Abstract
Natural killer (NK) cell function is regulated by NK cell receptors that bind classical MHC class I molecules or their structural relatives. The latter group includes self-ligands (MICA, RAE-I, H-60), as well as ligands encoded by viruses (UL18, m155, m157). Two distinct families of NK receptors have been identified: the immunoglobulin-like family (KIRs, LIRs) and the C-type lectin-like family (Ly49s, NKG2D, CD94/NKG2). Here we describe the crystal structures of NK receptors that have been determined to date, both in free form and bound to MHC class I or MHC class I-like molecules.
Keywords: Natural killer cell receptor, MHC, Virus, Crystal structure
1. Introduction
Natural killer (NK) cells are an essential component of innate immunity against tumors and virally infected cells [1,2]. Early studies by Kärre et al. [3] revealed that cell lines deficient in MHC class I expression showed increased susceptibility to lysis by NK cells, compared to their normal parental counter-parts, and that reconstitution of MHC expression restored lysis. These observations gave rise to the “missing self” hypothesis [4], according to which NK receptor engagement by MHC class I inhibits NK cell-mediated lysis of target cells expressing MHC class I (self), thereby directing the cytolytic activity of NK cells against virally infected or tumor cells that have lost MHC class I expression (non-self).
Several receptor families have been identified on primate and rodent NK cells that monitor MHC class I expression on surrounding cells [1,2,5]. These include the killer immunoglobulin-like receptors (KIRs), leukocyte Ig-like receptors (LIRs), members of the Ly49 family (Ly49s), and the CD94/NKG2 family of receptors. Additionally, the activating receptor NKG2D recognizes MHC-like molecules, such as MICA and RAE-Iβ, that are upregulated in stressed tissues [6]. A role for Ly49 receptors in viral immunity has been highlighted by the interaction of activating Ly49s with gene products of mouse cytomegalovirus (MCMV) [7-9].
NK receptors belong to two structurally distinct families, the immunoglobulin (Ig) superfamily and the C-type lectin superfamily [5]. Over the past several years, considerable progress has been made in determining the crystal structures of representative NK receptors, both in isolation and bound to MHC or MHC-like ligands. These include KIRs and LIRs, as well as C-type lectin-like receptors (Ly49s and NKG2D). These structures reveal the multiplicity of solutions that NK receptors have evolved to accomplish the task of recognizing MHC or MHC-like molecules, a crucial interaction for the regulation NK cytolytic activity by host or viral ligands.
2. MHC class I recognition by KIRs
KIRs are type I transmembrane glycoproteins comprising two or three extracellular C2-type Ig-like domains, and hence are designated KIR2D and KIR3D, respectively [5]. In general, KIR2D receptors recognize HLA-C alleles, whereas KIR3D receptors recognize HLA-A and HLA-B alleles. Our understanding of the specificity of KIR2D receptors for HLA-C comes from crystallographic studies of KIR2D molecules, both in free form [10-13] and bound to their respective HLA-C ligands [14,15]. To date, no crystal structures have been reported for KIR3D receptors. KIR2D molecules have two tandem N-terminal domains (D1 and D2) connected by a linker of 3-5 amino acids (Fig. 1a). In common with other Ig superfamily proteins, two anti-parallel β-sheets form each domain, in which a β-sheet of three (in D2) or four (in D1) anti-parallel strands (ABE or ABED, respectively) packs against another β-sheet of four anti-parallel strands (C’CFG) with one extra A’ strand. The topology and disposition of the domains is similar to those observed in hematopoietic receptors [16,17]. The angle between the D1 and D2 varies from about 60° to 80° in different KIRD2 receptors, despite a relatively conserved set of hydrophobic residues at the D1-D2 interface.
Fig. 1.
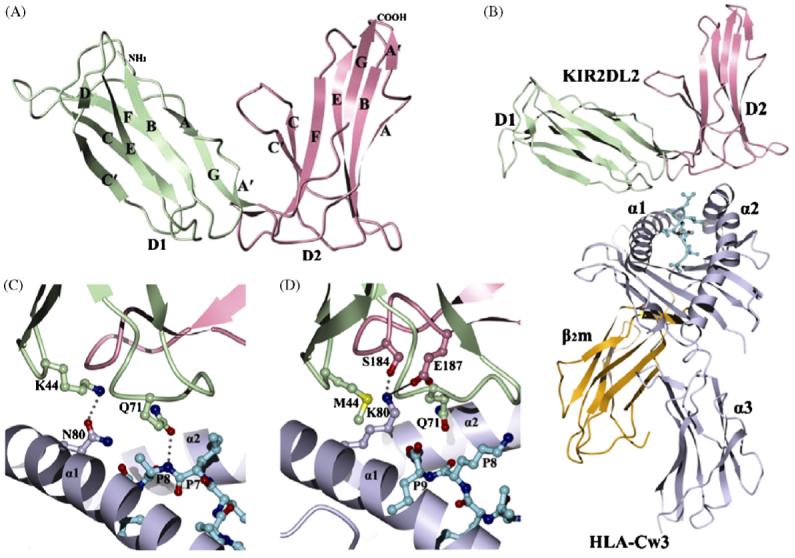
Three-dimensional structures of KIR2DL and KIR2DL/HLA-C complexes. (A) Ribbons diagram of KIR2DL1 (PDB accession code 1NKR). The D1 domain is green; D2 is pink. The secondary structural elements are labeled. (B) Ribbons diagram of KIR2DL2 bound to HLA-Cw3 (PDB accession code 1EFX). The α1, α2 and α3 domains of the HLA-Cw3 heavy chain are blue-grey; β2m is gold; the peptide in ball-and-stick representation is cyan. (C) Cartoon drawing illustrating the allelic specificity and peptide selectivity of KIR2D receptors. The dotted lines represent hydrogen bonds formed by Asn80 of HLA-Cw3 with Lys44 of KIR2DL2, and by Gln71 of HLA-Cw3 with P8 of the peptide. (D) Interactions of Lys80 of HLA-Cw4 (blue-grey) with specificity-determining residues of KIR2DL1 (D1 domain in green, D2 domain in pink) in the KIR2DL1/HLA-Cw4 complex (PDB accession code 1IM9). The solid line represents a salt bridge.
In the crystal structures of the KIR2DL2/HLA-Cw3 [14] and KIR2DL1/HLA-Cw4 [15] complexes, the KIRs bind their corresponding HLA-C ligands through the α1 and α2 helices and the C-terminal portion of the peptide (Fig. 1b). In both complexes, the D1D2 axis is approximately orthogonal to the axis of the bound peptide and the HLA-C α1 helix, aligning D1 with the α1 helix and D2 with α2. This orientation resembles the docking mode of T cell receptors (TCRs) onto MHC, but is completely distinct from those of LIR-1 or Ly49 NK receptors (see below). Each KIR contacts HLA-C through six loops in its interdomain hinge region: loops A’B, CC’, and EF of D1 contact the HLA-C α1 helix, while the hinge and loops BC and FG of D2 contact α2. Surprisingly, a comparison of the KIR2DL2/HLA-Cw3 and KIR2DL1/HLA-Cw4 complexes revealed that many conserved residues in KIR2 and HLA-C mediate different interactions in the two structures, apparently due to side chain rearrangements [15]. Both interfaces are highly charged, and comprise a number of salt bridges that are critical for binding.
The allelic specificity of KIR2DLs is readily explained by the structures of the KIR2DL2/HLA-Cw3 [14] and KIR2DL1/HLA-Cw4 [15] complexes. Thus, 11 of 12 HLA-Cw3 residues in contact with KIR2DL2 are invariant in all HLA-C alleles, including HLA-Cw4. The sole exception is Asn80, which site-directed mutagenesis has shown to be essential in defining allelic specificity. Of the 16 KIR2DL2 residues that contact HLA-Cw3, only those at positions 44 and 70 differ in KIR2DL1. In the KIR2DL2/HLA-Cw3 complex, Lys44 of KIR2DL2 makes a hydrogen bond with Asn80 of HLA-Cw3 (Fig. 1c). This hydrogen bond could not form if Lys44 were replaced by methionine, as in KIR2DL1, or if Asn80 were replaced by lysine, as in HLA-Cw2, 4, 5, 6 or 15. By contrast, KIR2DL1 uses a different mechanism to achieve allelic specificity. In the KIR2DL1/HLA-Cw4 complex, the side chain of HLA-Cw4 residue Lys80 is situated in a negatively charge pocket formed by a channel between D1 and D2 of KIR2DL1. This pocket includes the polymorphic Met44 residue of the KIR that forms both hydrophobic and polar interactions with Lys80 of HLA-Cw4 (Fig. 1d). In addition, Lys80 makes a hydrogen bond with Ser184 and a salt bridge with Glu187. Replacement of Met44 by lysine, as in KIR2DL2, would result in charge repulsion and steric hindrance with Lys80 of HLA-Cw4, leading to loss of binding. Conversely, substitution of Lys80 of HLA-Cw4 with asparagine, as in HLA-Cw3 and related alleles, would cause a loss of both the hydrogen bond and salt bridge, also resulting in loss of affinity.
Peptide preferences have been documented in KIR2D binding to HLA-C molecules [18,19], although it is unknown whether peptide selectivity has a role in NK receptor function (see, however, the discussion of Ly49 receptors). The KIR/HLA interaction is most sensitive to substitutions at peptide positions 7 and 8, in agreement with the finding that the KIR binding site is centered near C-terminal residues 7 and 8 of the peptide. By comparison, TCRs, which exhibit much greater peptide selectivity than KIRs, generally focus on the central portion of the MHC-bound peptide, at and around the P5 position [20]. In the KIR2DL1/HLA-Cw4 complex, the surface of KIR2DL1 around P8 Lys of the peptide is electronegative overall, such that substitution of P8 Lys with negatively charged residues abolishes binding [15]. In the corresponding region of the KIR2DL2/HLA-Cw3 complex, Gln71 of KIR2DL2 is hydrogen bonded to the main-chain nitrogen atom of P8 Ala [14]. This hydrogen bond may restrict the size of the side chain that can be accommodated, consistent with the observed preference for alanine or serine at the P8 position.
3. MHC class I recognition by LIRs
The LIR family of immunoreceptors (also known as immunoglobulin-like transcripts, or ILTs) is widely distributed on monocytes, B cells, dendritic cells, and some NK and T cells [21,22]. Like KIRs, LIR proteins contain various numbers of extracellular Ig-like domains: LIR-1, -2, -3, -4, -6a, -7 and -8 have four domains, whereas LIR-5 and -6b have only two. LIR-1, the most broadly expressed LIR family member, is an inhibitory receptor that binds multiple MHC class I molecules, both classical (HLA-A, -B and -C) and non-classical (HLA-E, -F and -G), with similar affinities and kinetics [23,24]. Individual KIRs, by contrast, exhibit allelic specificity, as discussed above. In addition, LIR-1 is a receptor for UL18, an MHC class I homolog encoded by human cytomegalovirus [25].
The crystal structure of LIR-1 (domains D1 and D2 only) has been determined in free form [26], and bound to HLA-A2 [27]. The two tandem Ig domains of LIR-1 form a bent structure with an acute interdomain angle (Fig. 2), similar to KIR2D (Fig. 1a). Each domain is composed of two anti-parallel β-sheets arranged in a KIR-like topology. Unlike KIRD2, however, LIR-1 includes several 310 helical regions interspersed in the mainly β structure. In the LIR-1/HLA-A2 complex [27], LIR-1 D1D2 binds the side of HLA-A2, forming two contact areas that comprise residues from the HLA-A2 α3 domain, which is polymorphic, and β2m, which is invariant. The tip of LIR-1 D1 contacts the HLA-A3 α3 domain, and the D1-D2 interdomain hinge region contacts (β2-microglobulin) (β2m) (Fig. 2). This docking mode, which differs completely from that of KIRs (Fig. 1b), is consistent with recognition by LIR-1 of a broad array of MHC class I molecules in a peptide-independent manner [21,22].
Fig. 2.

Ribbons diagram showing the crystal structure of LIR-1 bound to HLA-A2 (PDB accession code 1P7Q). Domains are labeled. The α1, α2 and α3 domains of the HLA-A2 heavy chain are cyan; β2m is blue; the peptide in ball-and-stick representation is grey. The D1 and D2 domains of LIR-1 are colored in orange and rose, respectively.
Of the LIR-1 binding surface buried in the complex, ∼70% is contributed by the interface with β2m, which helps explain the broad recognition properties of LIR-1. Concerning residues of the HLA-A2 α3 domain that contact LIR-1, most are conserved in MHC class I ligands, both classical and non-classical. On the receptor side of the interface, comparison of LIR-1 residues that interact with HLA-A2 with corresponding residues in other LIRs appears to divide LIR family members into two groups [27]. The first group comprises members showing ∼70% conservation of these residues, and includes the inhibitory receptor LIR-2, the soluble receptor LIR-4, and the activating receptors LIR-6a, -6b and -7. Ligands for this group are likely to resemble those for LIR-1. The second group, with only ∼30% conservation of HLA-A2-contacting residues, includes LIR-3, -5 and -8. These inhibitory receptors may engage a set of ligands different from LIR-1, possibly via another binding mode.
4. MHC class I recognition by Ly49 receptors
Ly49 receptors, of which there are at least 23 members (Ly49A-W), constitute the main MHC class I-monitoring receptor family on mouse NK cell [2,28]. Although most Ly49s inhibit NK cell-mediated cytolysis after binding to MHC class I ligands, some are activating. Individual Ly49s recognize multiple, but not all, H-2D and H-2K alleles [28]. In most cases, recognition appears to be independent of the bound peptide. Notable exceptions are Ly49C and Ly49I, which exhibit a degree of peptide specificity in their recognition of H-2Kb and H-2Kd, respectively [29,30]. Recently, crucial roles for Ly49 receptors in antiviral immunity have been discovered. In one case, the m157 gene product of MCMV was shown to interact directly with an inhibitory Ly49 (Ly49I) in a susceptible mouse strain and with an activating Ly49 (Ly49H) in a resistant one [7,8]. In another case, the activating receptor Ly49P was found to confer resistance to MCMV by interacting with H-2Dk on MCMV-infected cells [9].
Ly49 receptors belong to the C-type lectin-like family of proteins, which also includes CD94/NKG2 and NKG2D NK receptors [5]. Like NKG2D, Ly49 receptors are homodimeric type II glycoproteins, with each chain composed of a C-type lectin-like domain (CTLD) connected by a stalk to the transmembrane and cytoplasmic domains. Crystal structures have been determined for Ly49A bound to H-2Dd [31], Ly49C bound to H-2Kb [32], Ly49I in free form [33], and Ly49C in free form (Deng L, Dam J, Schuck P, Mariuzza RA, unpublished data). The Ly49 monomer consists of two α-helices (α1 and α2), and two anti-parallel β-sheets formed by β-strands β0, β1, β5; β2, β2′, β3 and β4 (Fig. 3a). To form the Ly49 homodimer, the monomers associate through strand β0, creating an extended anti-parallel β-sheet. In Ly49A, the C-terminal ends of the α2 helices pack against one another, creating a ‘closed’ dimer (Fig. 3b). In Ly49C and Ly49I, however, the α2 helices are not juxtaposed in the interface, opening these dimers by ∼20° compared with Ly49A (Fig. 3c). An important consequence of this variability in Ly49 dimerization geometry is to modulate the way these NK receptors bind MHC, as revealed by the Ly49A/H-2Dd and Ly49C/H-2Kb structures [31,32].
Fig. 3.
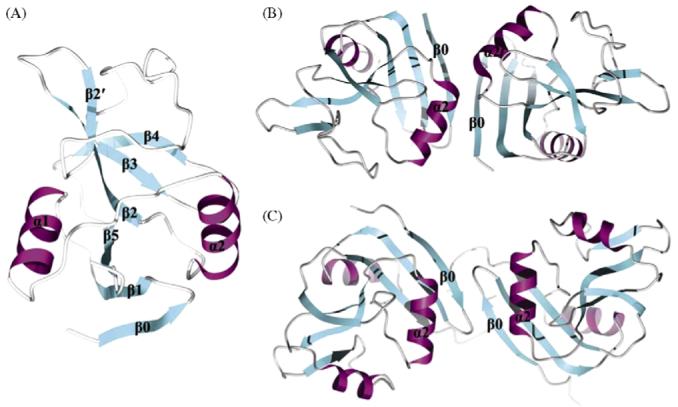
Crystal structures of Ly49 NK receptors. (A) Ribbons drawing of the Ly49A C-type lectin-like domain (PDB accession code 1QO3). Secondary structure elements are labeled. β-strands are shown in cyan, α-helices in magenta, and loops in grey. (B) Structure of the Ly49A homodimer. Secondary structure elements that participate in formation of the dimer interface are labeled. (C) Structure of the Ly49C homodimer (Deng L, Dam J, Schuck P, Mariuzza RA, unpublished data).
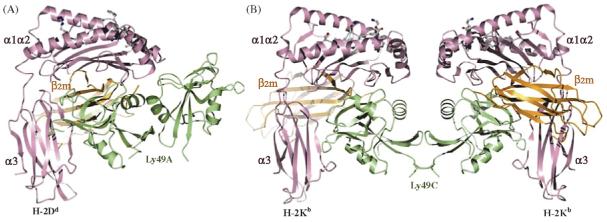
In the Ly49A/H-2Dd complex (Fig. 4a), the Ly49A homodimer engages H-2Dd asymmetrically at a broad cavity under-neath the peptide-binding platform of the MHC in a region that partially overlaps the LIR-1 binding site (Fig. 2). Here the Ly49A dimer makes contacts, through one of its subunits, with the α1/α2, α3 and β2m domains of H-2Dd. In the crystal, the second Ly49A subunit was observed to contact a neighboring H-2Dd molecule at a different site, at one end of the peptide-binding platform (not shown) [31]. However, subsequent mutagenesis studies of both Ly49A and H-2Dd unambiguously identified the region beneath the peptide-binding platform as the functional binding site for Ly49A on MHC that leads to inhibition of NK cell cytotoxicity [34-36]. By contrast, in the Ly49C/H-2Kb complex [32], the Ly49C dimer engages H-2Kb bivalently, such that each subunit makes identical interactions with MHC class I at a site overlapping the Ly49A binding site on H-2Dd, to form a symmetrical, butterfly-shaped assembly (Fig. 4b). In this view, the complex is oriented as if the two H-2Kb molecules stand on the target cell at the bottom and the Ly49C homodimer reaches H-2Kb from an opposing NK cell above, to which it is tethered through stalk regions projecting down to the N-termini of the subunits.
Fig. 4.
Structures of Ly49/MHC class I complexes. Domains are labeled. The α1, α2 and α3 domains of the MHC class I heavy chain are rose; β2m is orange; the peptide in ball-and-stick representation is grey; the Ly49 molecules are green. (A) Ribbon diagram of Ly49A bound to H-2Dd (PDB accession code 1QO3). (B) Structure of Ly49C in complex with H-2Kb (Deng L, Dam J, Schuck P, Mariuzza RA, unpublished data).
The structural basis for the very different modes of MHC engagement seen in the Ly49A/H-2Dd and Ly49C/H-2Kb complexes is the different dispositions of the Ly49A and Ly49C dimers. Major steric clashes between MHC molecules would preclude the closed Ly49A dimer from simultaneously binding two MHC in the manner of the open Ly49C dimer. This would require a conformational switch from closed to open forms. In this regard, we recently found that the Ly49A dimer can adopt the open conformation in solution (Baber J, Grishaev A, Dam J, Mariuzza RA, Bax A, unpublished data). Thus, it appears that Ly49s may exist on the cell surface in dynamic equilibrium between a closed form, which only allows engagement of one MHC, and an open form, which permits bivalent binding. Ly49-mediated MHC cross-linking may serve to stabilize receptor-ligand interactions at the NK cell-target cell interface, facilitating signal transmission to the NK cell.
Like KIRs, Ly49 receptors exhibit MHC allelic specificity. For example, although Ly49A and Ly49C both recognize H-2Dd and H-2Dk, the more promiscuous Ly49C also binds H-2Kb, H-2Kd and H-2Db [28,30]. The Ly49A/H-2Dd and Ly49C/H-2Kb structures indicate that Ly49 receptors have evolved a two-tiered strategy for recognizing MHC. Primary recognition is mediated by a small number of conserved residues in nearly superimposable regions of the MHC-binding site that together contribute most of the binding free energy. Overlayed onto these primary interactions are secondary ones involving more variable portions of the binding site that confer different MHC specificities. Most importantly, the contacting element formed by residues 218-231, which display high sequence variability across the Ly49 family, adopts markedly different conformations in Ly49A (Fig. 5a) and Ly49C (Fig. 5b). The result is completely different sets of interactions with the MHC heavy chain at this region of the interface that mediate allelic specificity.
Fig. 5.
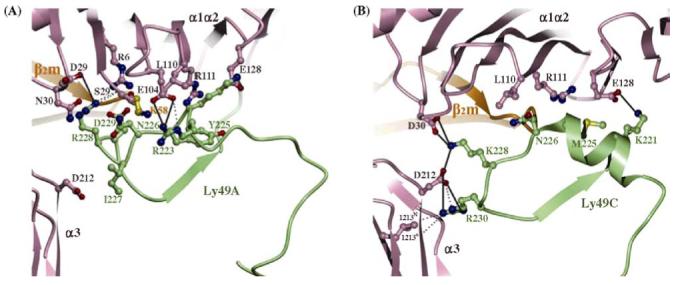
Close-up view of Ly49/MHC class I interfaces. Domains are labeled. The α1, α2 and α3 domains of the MHC class I heavy chain are rose; the β2m is orange; the Ly49 molecules are green. Salt bridges and hydrogen bonds are represented by solid and dotted lines, respectively. (A) The Ly49A/H-2Dd interface, highlighting interactions made by residue 211-231 of Ly49A. (B) The Ly49C/H-2Kb interface, showing interactions made by the corresponding region of Ly49C (Deng L, Dam J, Schuck P, Mariuzza RA, unpublished data).
The total absence of direct contacts between Ly49A and the MHC-bound peptide in the Ly49A/H-2Dd structure (Fig. 4a) explains why MHC recognition by Ly49A is independent of peptide sequence [31]. However, this same lack of contacts between Ly49C and peptide in the Ly49C/H-2Kb complex (Fig. 4b) precludes a simple explanation for the remarkable peptide selectivity of Ly49C in both cellular and binding assays [29,32]. One possibility is that peptides indirectly modulate Ly49C binding by influencing the conformation and/or mobility of Ly49C-contacting residues. Alternatively, because binding of Ly49C to H-2Kb requires the α1/α2, α3 and β2m domains to adopt a relative orientation that differs considerably from that found in free H-2Kb structures, certain peptides could affect receptor binding by altering the energetic cost of distorting H-2Kb from its ground state conformation to the Ly49C-bound quaternary structure.
The biological role, if any, of the peptide selectivity of certain Ly49s (and KIRs) is unknown. However, an intriguing possibility is suggested by the recent discovery of a functional interaction between Ly49P and H-2Dk on MCMV-infected cells that confers resistance to MCMV [9]. Although the molecular nature of this interaction has not been defined, one possibility is peptide-dependent recognition of MCMV by Ly49P. If so, peptide selectivity could confer on NK cells a degree of specificity for viral pathogens previously thought to be exclusive to cytotoxic T cells.
5. Ligand recognition by NKG2D
The homodimeric C-type lectin-like NK receptor NKG2D binds multiple protein ligands that are distant structural homologs of MHC class I, including MICA, MICB, ULBP3, and RAE-1β [6,36]. However, unlike true MHC class I molecules, NKG2D ligands bind neither peptides (or other small molecules) nor β2m, and ULBP3 and RAE-1β even lack the heavy chain α3 domain, existing on the cell surface as glycophosphatidylinositol-linked α1/α2 platform domains. In humans, MICA and MICB are minimally expressed in normal tissues, but are upregulated in stressed cells and epithelial tumors [37,38]. Rodent homologs of MICA and MICB have not been identified, but other molecules with weak homology to MHC class I, notably RAE-I and H-60, serve as ligands for mouse NKG2D [39,40]. Like MICA and MICB, RAE-1 and H-60 are frequently expressed on tumor but not normal cells, suggesting that NKG2D functions as a receptor for tumor surveillance by NK cells. In addition to MICA and MICB, a group of proteins designated ULBPs, which bind CMV glycoprotein UL16, have been identified as ligands for human NKG2D [41]. UL16 is believed to act as a decoy receptor for NKG2D ligands, facilitating viral evasion of the immune system. The interaction of mouse NKG2D with RAE-1, H-60 and MULT-1 is disrupted by the HCMV-encoded MHC class I homologs m152, m155 and m145. By preventing NK cell activation, this disruption promotes viral survival [42-44]. Crystal structures have been reported for mouse and human NKG2D in free form [36,45], for mouse NKG2D in complex with RAE-1β [46], and for human NKG2D bound to MICA and ULBP3 [47,48].
MICA is composed of two structural domains, an α1/α2 platform domain and a C-type Ig-like α3 domain (Fig. 6a). The α1/α2 platform domain contains the α1 and α2 helices that define the peptide-binding groove in classical MHC class I molecules. In the NKG2D/MICA complex [47], the NKG2D homodimer binds orthogonally to the axes of the α1 and α2 helices of the MICA platform in a manner resembling the docking mode of TCR onto MHC class I, but distinct from that of Ly49C, each of whose subunits comprises an independent binding site for MHC (Fig. 4b). This recognition of an asymmetric ligand by a symmetric receptor is mediated by very similar surfaces on the NKG2D monomers that interact with two distinct surfaces on MICA. Specifically, 7 of the 11 contact residues contributed by each NKG2D subunit are common to both MICA binding surfaces, although six are engaged in distinct interactions at the two interfaces. In contrast to the KIR2D/HLA-C and Ly49/MHC interfaces, which are highly hydrophilic and dominated by polar and charged interactions, the NKG2D/MICA interface comprises a mixture of hydrophobic, polar, and charged interactions more typical of protein-protein complexes. In the crystal structure of unbound MICA [49], 10 residues in the central portion of the α2 helix were found to be disordered and presumed to constitute a flexible loop. Upon binding NKG2D, however, these residues become ordered into two turns of helix, restoring the canonical α2 helix of classical MHC class I molecules.
Fig. 6.
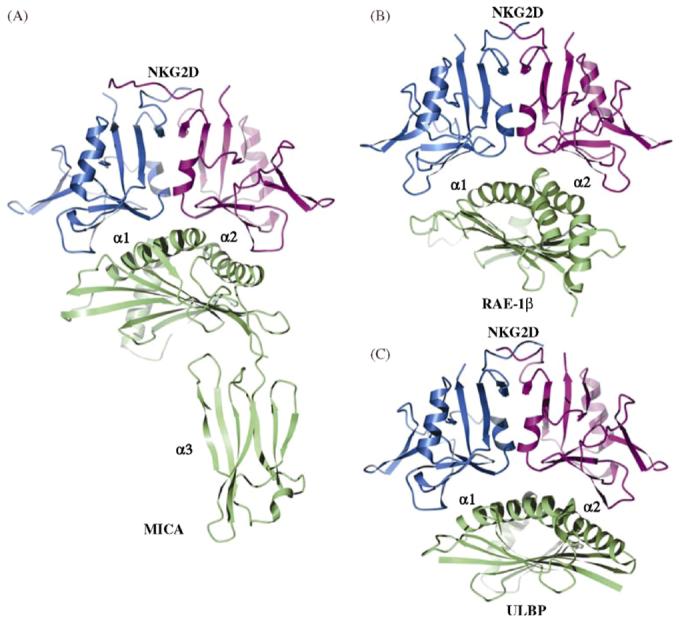
Ribbon diagram showing crystal structures of NKG2D bound to MHC class I homologs. Domains are labeled. The two subunits of NKG2D homodimer are colored in blue and magenta. MICA, RAE-1β and ULBP3 are all in green. (A) The human NKG2D/MICA complex (PDB accession code 1HYR). (B) The mouse NKG2D/RAE-1β complex (1JSK). (C) The human NKG2D/ULBP3 complex (1KCG).
As in the NKG2D/MICA complex, the NKG2D homodimer binds ULBP3 and RAE-1β orthogonally to the α1/α2 platform domain of these other ligands (Fig. 6b and c) [46,48]. However, the binding interfaces exhibit significant differences between complexes. Upon superposition of the three complexes through their common NKG2D element, there is an approximately 10° difference in orientation between MICA and RAE-1β, 5° between MICA and ULBP3, and 10° between ULBP3 and RAE-1β. The interfaces contain 7-13 hydrogen bonds and 1-3 salt bridges, of which only two hydrogen bonds and one salt bridge are conserved in all three complexes. However, a shared subset of NKG2D residues, forming two large patches on the receptor surface, contributes to hydrophobic interactions with each of the ligands.
Taken together, these results demonstrate that the single NKG2D binding site has evolved to recognize six different surfaces on the α1/α2 domains of MICA, RAE-1β, and ULBP3, which are distantly related in terms of both sequence identity (∼25%) and detailed structure. Two mechanisms may be considered to explain multispecific ligand recognition by the NKG2D receptor [46,48]. NKG2D may possess a degree of conformational flexibility that allows a single receptor to recon-figure its binding site to accommodate diverse ligands (induced fit). Alternatively, an essentially rigid binding site on NKG2D may make different interactions with structurally distinct ligand surfaces, without significant conformational changes in the receptor (rigid adaptation). To distinguish between these possibilities, the energetic contributions made by different NKG2D residues to binding MICA, RAE-1β, and ULBP3 was evaluated by in silico site-directed mutagenesis [36]. This analysis suggested that the binding free energy is unevenly distributed across the NKG2D/ligand interfaces, resulting in obvious hot spots and the energetic dominance of one of the NKG2D subunits. Importantly, the hot spots were associated with structurally conserved receptor elements that interact with relatively conserved residues of the ligands, which argues in favor of a rigid adaptation recognition mechanism. Arguing against induced fit is the absence of large conformational changes in NKG2D on ligand binding, based on comparisons between free and bound forms of the receptor [36]. In addition, experimentally measured heat capacity changes (ΔCp) for the binding of NKG2D to MICA, RAE-1β, and ULBP3 closely matched values calculated using empirical relationships between ΔCp and buried surface area that assume no significant conformational changes in the interacting species on complex formation [50]. Thus, multispecific ligand recognition by NKG2D is explained by rigid adaptation, rather than induced fit.
6. Perspective
Structural studies of NK receptors and their complexes with MHC class I or MHC class I-like ligands have provided insights into how cells of the innate immune system discriminate between normal and tumor or virus-infected cells of the host. They have revealed that NK receptors have developed multiple solutions for recognizing the MHC class I scaffold, involving interactions with several distinct faces of this molecule. Future studies will be directed at understanding how NK receptors bind viral homologs of MHC class I molecules, such as m157 [7,8] and m144 [51], as well as establishing the structural basis for other mechanisms of NK cell-mediated resistance to viral infections [9].
Acknowledgement
This work was supported by National Institutes of Health grant AI47990 (to RAM).
References
- [1].McQueen KL, Parham P. Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol. 2002;14:615–21. doi: 10.1016/s0952-7915(02)00380-1. [DOI] [PubMed] [Google Scholar]
- [2].Plougastel BFM, Yokoyama WM. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–16. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- [3].Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defense strategy. Nature. 1986;319:675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- [4].Ljunggren HG, Käarre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- [5].Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–85. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- [6].Vivier E, Tomasello E, Paul P. Lymphocyte activation via NKG2D: towards a new paradigm in immune recognition? Curr Opin Immunol. 2002;14:306–11. doi: 10.1016/s0952-7915(02)00337-0. [DOI] [PubMed] [Google Scholar]
- [7].Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- [8].Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–31. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Derosiers M-P, Kielczewska A, Loredi-Osti J-C, Adam SG, Makrigiannis AP, Lemiex S, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–9. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fan QR, Mosyak L, Winter CC, Wagtmann N, Long EO, Wiley DC. Structure of the inhibitory receptor for human natural killer cells resembles hematopoietic receptors. Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- [11].Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci USA. 1999;96:3864–9. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure. 1999;7:391–8. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- [13].Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) J Exp Med. 2003;197:933–8. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–43. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- [15].Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–60. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- [16].de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–12. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- [17].Somers W, Ultsch M, De Vos AM, Kossiakoff AA. The X-ray structure of a growth hormone-prolactin receptor complex. Nature. 1994;372:478–81. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- [18].Rajakopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523–8. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci USA. 1997;94:6313–8. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rudolph MG, Luz JG, Wilson IA. Structural and thermodynamic correlates of T cell signaling. Annu Rev Biophys Biomol Struct. 2002;31:121–49. doi: 10.1146/annurev.biophys.31.082901.134423. [DOI] [PubMed] [Google Scholar]
- [21].Borges L, Cosman D. LIRs/ILTs/MIRs, inhibitory and stimulatory Ig-superfamily receptors expressed in myeloid and lymphoid cells. Cytokine Growth Factor Rev. 2000;11:209–17. doi: 10.1016/s1359-6101(00)00007-1. [DOI] [PubMed] [Google Scholar]
- [22].Cella M, Nakajima H, Facchetti F, Hoffmann T, Colonna M. ILT receptors at the interface between lymphoid and myeloid cells. Curr Top Microbiol Immunol. 2000;251:161–6. doi: 10.1007/978-3-642-57276-0_20. [DOI] [PubMed] [Google Scholar]
- [23].Chapman TL, Haikema AP, Bjorkmann PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral MHC homolog UL18. Immunity. 1999;11:603–11. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- [24].Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human inhibitory receptors ILT2 and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–61. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–82. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- [26].Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT-2) Immunity. 2000;13:727–36. doi: 10.1016/s1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- [27].Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–9. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- [28].Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- [29].Franksson L, Sundbäck J, Achour A, Bernlind J, Glas R, Kärre K. Peptide dependency and selectivity of the NK inhibitory receptor Ly49C. Eur J Immunol. 1999;29:2748–58. doi: 10.1002/(SICI)1521-4141(199909)29:09<2748::AID-IMMU2748>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [30].Hanke T, Takizawa H, McMahon CW, Busch DH, Pamer EG, Miller JD, et al. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity. 1999;11:67–77. doi: 10.1016/s1074-7613(00)80082-5. [DOI] [PubMed] [Google Scholar]
- [31].Tormo J, Natarajan K, Margulies DH, Mariuzza RA. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 1999;402:623–31. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- [32].Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, et al. Variable MHC class I engagement by Ly49 NK receptors revealed by the crystal structure of Ly49C bound to H-2Kb. Nat Immunol. 2003;4:1213–22. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- [33].Dimasi N, Sawicki MW, Reineck LA, Li Y, Natarajan K, Margulies DH, et al. Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family. J Mol Biol. 2002;320:573–85. doi: 10.1016/s0022-2836(02)00498-9. [DOI] [PubMed] [Google Scholar]
- [34].Matsumoto N, Mitsuki M, Tajima K, Yokoyama WM, Yamamoto K. The functional binding site for the C-type lectin-like natural killer cell receptor Ly49A spans three domains of its major histocompatibility complex class I ligand. J Exp Med. 2001;193:147–58. doi: 10.1084/jem.193.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang J, Whitman MC, Natarajan K, Tormo J, Mariuzza RA, Margulies DH. Binding of the NK inhibitory receptor Ly49A to its MHC-I ligand: crucial contacts include both H-2Dd and β2-microglobulin. J Biol Chem. 2002;277:1433–42. doi: 10.1074/jbc.M110316200. [DOI] [PubMed] [Google Scholar]
- [36].McFarland BJ, Kortemme T, Yu SF, Baker D, Strong RK. Symmetry recognizing asymmetry: analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure. 2003;11:411–22. doi: 10.1016/s0969-2126(03)00047-9. [DOI] [PubMed] [Google Scholar]
- [37].Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science. 1998;279:1737–40. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- [38].Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–84. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–7. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- [40].Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- [41].Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–33. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- [42].Lodoen M, Ogasawara K, Hamerman JA, Arase A, Houchins JP, Mocarski ES, et al. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–53. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med. 2004;200:1075–81. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–20. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wolan DW, Teyton L, Rudolph MG, Villmow B, Bauer S, Busch DH, et al. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 Å. Nat Immunol. 2001;2:248–54. doi: 10.1038/85311. [DOI] [PubMed] [Google Scholar]
- [46].Li P, McDermott G, Strong RK. Crystal structure of RAE-1β and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16:77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- [47].Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–51. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- [48].Radaev S, Rostro B, Brooks AG, Colonna M, Sun P. Conformational plasticity revealed by the co-crystal structure of the activating NK receptor NKG2D and its MHC-like ligand ULBP. Immunity. 2001;15:1039–49. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- [49].Li P, Willie ST, Bauer S, Morris DL, Spies T, Strong RK. Crystal structure of the MHC class I homolog MICA, a γδ T cell ligand. Immunity. 1999;10:577–84. doi: 10.1016/s1074-7613(00)80057-6. [DOI] [PubMed] [Google Scholar]
- [50].McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: not induced fit but rigid adaptation. Immunity. 2003;19:803–12. doi: 10.1016/s1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- [51].Natarajan K, Hicks A, Mans J, Robinson H, Guan R, Mariuzza RA, et al. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J Mol Biol. 2006;358:157–71. doi: 10.1016/j.jmb.2006.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]