Abstract
Glycogen synthase kinase 3 (GSK3) is a unique serine/threonine kinase that is implicated in a variety of cellular processes and is regulated by phosphorylation or protein–protein interaction in animal cells. BIN2 is an Arabidopsis GSK3-like kinase that negatively regulates brassinosteroid (BR) signaling. Genetic studies suggested that BIN2 is inhibited in response to BR perception at the cell surface to relieve its inhibitory effects on downstream targets; however, little is known about biochemical mechanisms of its inhibition. Here, we show that BIN2 is regulated by proteasome-mediated protein degradation. Exogenous application of a BR biosynthesis inhibitor and an active BR increased and decreased the amount of BIN2 proteins, respectively. Interestingly, the gain-of-function bin2-1 mutation significantly stabilizes BIN2, making it unresponsive to BR-induced BIN2 depletion. Exogenous application of different plant growth hormones revealed that BIN2 depletion is specifically induced by BR through a functional BR receptor, while treatment of a proteasome inhibitor, MG132, not only prevented the BR-induced BIN2 depletion but also nullified the inhibitory effect of BR on the BIN2 kinase activity. Taken together, our results strongly suggest that proteasome-mediated protein degradation constitutes an important regulatory mechanism for restricting the BIN2 activity.
INTRODUCTION
Originally identified as a kinase that phosphorylates and inactivates glycogen synthase, glycogen synthase kinase 3 (GSK3) is a highly conserved serine/threonine kinase that is implicated in a wide variety of cellular processes controlling cell proliferation, cell differentiation, cytoskeleton dynamics, and programmed cell death (Frame and Cohen, 2001). In resting cells, GSK3 is a constitutively active kinase that phosphorylates a wide array of protein substrates to directly inhibit their biochemical activities, interfere with their sub-cellular localization, or promote their degradation (Ali et al., 2001). In response to environmental stimuli or developmental cues, GSK3 is inactivated, thus relieving its inhibitory effect on its targets.
BIN2 is one of the 10 GSK3-like kinases in Arabidopsis (Jonak and Hirt, 2002) and is thought to play a negative role in the signal transduction pathway of brassinosteroids (BRs) (Li and Nam, 2002)—a plant-specific class of polyhydroxysteroids critical for plant growth and development (Clouse and Sasse, 1998). Gain-of-function bin2 mutations or overexpression of the wild-type BIN2 gene result in a morphological phenotype resembling that of BR-deficient or BR-signaling mutants (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), whereas a triple mutant lacking BIN2 and its two closest homologs is morphologically similar to mutants with a constitutively active BR signaling pathway or transgenic plants that overexpress the BR receptor BRI1 or a rate-limiting BR biosynthetic enzyme (Vert and Chory, 2006). Genetic and biochemical studies suggested that BIN2 is a constitutively active kinase that phosphorylates BES1 and BZR1—two closely related members of a plant-specific transcriptional factor family (He et al., 2005; Yin et al., 2005)—to promote their protein degradation (He et al., 2002; Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002), to affect their nuclear localization (Gampala et al., 2007; Ryu et al., 2007), and/or to inhibit their DNA-binding activities (Vert and Chory, 2006), thus blocking the transduction of the plant steroid signal into the nucleus. BR binding to its receptor BRI1 leads to dimerization and activation of BRI1 and its presumed co-receptor, BAK1 (Li et al., 2002; Nam and Li, 2002). As a result, BIN2 is inhibited while BES1 and BZR1 become dephosphorylated and accumulate in the nucleus, where the two transcription factors bind to DNA to regulate expression of many known BR-responsive genes (Li and Jin, 2007).
Extensive biochemical studies have revealed that animal GSK3 kinases are generally regulated by two different mechanisms, one involving phosphorylation of a conserved N-terminal serine residue and the other through sequestration of GSK3 by protein–protein interaction (Jope and Johnson, 2004). For instance, in the insulin-signaling pathway, insulin activates a serine/threonine kinase PKB/Akt, which phosphorylates Ser21 of GSK3α or Ser9 of GSK3β, leading to GSK3 auto-inhibition and activation of glycogen synthase (Cross et al., 1995; Frame et al., 2001). In the canonical Wnt pathway, the binding of Wnts to their receptor Frizzled results in GBP/Frat binding to GSK3, leading to disruption of the APC–Axin–GSK3–β-catenin scaffolding complex and accumulation of the non-phosphorylated β-catenin in the nucleus (Cadigan and Liu, 2006). Recent studies suggested that other mechanisms, such as proteolytic truncation (Goni-Oliver et al., 2007), ubiquitin/proteasome-mediated protein degradation (Failor et al., 2007), and differential sub-cellular localization (Diehl et al., 1998; Bijur and Jope, 2001; Meares and Jope, 2007), might also be involved in regulating the GSK3 kinase.
By contrast, little is known about the biochemical mechanism of BIN2 inhibition in response to activation of BRI1 and BAK1 at the cell surface. BIN2 lacks the conserved N-terminal serine residue critical for GSK3 regulation in animal cells. Neither BRI1 nor BAK1 was able to interact directly with or phosphorylate BIN2, suggesting that additional BR-signaling proteins are involved to inactivate the Arabidopsis GSK3 kinase (Li, 2005). A recent study revealed that a mutated bin2-1:GFP fusion protein carrying the gain-of-function bin2-1 (E263K) mutation mainly accumulates inside the nucleus while the wild-type BIN2:GFP distributes more or less evenly among the plasma membrane, cytosol, and the nucleus (Vert and Chory, 2006), suggesting that differential sub-cellular localization might be an important mechanism to regulate the BIN2 activity through physical separation of BIN2 from its two nuclear substrates. In this report, we present compelling evidence to show that BIN2 is regulated by a proteasome-mediated protein degradation mechanism to keep the negative regulator at a very low level.
RESULTS
The Mutated bin2-1 Protein is More Stable than its Wild-Type Form
To investigate how BIN2 is regulated by the plant steroid hormone, we translationally fused a green fluorescent protein (GFP) tag with the pea Rubisco E9 terminator to the last codon of a 3.8-kb genomic fragment containing the native promoter and the entire coding region of the Arabidopsis BIN2 gene. The resulting gBIN2:GFP transgene was transformed into wild-type Arabidopsis plants. Consistent with our previous results (Li and Nam, 2002), ~5% of the gBIN2:GFP transgenic plants exhibited bin2-like phenotypes (Figure 1A). We have also introduced the bin2-1 (E263K) mutation via site-directed mutagenesis into the gBIN2:GFP transgene and transformed the mutated gbin2-1:GFP construct into the wild-type Arabidopsis plants. A majority of the resulting gbin2-1:GFP transgenic plants recapitulated the phenotypes of heterozygous and homozygous bin2 mutants (Figure 1B). These experiments revealed that the GFP tag did not affect the BIN2 kinase activity and that the BIN2:GFP fusion protein behaves similarly to the endogenous BIN2 in regulating BR signaling.
Figure 1. bin2-1 is a More Stable Protein than BIN2.
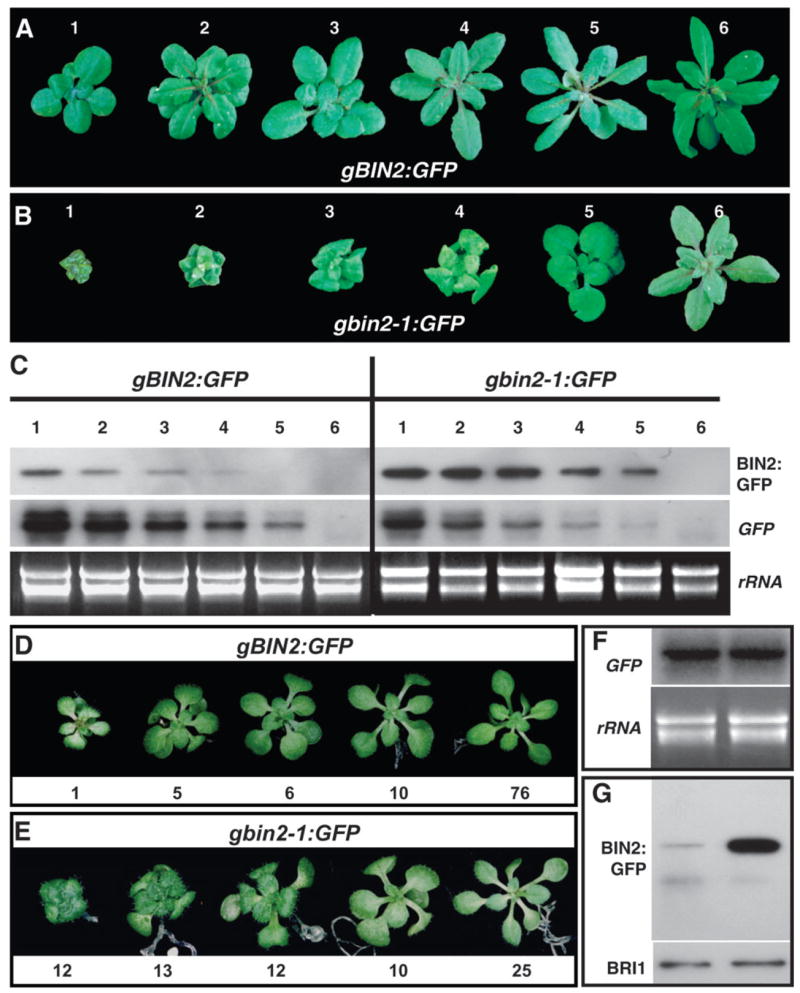
(A) and (B) Six representative transgenic plants expressing the wild-type BIN2:GFP (A) and bin2-1:GFP (B) fusion proteins with varying phenotypes.
(C) Western (upper strip) and Northern blot (middle) analysis of accumulation of the GFP fusion proteins and expression of the transgenes in 12 chosen transgenic lines. GFP fusion proteins were immunoprecipitated by a polyclonal anti-GFP antiserum, separated by SDS-PAGE, and analyzed by immunoblotting with a monoclonal anti-GFP antibody. For the Northern blot analysis, total RNAs were isolated from 4-week-old seedlings, separated by a denaturing agarose gel, transferred to a nylon membrane, and hybridized with a GFP-derived probe (middle strip). The lower strip shows ethidium bromide-staining of rRNAs.
(D) and (E) Grouping of two collections of T1 gBIN2:GFP (D) and gbin2-1:GFP (E) transgenic plants. The numerical values indicate the numbers of transgenic plants exhibiting similar morphologies to the pictured seedlings.
(F) Northern blot analysis of the transgene expression of the two T1 transgenic plant collections. The lower panel shows ethidium bromide-staining of rRNAs.
(G) IP/Western analysis of the GFP fusion proteins in the two T1 collections. The amounts of total protein crude extracts used for the assay were compared by immunoblotting with an anti-BRI1 antibody (lower strip).
We have selected six transgenic lines of varying phenotypes for each construct (Figure 1A and 1B), and examined transgene expression and GFP accumulation by Northern and Western blotting analyses using a GFP probeand anti-GFP antibodies, respectively. Because direct Western blotting failed to detect the presenceofBIN2:GFPorbin2-1:GFPinall tested transgeniclines, we first immunoprecipitated the GFP fusion proteins using a polyclonal GFP antiserum and then analyzed the amount of immunoprecipitated BIN2:GFPorbin2-1:GFP with a monoclonal GFP antibody. As shown in Figure 1C, while the transcript levels of the gBIN2:GFP transgene are comparable to those of the gbin2-1:GFP transgene, the amounts of wild-type BIN2:GFP fusion proteins are significantly lower than those of the mutated bin2-1:GFP proteins. Notably, the two morphologically similar transgenic lines, gBIN2:GFP line 1 and gbin2-1:GFP line 5, accumulated similar levels of GFP fusion proteins; however, the transcript level of the gBIN2:GFP transgene is much higher than that of the gbin2-1:GFP transgene, strongly suggesting that bin2-1 is a much more stable protein than its wild-type form.
To further confirm our observation, we collected 98 T1 gBIN2:GFP transgenic seedlings and 72 T1 gbin2-1:GFP transgenic seedlings (Figure 1D and 1E) and ground the two collections separately into fine powder in liquid nitrogen. We divided each sample into two fractions: one for total RNA isolation and the other for extraction of total proteins. Northern blot analysis using a GFP-derived probe revealed that the two collections of T1 transgenic seedlings accumulated similar amounts of transgene transcripts (Figure 1F), while immunoprecipitation (IP)/Western blotting analysis showed that the gbin2-1:GFP transgenic seedlings accumulated a much higher level of GFP fusion proteins than the combined gBIN2:GFP transgenic plants (Figure 1G). Because the two transgenes differ only by a single-nucleotide change in the coding region, the observed different accumulation of GFP fusion proteins is likely caused by their different protein stability.
The BIN2 Protein Level is Regulated by BR
To directly test whether BIN2 is regulated at the protein level, we selected a gBIN2:GFP line that exhibits a weak bin2 phenotype. Exogenous application of brassinolide (BL)—the most active member of the BR family—was able to suppress its weak dwarf phenotype, whereas a similar BL treatment had little effect on a severe gbin2-1:GFP transgenic line (Figure 2A). This experiment revealed that the BIN2:GFP fusion protein is still responsive to the plant steroid hormone. To test whether the amount of BIN2 is influenced by the level of BRs, we grew the gBIN2:GFP line and a strong gbin2-1:GFP line on synthetic medium containing 2 μM brassinazol (BRZ)—a specific inhibitor of BR biosynthesis (Asami et al., 2000). As shown in Figure 2B, the BRZ treatment enhanced the bin2-like phenotype of the gBIN2:GFP line but had no obvious effect on the gbin2-1:GFP seedlings. Consistent with the morphological change, the BRZ treatment significantly increased the amount of BIN2:GFP but had little effect on the accumulation of bin2-1:GFP.
Figure 2. BRZ Enhances the Protein Stability of BIN2.
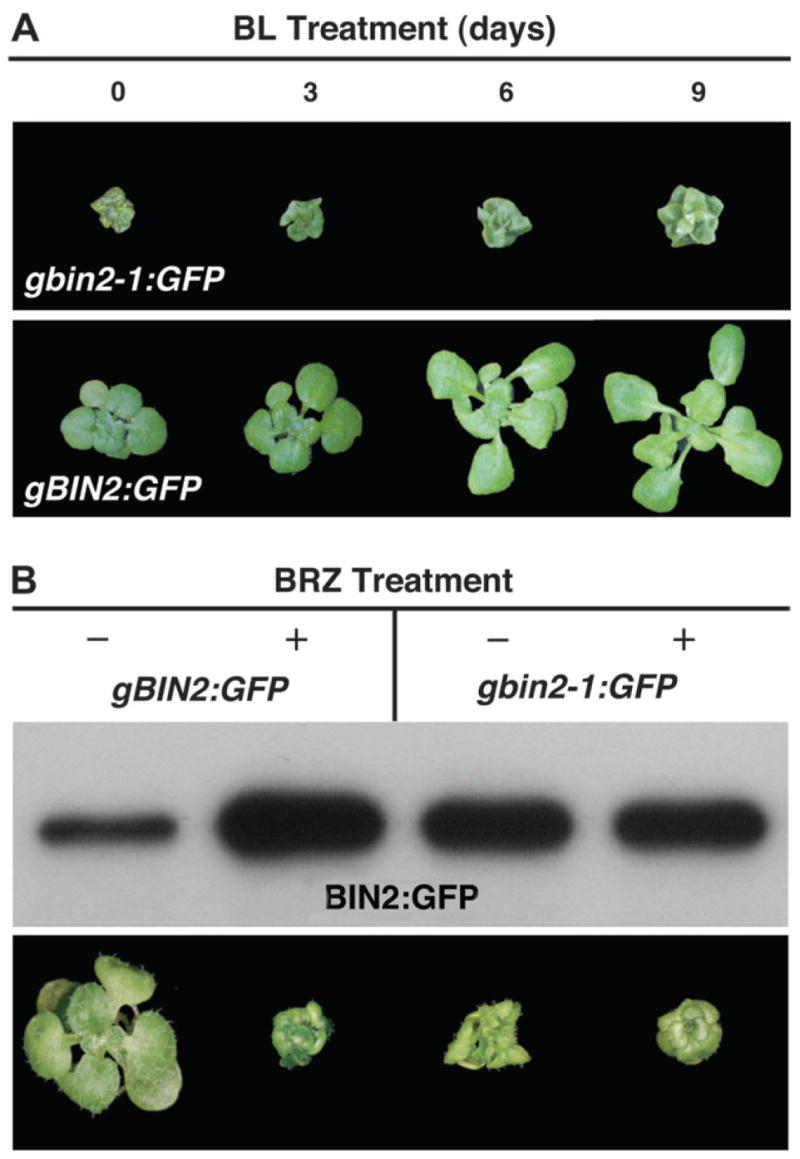
(A) A gBIN2:GFP transgenic line responds to BL treatment whereas a strongly dwarfed gbin2-1:GFP line is BL-insensitive. From left to right are gbin2-1:GFP (top panel) and gBIN2:GFP (lower panel) transgenic seedlings grown on 1 μM BL-containing medium for 0, 3, 6, and 9 d.
(B) BRZ treatment significantly increases the steady-state level of BIN2:GFP but has a little effect on bin2-1:GFP. The top panel is the result of the IP/Western analysis while the lower panel shows the effect of BRZ treatment on the morphology of the selected transgenic lines.
Next, we tested whether the BRZ-stabilized BIN2:GFP protein can be depleted by BL treatment. The BRZ-treated gBIN2:GFP seedlings were transferred to liquid medium containing different concentrations of BL and incubated for different periods of time. As shown in Figure 3A and 3B, the BRZ-stabilized BIN2:GFP is rapidly depleted after 20–30 min treatment with 0.1 or 1 μM BL, whereas similar treatments had little effect on the abundance of the bin2-1:GFP protein (Figure 3C and 3D).
Figure 3. BL Induces Rapid BIN2 Depletion in a Dosage-Dependent Manner.
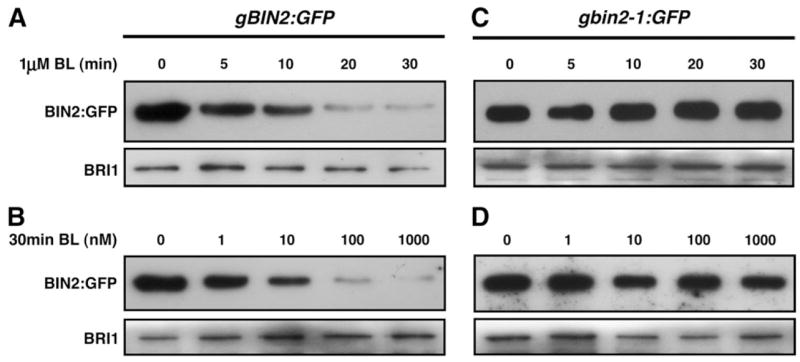
The gBIN2:GFP or gbin2-1:GFP transgenic seedlings were germinated and grown on BRZ-containing medium for 14 d and transferred to liquid medium containing various concentrations of BL. The seedlings were removed at different time points for the IP/Western analysis of BIN2:GFP (upper strip). The relative amounts of total protein extracts of each sample were analyzed by Western blot using the anti-BRI1 antibody (lower strip).
(A) The time course of BL-induced BIN2 depletion.
(B) The dosage-dependency of the BL-induced BIN2 disappearance.
The time-course (C) and dosage-dependency (D) tests of the effect of BL on the accumulation of bin2-1:GFP.
To determine whether other plant hormones could induce a similar depletion of the Arabidopsis GSK3 kinase, we transferred the BRZ-treated gBIN2:GFP seedlings to liquid medium containing various other plant hormones at concentrations known to inhibit root growth of wild-type Arabidopsis seedlings (Li et al., 2001). The treated seedlings were collected after 30 min incubation for extraction of total proteins and the amount of BIN2:GFP fusion proteins was analyzed by the IP/Western blotting method. As shown in Figure 4, none of the hormone treatments resulted in significant depletion of the BRZ-stabilized BIN2:GFP fusion proteins. We thus concluded that the steady-state level of BIN2 is specifically regulated by the plant steroid hormone.
Figure 4. BIN2 Depletion is Specifically Induced by BL.
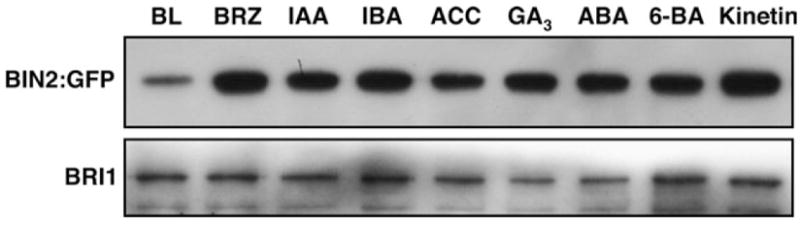
Similar amounts of total protein crude extracts (revealed in the lower strip by immunoblot using the BRI1 antibody) of treated gBIN2:GFP seedlings were used for the IP/Western blot analysis to reveal the effects of various plant hormones on the stability of BIN2:GFP.
The BL-Induced BIN2 Depletion Requires a Functional BRI1 Receptor
To test whether the observed BL-induced BIN2 depletion is mediated by the Arabidopsis BR receptor BRI1, we crossed the gBIN2:GFP transgene into bri1-9—a previously characterized weak bri1 mutant that is caused by ER retention of a structurally defective but biochemically competent BRI1 (Jin et al., 2007). The resulting gBIN2:GFPbri1-9 transgenic seedlings were grown side by side with the parental gBIN2:GFP seedlings on synthetic medium containing 2 μM BRZ for 2 weeks. The seedlings were then transferred to liquid medium containing 1 μM BL and collected after 30 min incubation for the IP/Western blotting analysis. As shown in Figure 5, the amount of BIN2:GFP in BRZ-treated gBIN2:GFP bri1-9 seedlings is similar to that of the BRZ-treated gBIN2:GFP plants and that further treatment of 1 μM BL did not lead to BIN2:GFP depletion, indicating that a functional BR receptor is required for the BL-induced BIN2 depletion.
Figure 5. The BL-Induced BIN2 Depletion Requires a Functional BR Receptor.
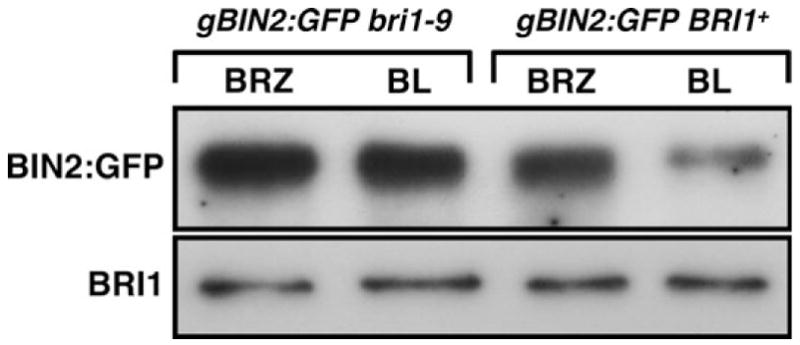
The steady-state levels of BIN2:GFP after treatment of BRZ and BL were analyzed by the IP/Western assay and the relative amounts of total protein extracts were estimated by immunoblotting with the anti-BRI1 antibody.
The BL-Induced BIN2 Depletion Involves Proteasome-Mediated Protein Degradation
Proteasome-mediated protein degradation is an important regulatory mechanism for a wide variety of plant signaling pathways (Smalle and Vierstra, 2004), including the BR-signaling process (He et al., 2002; Yin et al., 2002). To directly test whether the BL-induced BIN2 depletion is caused by proteasome-mediated protein degradation, we first grew gBIN2:GFP transgenic seedlings on BRZ-containing medium for 14 days and then transferred them to liquid medium containing 1 μM BL and/or 10 μM MG132—a well known proteasome inhibitor (Rock et al., 1994). The seedlings were collected after 30 min incubation for the IP/Western blotting analysis. As shown in Figure 6, MG132 treatment effectively blocked the BL-induced depletion of BIN2:GFP, whereas a similar treatment with a cocktail of common protease inhibitors had no effect on BIN2 stability. This result not only ruled out the possibility that the BL-induced depletion of BIN2 is due to low immunoprecipitation efficiency of the wild-type BIN2:GFP fusion protein by the polyclonal GFP antiserum, but also confirmed that a proteasome-mediated protein degradation process is involved in the BL-induced BIN2 disapperance.
Figure 6. A Proteosome-Mediated Protein Degradation is Responsible for the BL-Induced BIN2 Depletion.
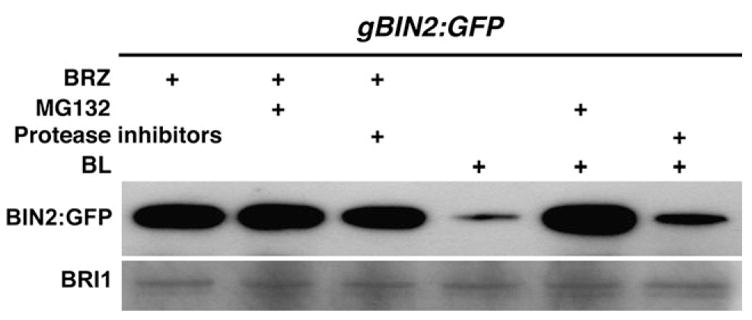
gBIN2:GFP seedlings were grown on BRZ-containing medium for 2 weeks and transferred to liquid medium containing 1 μM BRZ or 1 μM BL supplemented with 10 μM MG132 or a mixture of protease inhibitors. The amount of BIN2:GFP fusion protein in various treated seedlings was analyzed by the IP/Western analysis and the relative amount of total proteins was estimated by immunoblotting with the BRI1 antibody.
The BL-Induced BIN2 Degradation Correlated Well with the BL-Induced Reduction in BIN2 Activity
To test whether the BL-induced BIN2 degradation is responsible for the previously observed BL-induced reduction in BIN2 kinase activity (Mora-Garcia et al., 2004; Vert and Chory, 2006), we carried out an immuno-kinase assay. Anti-GFP immunoprecipitates of gBIN2:GFP seedlings were incubated with 32P-labeled ATP and E. coli-expressed GST:BZR1 fusion protein for 1 h, and the reaction mixtures were separated by gel electrophoresis. The amount of immunoprecipitated BIN2:GFP was analyzed by Western blot analysis, while the phosphorylation level of GST:BZR1 was visualized by autoradiography. As shown in Figure 7, BL treatment led to a significant reduction in the BZR1 phosphorylation activity, similar to that caused by treatment of lithium, known to be a specific inhibitor of GSK3 kinase (Zhao et al., 2002). More importantly, addition of MG132 was able to nullify the inhibitory effect of BL on the BIN2 kinase activity, since the GST:BZR1 phosphorylation level of immunoprecipitated BIN2:GFP from the BL/MG132-treated seedlings is similar to that of mock-treated seedlings. It is worthy mentioning that a longer exposure of the Western blot revealed a ladder of weak bands above the BIN2:GFP signal in the MG132/BL lane, which likely represents ubiquitinated forms of BIN2. These results strongly suggest that the ubiquitin/proteasome-mediated protein degradation constitutes a major regulatory mechanism for reducing the BIN2 kinase activity.
Figure 7. The BL-Induced BIN2 Degradation is an Important Regulatory Mechanism for BIN2 Regulation.
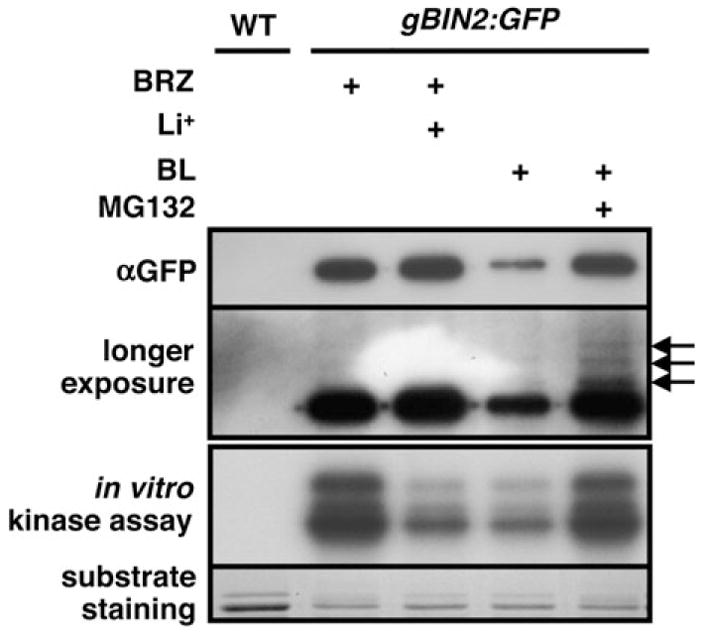
gBIN2:GFP transgenic seedlings were grown on BRZ-containing medium for 2 weeks and transferred to liquid medium containing 100 mM LiCl, 1 μM BL or 1 μM BL plus 10 μM MG132. After immunoprecipitation, BIN2:GFP was used as the kinase for an in vitro phosphorylation assay with GST:BZR1. The amount of immunoprecipitated BIN2:GFP proteins was examined by Western blotting with the monoclonal GFP antibody (top panel). Longer exposure of the blot revealed a ladder of weak bands (indicated by three arrows in the second panel). The phosphorylation levels of GST:BZR1 were visualized by autoradiography (the third panel), and the amounts of the substrate used in the assay were shown by Coomassie Blue staining (bottom panel).
DISCUSSION
One of the major deficiencies in our current knowledge of the BR signaling pathway is lack of understanding of the inhibitory mechanism of BIN2 in response to activation of BRI1 and BAK1 at the cell surface (Li and Jin, 2007). In this report, we presented biochemical evidence to implicate an ubiquitin/proteasome-mediated protein degradation process in reducing the amount of this negative regulator of the BR signaling pathway. The idea that BIN2 might be regulated at the protein level was raised by our unexpected discoveries that bin2-1 is a much more stable protein than its wild-type form, which was confirmed by our observation that the stead-state level of the wild-type BIN2:GFP can be significantly enhanced by treatment with BRZ and that the BRZ-stabilized BIN2:GFP can be rapidly depleted by BL in a concentration-dependent manner. We have shown here that the reduction in BIN2 accumulation is specifically induced by BR but not by other plant hormones and that the BL-induced BIN2 disappearance requires a functional BR receptor, since no BL-induced BIN2 degradation was observed in bri1-9 mutant that lacks a functional BR receptor at the cell surface. Moreover, treatment of MG132—a well known proteasome inhibitor—effectively blocked the BL-induced BIN2 depletion and led to the detection of a potential ladder of ubiquitinated BIN2 species, suggesting the involvement of an ubiquitin/proteasome-mediated protein degradation process in the observed BIN2 depletion. More importantly, our data revealed that MG132 co-treatment nullified the inhibitory effect of BL on the BIN2 phosphorylation activity, indicating that the BL-induced BIN2 degradation constitutes an important regulatory step in the BR signaling pathway. However, a previous study showed that a pretreatment of 10 μM MG132 had little effect on the kinetics of the BR-induced BZR1 dephosphorylation in vivo (He et al., 2002), possibly due to activation of a protein Ser/Thr phosphatase, such as BSU1 (Mora-Garcia et al., 2004). On the other hand, a 2-h BR treatment of the homozygous bin2-1 mutant only slightly dephosphorylated BZR1 (J. Zhao and J. Li, unpublished data). It is thus likely that BR-induced dephosphorylation of BZR1 or BES1 involves atleast two different mechanisms: BIN2 degradation and phosphatase activation.
Our results contradict with that of a recent study showing that the level or sub-cellular localization of BIN2 is not affected by BL treatment (Vert and Chory, 2006). Such a contradiction is likely caused by different constructs used in the two studies. The Vert and-Chory study used a BIN2:GFP minigene containing the BIN2 promoter and a BIN2 cDNA fused to GFP, while this study used a genomic BIN2:GFP construct that contains the BIN2 native promoter plus all the introns, including the first intron of 591 bp long. It was generally thought that long introns often contain important regulatory sequences critical for transcriptional initiation, RNA processing, or translation efficiency (Le Hir et al., 2003). For example, the first intron of the Arabidopsis SUVH3 gene harbors cis-acting DNA sequences that control the tissue/development-specific gene expression (Casas-Mollano et al., 2006). We suspected that the first BIN2 intron contains crucial regulatory sequences that not only direct the tissue/cell-specific expression of BIN2, but also affect the translational efficiency of the BIN2 transcript. Consistent with this interpretation, the expression of the BIN2 minigene in transgenic Arabidopsis plants led to easy detection of the BIN2:GFP fusion protein by both Western blotting analysis and confocal microscopy (Vert and Chory, 2006). By contrast, no GFP signal was detected by either method from the transgenic plants containing the BIN2:GFP genomic construct, despite the fact that expression of gBIN2:GFP and gbin2-1:GFP transgenes resulted in a higher percentage of bin2-like transgenic plants than the BIN2 and bin2-1 minigenes, respectively. A further support for our explanation came from our recent discovery that 35S-BIN2:GFP transgenic lines, which exhibit GFP signals by both direct Western blotting and confocal microscopy, did not show the BR-induced BIN2 degradation (Z. Yan and J. Li, unpublished data).
Our data provided another example of the involvement of protein degradation in GSK3 regulation. GSK3 depletion was previously discovered as an early molecular consequence of cortical rotation in early Xenopus embryos and could be induced by overexpression of the Xenopus GSK3-binding protein GBP (Dominguez and Green, 2000). A recent study revealed that glucocorticoids can also induce the ubiquitin/proteasome-mediated degradation of GSK3 in rat mammary epithelial tumor cells through activation of Sgk and Akt that phosphorylate GSK3β at Ser-9 and that mutation of this regulatory residue prevented the glucocorticoid-induced GSK3 degradation (Failor et al., 2007). In plants, a Medicago GSK3-like kinase, MsK1, was recently shown to be degraded by proteasomes to activate a MAP kinase-mediated plant defense signaling pathway (Wrzaczek et al., 2007). In addition to proteasome-mediated degradation that eliminates the GSK3 kinase, site-specific proteolysis can activate the GSK3 kinase activity. Goni-Oliver et al. recently showed that the N-terminal cleavage of GSK3β by calpain—a calcium-dependent cysteine protease—resulted in a significant increase in GSK3 activity, most likely by removing the N-terminal autoinhibitory domain (Goni-Oliver et al., 2007).
It remains to be determined what triggers the BIN2 degradation. It is well known that phosphorylation is a common signal in animal systems for targeting a given protein for ubiquitin/proteasome-mediated protein degradation (Gao and Karin, 2005). For example, GSK3 phosphorylation of β-catenin generates a recognition site for a WD40-containing F-box protein called β-TrCP that interacts with Skp1 and Cullin to form an SCF-type E3 ligase (Kitagawa et al., 1999), and mutations of GSK3-phosphorylation sites on β-catenin can significantly stabilize the GSK3 substrate, thus activating the Wnt signaling pathway (Liu et al., 1999). In plants, there are many examples of ubiquitin/proteasome-mediated protein degradation (Smalle and Vierstra, 2004), but the relationship between phosphorylation and protein degradation is not well established. For example, CRY2—one of the two Arabidopsis blue/ultraviolet-A light receptors for mediating light regulation of seedling development and photoperiodic flowering (Guo et al., 1998)—was known to undergo the blue-light-induced phosphorylation and protein degradation (Shalitin et al., 2002). A more relevant example is BZR1 that is thought to be phosphorylated by BIN2 and degraded in the absence of the plant steroid hormone (He et al., 2002). It remains to be determined whether phosphorylation is required for CRY2 or BZR1 degradation.
Given the fact that BIN2 is inhibited in response to activation of two cell surface receptor kinases, it was thought previously that BIN2 might be phosphorylated directly by either BRI1 or BAK1. However, yeast two-hybrid and in vitro phosphorylation assays revealed that neither receptor kinase was able to directly interact with or phosphorylate the Arabidopsis GSK3-like kinase, suggesting the involvement of one or more other protein kinases in BIN2 regulation (Li and Nam, 2002; Li and Jin, 2007). One potential candidate for such a kinase is casein kinase II that recognizes the consensus phosphorylation site S/TxxD/E (Meggio and Pinna, 2003). The S/TxxD/E site matches a highly conserved four-amino-acid T261R262E263E264 motif of BIN2 that was mutated in seven out of eight known hypermorphic bin2/ucu1/dwarf12 mutations (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002). Although one can argue that these mutations might simply block the protein–protein interaction between BIN2 and a critical component of a BR-activated BIN2 regulatory machinery, since the TREE motif is part of a short α-helix exposed on the surface of the protein (Dajani et al., 2001), the lack of mutations at other positions within this short α-helix favors the phosphorylation hypothesis. We suspected that mutations of T261 (bin2-2), E263 (bin2-1 and dwf12-2D), or E264 (ucu1-1/2 and dwf12-1D) within this TREE motif would prevent the phosphorylation of BIN2 by CKII or other acidic-residue-directed kinases and its recognition by an E3 ligase, thus making bin2-1 or bin2-2 a much more stable protein than its wild-type form. Indeed, preliminary results with gbin2-2:GFP transgenetic plants revealed that the T261K mutation also significantly stabilizes the GFP-tagged kinase. Determination of whether and where BIN2 is phosphorylated in response to BR and identification of a BL-regulated protein kinase that phosphorylates BIN2 are required for a complete understanding of the biochemical mechanism that regulates the Arabidopsis BIN2 kinase.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was the wild-type control and the parental strain for all transgenic lines. Methods for seed sterilization and conditions for plant growth were described previously (Li et al., 2001).
Generation of Transgenic Plants
A 3.8-kb BIN2 genomic fragment containing its native promoter and the entire coding sequence was cloned into a modified pPZP212 vector (Hajdukiewicz et al., 1994) containing GFP and the E9 terminator of the pea Rubisco small subunit to generate pPZP-gBIN2:GFP construct. The bin2-1 mutation (E263K) was introduced into the pPZP-gBIN2:GFP transgene by site-directed mutagenesis using the Stratagene’s QuickChange II XL site-directed mutagenesis kit to generate the pPZP-gbin2-1:GFP construct. Both constructs were individually transformed into the wild-type Arabidopsis plants using the Agrobacterium-mediated method (Bechtold and Pelletier, 1998). The pPZP-gBIN2:GFP bri1-9 line was generated by crossing bri1-9 with a chosen pPZP-gBIN2:GFP transgenic line that exhibits weak bin2-like dwarf phenotype and responds to BL treatment.
Chemical Treatment of Arabidopsis Seedlings
Seeds of various transgenic lines were germinated on a synthetic medium (1/2 MS plus 1% sucrose) supplemented with 2 μM BRZ and grown under a 16-h light/8-h dark long day growth condition at 22°C for 2 weeks. The seedlings were then removed from the agar plates, transferred to liquid synthetic medium (1/2 MS plus 1% sucrose) containing different concentrations of BL, and incubated for various times before being removed for immunoblotting analysis. To investigate a possible mechanism of BIN2 degradation, BRZ-treated seedlings were further incubated in liquid medium containing 10 μM MG132 (Sigma) for 1 h before transferring into 1 μM BL-containing medium. To determine the effect of other plant hormones on BIN2 stability, BRZ-treated transgenic seedlings were transferred to liquid medium for a 30-min incubation with 1 μM gibberellic acid (GA3), 1 μM auxin (indole-3-acetic acid [IAA] or indole-3-butyric acid [IBA]), 1 μM cytokinin (kinetin or 6-benzylaminopurine [6-BA]), 0.5 μM abscisic acid, or 1 μM ACC (a precursor of ethylene).
RNA Isolation and Northern Blot Analysis
Tissues of 4-week-old soil-grown wild-type Arabidopsis plants and various transgenic lines were harvested into liquid nitrogen. Methods for total RNA isolation and Northern blot analysis were described previously (Li et al., 2001). A 0.7-kb DNA fragment derived from GFP was used as the probe in the Northern blot assay.
Immunoprecipitation and Western Blot Analysis
Arabidopsis seedlings were collected and ground into fine powders in liquid nitrogen. Proteins were dissolved in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40, and a protease inhibitor cocktail containing 1 mM phenylmethylsulphonyl fluoride plus 2 μg mL−1 each of aprotinin, leupeptin, and pepstatin A (all purchased from Fisher Scientific)). Total protein crude extracts were prepared by centrifuging the dissolved plant powders at 8000 g for 10 min at 4°C. The resulting supernatants were mixed with a polyclonal GFP antibody (1:1000, Torrey Pines Biolabs, TX) followed by incubation with 50 μl 50% slurry of Protein-A beads (Amersham Biosciences). After three-time washing with the lysis buffer, immunoprecipitates were resuspended in 90 μl 2X SDS sample buffer, boiled at 100°C for 5 min, and separated by 7.5% SDS-PAGE. The amounts of BIN2:GFP or bin2-1:GFP protein in various immunoprecipitates were analyzed by immunoblotting using a monoclonal GFP antibody (Covance Research Products, CA).
Expression of Fusion Protein and Immunokinase Assay
Expression and purification of the GST:BZR1 fusion protein were carried out according to the previously described procedures (Zhao et al., 2002). Equal amounts of total protein extracts of various gBIN2:GFP seedlings were used to immunoprecipitate BIN2:GFP as described above. The resulting immunoprecipitates were washed three times with the lysis buffer and twice with the GSK3 kinase buffer (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM dithiothreitol), and resuspended in 50 μl GSK3 kinase buffer. The kinase reaction was initiated at room temperature by adding 1 μg of purified GST:BZR1, 1 μl 1 mM ATP and 1 μl [γ-32P]ATP (3000 Ci mmol−1, MP Biomedical), and was terminated after 1 h incubation at room temperature by adding equal volume of 2X SDS sample buffer. The reaction mixtures were boiled for 3 min and separated by 8% SDS-PAGE. The amount of immunoprecipitated BIN2:GFP fusion protein was determined by Western blot with the monoclonal GFP antibody, while phosphorylation level of GST:BZR1 was visualized by overnight autoradiography at −70°C.
Acknowledgments
We thank Dr Tadoa Asami for his generous supply of brassinazole and members of the Li lab for stimulating discussion. This work was supported by a National Institutes of Health grant (GM060519) to JL. No conflict of interest declared.
References
- Ali A, Hoeflich KP, Woodgett JR. Glycogen synthase kinase-3: properties, functions, and regulation. Chem Rev. 2001;101:2527–2540. doi: 10.1021/cr000110o. [DOI] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bijur GN, Jope RS. Proapoptotic stimuli induce nuclear accumulation of glycogen synthase kinase-3 beta. J Biol Chem. 2001;276:37436–37442. doi: 10.1074/jbc.M105725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Liu YI. Wnt signaling: complexity at the surface. J Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- Casas-Mollano JA, Lao NT, Kavanagh TA. Intron-regulated expression of SUVH3, an Arabidopsis Su(var)3–9 homologue. J Exp Bot. 2006;57:3301–3311. doi: 10.1093/jxb/erl093. [DOI] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I, Green JB. Dorsal downregulation of GSK3beta by a non-Wnt-like mechanism is an early molecular consequence of cortical rotation in early Xenopus embryos. Development. 2000;127:861–868. doi: 10.1242/dev.127.4.861. [DOI] [PubMed] [Google Scholar]
- Failor KL, Desyatnikov Y, Finger LA, Firestone GL. Glucocorticoid-induced degradation of GSK3 protein is triggered by Sgk and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Mol Endocrinol. 2007;21 doi: 10.1210/me.2007-0143. in press. [DOI] [PubMed] [Google Scholar]
- Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- Gampala SS, et al. An essential role for 14–3–3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Goni-Oliver P, Lucas JJ, Avila J, Hernandez F. N-terminal cleavage of GSK-3 by calpain: a new form of GSK-3 regulation. J Biol Chem. 2007;282:22406–22413. doi: 10.1074/jbc.M702793200. [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Li J. Brassinosteroid signaling: from receptor kinases to transcription factors. Curr Opin Plant Biol. 2005;8:526–531. doi: 10.1016/j.pbi.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plant Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meares GP, Jope RS. Resolution of the nuclear localization mechanism of glycogen synthase kinase-3: functional effects in apoptosis. J Biol Chem. 2007;282:16989–17001. doi: 10.1074/jbc.M700610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsisgene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. Nucleo cytoplasmic Shuttling of BZR1 Mediated by Phosphorylation Is Essential in Arabidopsis Brassinosteroid Signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Rozhon W, Jonak C. A Proteasome-regulated glycogen synthase kinase-3 modulates disease response in plants. J Biol Chem. 2007;282:5249–5255. doi: 10.1074/jbc.M610135200. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]


