobiotic-metabolizing enzymes by selenium-enriched garlic in rats. Nutr Cancer. 1997;28(2):184–8. doi: 10.1080/01635589709514573. [DOI] [PubMed] [Google Scholar]
- 13.Ip C, Lisk DJ, Ganther HE. Chemoprevention with triphenylselenonium chloride in selenium-deficient rats. Anticancer Res. 2000;20(6B):4179–82. [PubMed] [Google Scholar]
- 14.Yang JG, Hill KE, Burk RF. Dietary selenium intake controls rat plasma selenoprotein P concentration. J Nutr. 1989;119(7):1010–2. doi: 10.1093/jn/119.7.1010. [DOI] [PubMed] [Google Scholar]
- 15.Lane HW, Tracey CK, Medina D. Growth, reproduction rates and mammary gland selenium concentration and glutathione-peroxidase activity of BALB/c female mice fed two dietary levels of selenium. J Nutr. 1984;114(2):323–31. doi: 10.1093/jn/114.2.323. [DOI] [PubMed] [Google Scholar]
- 16.Medina D, Lane HW, Shepherd F. Effect of dietary selenium levels on 7,12-dimethylbenzanthracene-induced mouse mammary tumorigenesis. Carcinogenesis. 1983;4(9):1159–63. doi: 10.1093/carcin/4.9.1159. [DOI] [PubMed] [Google Scholar]
- 17.Whanger PD. Selenium and its relationship to cancer: an update dagger. Br J Nutr. 2004;91(1):11–28. doi: 10.1079/bjn20031015. [DOI] [PubMed] [Google Scholar]
- 18.El-Bayoumy K, Sinha R. Molecular chemoprevention by selenium: a genomic approach. Mutat Res. 2005;591(1-2):224–36. doi: 10.1016/j.mrfmmm.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111(1):41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 20.Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17(4):539–45. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 21.Hansen J. Risk of breast cancer after night- and shift work: current evidence and ongoing studies in Denmark. Cancer Causes Control. 2006;17(4):531–7. doi: 10.1007/s10552-005-9006-5. [DOI] [PubMed] [Google Scholar]
- 22.O’Leary ES, Schoenfeld ER, Stevens RG, et al. Shift work, light at night, and breast cancer on Long Island, New York. Am J Epidemiol. 2006;164(4):358–66. doi: 10.1093/aje/kwj211. [DOI] [PubMed] [Google Scholar]
- 23.Zeeberg BR, Qin H, Narasimhan S, et al. High-Throughput GoMiner, an ’industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ip C, Lisk DJ, Thompson HJ. Selenium-enriched garlic inhibits the early stage but not the late stage of mammary carcinogenesis. Carcinogenesis. 1996;17(9):1979–82. doi: 10.1093/carcin/17.9.1979. [DOI] [PubMed] [Google Scholar]
- 25.Dong Y, Lisk D, Block E, Ip C. Characterization of the biological activity of gamma-glutamyl-Se-methylselenocysteine: a novel, naturally occurring anticancer agent from garlic. Cancer Research. 2001;61(7):2923–8. [PubMed] [Google Scholar]
- 26.Ip C, Birringer M, Block E, et al. Chemical speciation influences comparative activity of selenium-enriched garlic and yeast in mammary cancer prevention. Journal of Agricultural & Food Chemistry. 2000;48(6):2062–70. doi: 10.1021/jf000051f. erratum appears in J Agric Food Chem 2000 Sep;48(9):4452. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Pei H, Ip C, Lisk DJ, Ganther H, Thompson HJ. Effect on an aqueous extract of selenium-enriched garlic on in vitro markers and in vivo efficacy in cancer prevention. Carcinogenesis. 1996;17(9):1903–7. doi: 10.1093/carcin/17.9.1903. [DOI] [PubMed] [Google Scholar]
- 28.Rosbash M, Takahashi JS. Circadian rhythms: the cancer connection. Nature. 2002;420(6914):373–4. doi: 10.1038/420373a. [DOI] [PubMed] [Google Scholar]
- 29.Holzberg D, Albrecht U. The circadian clock: a manager of biochemical processes within the organism. J Neuroendocrinol. 2003;15(4):339–43. doi: 10.1046/j.1365-2826.2003.00992.x. [DOI] [PubMed] [Google Scholar]
- 30.Cha RS, Thilly WG, Zarbl H. N-nitroso-N-methylurea-induced rat mammary tumors arise from cells with preexisting oncogenic Hras1 gene mutations. Proc Natl Acad Sci U S A. 1994;91(9):3749–53. doi: 10.1073/pnas.91.9.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Z, Houle B, Mikheev AM, Cha RS, Zarbl H. Alterations in H-ras1 promoter conformation during N-nitroso-N-methylurea-induced mammary carcinogenesis and pregnancy. Cancer Res. 1996;56(21):4927–35. [PubMed] [Google Scholar]
- 32.Cha RS, Guerra L, Thilly WG, Zarbl H. Ha-ras-1 oncogene mutations in mammary epithelial cells do not contribute to initiation of spontaneous mammary tumorigenesis in rats. Carcinogenesis. 1996;17(11):2519–24. doi: 10.1093/carcin/17.11.2519. [DOI] [PubMed] [Google Scholar]
- 33.Mikheev AM, Inoue A, Jing L, et al. Frequent activation of CArG binding factor-A expression and binding in N-methyl-N-nitrosourea-induced rat mammary carcinomas. Breast Cancer Res Treat. 2004;88(1):95–102. doi: 10.1007/s10549-004-1280-5. [DOI] [PubMed] [Google Scholar]
- 34.Nandi S, Guzman RC, Yang J. Hormones and mammary carcinogenesis in mice, rats, and humans: a unifying hypothesis. Proc Natl Acad Sci U S A. 1995;92(9):3650–7. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maffini MV, Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117(Pt 8):1495–502. doi: 10.1242/jcs.01000. [DOI] [PubMed] [Google Scholar]
- 36.Rajkumar L, Kittrell FS, Guzman RC, Brown PH, Nandi S, Medina D. Hormone-induced protection of mammary tumorigenesis in genetically engineered mouse models. Breast Cancer Res. 2007;9(1):R12. doi: 10.1186/bcr1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Y, Ganther HE, Stewart C, Ip C. Identification of molecular targets associated with selenium-induced growth inhibition in human breast cells using cDNA microarrays. Cancer Res. 2002;62(3):708–14. [PubMed] [Google Scholar]
- 38.Dong Y, Lisk D, Block E, Ip C. Characterization of the biological activity of gamma-glutamyl-Se-methylselenocysteine: a novel, naturally occurring anticancer agent from garlic. Cancer Res. 2001;61(7):2923–8. [PubMed] [Google Scholar]
- 39.Dong Y, Zhang H, Hawthorn L, Ganther HE, Ip C. Delineation of the molecular basis for selenium-induced growth arrest in human prostate cancer cells by oligonucleotide array. Cancer Res. 2003;63(1):52–9. [PubMed] [Google Scholar]
- 40.Unni E, Kittrell FS, Singh U, Sinha R. Osteopontin is a potential target gene in mouse mammary cancer chemoprevention by Se-methylselenocysteine. Breast Cancer Res. 2004;6(5):R586–92. doi: 10.1186/bcr914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3(5):350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 42.Metz RP, Qu X, Laffin B, Earnest D, Porter WW. Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev Dyn. 2006;235(1):263–71. doi: 10.1002/dvdy.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng B, Larkin DW, Albrecht U, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400(6740):169–73. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 44.Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17(4):525–30. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 45.Yeh KT, Yang MY, Liu TC, et al. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206(1):111–20. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 46.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26(7):1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 47.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007 doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schernhammer ES, Laden F, Speizer FE, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst. 2001;93(20):1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 50.Blask DE, Dauchy RT, Sauer LA, Krause JA. Melatonin uptake and growth prevention in rat hepatoma 7288CTC in response to dietary melatonin: melatonin receptor-mediated inhibition of tumor linoleic acid metabolism to the growth signaling molecule 13-hydroxyoctadecadienoic acid and the potential role of phytomelatonin. Carcinogenesis. 2004;25(6):951–60. doi: 10.1093/carcin/bgh090. [DOI] [PubMed] [Google Scholar]
- 51.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 52.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277(46):44244–51. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 53.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


