Abstract
The genomes of positive-strand RNA viruses undergo conformational shifts that complicate efforts to equate structures with function. We have initiated a detailed analysis of secondary and tertiary elements within the 3′ end of Turnip crinkle virus (TCV) that are required for viral accumulation in vivo. MPGAfold, a massively parallel genetic algorithm, suggested the presence of five hairpins (H4a, H4b, and previously identified hairpins H4, H5, and Pr) and one H-type pseudoknot (Ψ3) within the 3′-terminal 194 nucleotides (nt). In vivo compensatory mutagenesis analyses confirmed the existence of H4a, H4b, Ψ3 and a second pseudoknot (Ψ2) previously identified in a TCV satellite RNA. In-line structure probing of the 194-nt fragment supported the coexistence of H4, H4a, H4b, Ψ3 and a pseudoknot that connects H5 and the 3′ end (Ψ1). Stepwise replacements of TCV elements with the comparable elements from Cardamine chlorotic fleck virus indicated that the complete 142-nt 3′ end, and subsets containing Ψ3, H4a, and H4b or Ψ3, H4a, H4b, H5, and Ψ2, form functional domains for virus accumulation in vivo. A new 3-D molecular modeling protocol (RNA2D3D) predicted that H4a, H4b, H5, Ψ3, and Ψ2 are capable of simultaneous existence and bears some resemblance to a tRNA. The related Japanese iris necrotic ring virus does not have comparable domains. These results provide a framework for determining how interconnected elements participate in processes that require 3′ untranslated region sequences such as translation and replication.
Replication of plus-strand RNA viruses initially requires translation of the genomic RNA to produce the virus-encoded, RNA-dependent RNA polymerase (RdRp) and any auxiliary viral proteins necessary for transcription. In a process that is poorly defined but likely dictated by viral and/or cellular factors, translation is terminated and the genomic RNA becomes available for reiterative synthesis of complementary strands, a process that requires membrane association (1, 2, 26). For some viruses, subsequent viral plus-strand synthesis occurs in virus-specific membrane invaginations known as spherules, which contain a limited number of minus-sense genomes and whose formation is induced by specific viral proteins (1, 15, 26). Although the process of producing viral progeny has been extensively studied using many different viral systems, it remains poorly understood. For example, fundamental questions, such as the role that conformational shifts in RNA structure play in switching the template from translation to replication, the proteins required to enact such events, and if cis-acting core promoters, enhancers, and repressors are organized into functional, interacting modules, remain virtually unanswered.
Recent reports that portions of RNA viral genomes undergo conformational shifts to execute different functions (10, 13, 21, 28) complicate efforts to assign biological roles to groups of cis-acting elements that may not structurally coexist. The ability of viral RNAs to assume multiple conformations combined with evidence that newly synthesized plus strands may assume forms that suppress transcription (43) supports the “stamping model” hypothesis for viral replication. This model, based on the distribution of mutants arising from single bursts in the RNA bacteriophage ϕ6, suggests that most progeny plus strands are derived from minus strands synthesized from the original entering plus-strand genome (6). This increasingly complex image of RNA virus replication is requiring a reevaluation of conclusions reached using reductionist, cell-free approaches, where RNA elements are isolated from other cis-acting sequences with which they may normally communicate or form functional domains.
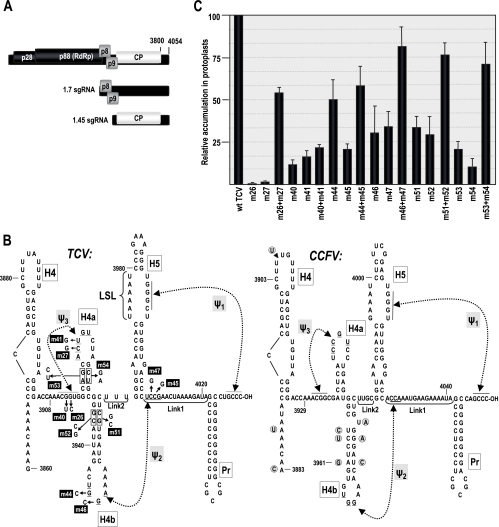
To gain a more accurate picture of the replication process and the switch between translation and replication, more complete secondary and tertiary structural maps of regions containing elements engaged in these activities must be developed, followed by a determination of which elements coexist in functional domains. Turnip crinkle virus (TCV), a member of the genus Carmovirus in the family Tombusviridae, is well suited for this analysis. TCV contains a small (4054-nucleotide [nt]), single genomic RNA that encodes five proteins involved in replication (p28 and p88), movement (p8 and p9), and encapsidation/suppression of RNA silencing (coat protein [CP]) (Fig. 1A) (11, 25). TCV participates in a mutualistic association with a dispensable, untranslated satellite RNA (satC; 356 nt) that contains 151 nt derived from the TCV 3′ region (34). This portion of satC assumes alternative conformations when transcriptionally active or inactive, mediated by an element shared with TCV known as the derepressor (DR) (43). The 5′ ends of TCV genomic RNA and satC are not capped, and the 3′ termini do not contain poly(A) tails or terminally located tRNA-like structures. As with other members of the Tombusviridae, TCV has a 3′ untranslated region (UTR) that contains a translational enhancer that requires the presence of its 5′ UTR for high-level activity (24, 41). While the precise location of the translational enhancer has not been reported, it is located within a region that likely overlaps with sequences required for initiation of minus-strand synthesis. This region is downstream of the mapped 3′ translational enhancer of carmovirus Hibiscus chlorotic ringspot virus (HCRSV), which contains an important 6-nt element missing from the 3′ UTR of TCV (14).
FIG. 1.
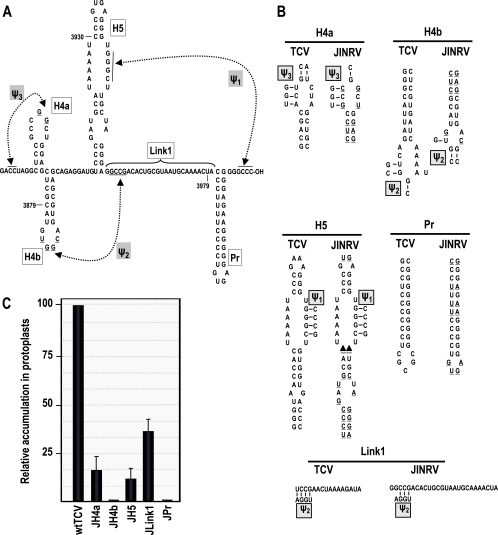
Genomic organization and 3′ UTR RNA structures in TCV. (A) TCV genomic and subgenomic RNAs encode proteins involved in replication (p28 and p88), movement (p8 and p9), and encapsidation/RNA silencing suppression (CP). Carmoviruses CCFV and JINRV have similar genomic arrangements, but open reading frames have not been analyzed. (B) Structure of the 3′-terminal regions of TCV (left) and CCFV (Blue Lake strain) (right). H5 and Pr hairpins are conserved in nearly all carmoviruses. Ψ1 is also conserved among carmoviruses and was previously shown to exist for TCV in vivo (46). H4a, H4b, and Ψ3 in TCV were predicted by MPGAfold. Ψ2 forms in the preactive structure of satC (44). Mutations constructed to examine the existence of these elements in TCV and their numeric designations are shown. Base differences found in the CCFV Clear Lake strain are circled. Putative CCFV elements are designated according to analogous structures in TCV. (C) Accumulation of mutant TCV in protoplasts at 40 hpi. All values were averages from at least three experiments. Standard deviation bars are indicated.
The 3′ UTRs of TCV and all carmoviruses, with the exception of Galinsoga mosaic virus, contain a very stable 3′-terminal hairpin (Pr) that has been tentatively identified as the core promoter for minus-strand synthesis based on analysis of the comparable hairpin in satC (Fig. 1B) (35, 36) Directly upstream of all carmoviral Pr hairpins is a structurally conserved and critically important hairpin (H5), which is proposed to function as an RdRp chaperone in TCV (19) and is similar to the SL3 hairpin in tombusviruses (23). The 3′ side of the H5 large symmetrical internal loop (LSL) forms an RNA-RNA interaction with bases at the 3′ terminus (termed Ψ1) that is important for efficient TCV accumulation in vivo (46) and is present throughout the Tombusviridae (Fig. 1B and C) (20, 22, 46). A third hairpin in the 3′ UTR, H4, is also critical for TCV accumulation in vivo, and fragments containing H4 and adjacent adenylates can enhance transcription and bind RdRp in both orientations in vitro (36). Curiously, Li and Wong (17) reported that virtually the entire TCV 3′ UTR, including H4 and H5, is dispensable for viral accumulation in Arabidopsis protoplasts but not Hibiscus protoplasts. This study contradicts previous reports on TCV using Arabidopsis or turnip protoplasts (4, 19, 46) and reports on related viruses (22).
To investigate these contradictory results and begin assigning known and newly discovered elements into functional coexisting domains within the TCV 3′ UTR, we have initiated a comprehensive evaluation of structures in TCV based on those previously found in satC and/or predicted by the massively parallel genetic algorithm MPGAfold (29, 32). In vitro structure mapping using in-line probing and/or in vivo compensatory mutation analyses confirmed the importance of two additional juxtaposed hairpins, H4a and H4b, as well as an H-type pseudoknot predicted by MPGAfold and a second pseudoknot previously found in the preactive structure of satC (45). Stepwise replacements of subsets of elements with the comparable elements from the related carmovirus Cardamine chlorotic fleck virus (CCFV) indicated several regions with interacting elements, including a 100-nt region encompassed by two pseudoknots and containing H4a, H4b, and H5. This 100-nt domain was subjected to the new 3-D molecular modeling protocol RNA2D3D (18, 40), which predicted that the elements are capable of simultaneous existence, and folds into a highly stable structural unit that bears some resemblance to a tRNA. These results are not consistent with the study by Li and Wong and provide a framework for determining how interconnected elements participate in the mutually exclusive processes of translation and replication.
MATERIALS AND METHODS
MPGAfold analysis of the TCV 3′ UTR.
MPGAfold (the massively parallel genetic algorithm for RNA folding) was used to generate the secondary structure predictions for a portion of the TCV 3′ UTR sequences (positions 3839 to 4054). The algorithm has been previously described (29, 32). MPGAfold results were analyzed using the RNA/DNA structure analysis workbench StructureLab (30, 31). Stem Trace (12), a component of StructureLab, was used to depict the results of 20 MPGAfold runs of the TCV 3′ UTR population.
Construction of TCV mutants.
All mutants were constructed by oligonucleotide-mediated, site-directed mutagenesis. The parental TCV clone for most element replacements was TCV-TSNL5, which contains the full-length TCV sequence (the Massachusetts strain) downstream from a T7 RNA polymerase promoter. The TCV sequence in TCV-TSNL5 was modified from the wild-type (wt) TCV by insertion of “CGU” just downstream from position 4014 in Link1 (the linker sequence between H5 and Pr), creating a SnaBI site to permit ease of cloning. All mutants were subjected to sequencing to confirm alterations.
Protoplast preparation, inoculation, and RNA gel blots.
TCV constructs were digested with SmaI and in vitro transcribed using T7 RNA polymerase, producing full-length viral RNA with exact 5′ and 3′ ends. Protoplasts were generated by digestion of callus derived from Arabidopsis thaliana ecotype Col-0 seeds with cellulase and pectinase as previously described (45). Twenty micrograms of genomic TCV RNA was inoculated onto protoplasts using polyethylene glycol (45). Total RNA was extracted at 40 h postinoculation (hpi), subjected to electrophoresis, and transferred to a nitrocellulose membrane, and viral genomic RNA was detected using a complementary 32P-labeled oligonucleotide. Blots were stripped and then reprobed with a fragment complementary to the ribosomal RNAs. Data were quantified using Quantity One software (Bio-Rad Laboratories), and viral genomic RNA levels were normalized to levels of rRNA.
Inoculation of turnip seedlings and extraction of viral progeny RNA.
Mutant and wt TCV plasmids were digested with SmaI, and transcripts were synthesized in vitro using T7 RNA polymerase. Twelve micrograms of transcripts was mixed with infection buffer and inoculated onto turnip (cv. Just Right) seedlings, and total RNA was extracted 20 days later as previously described (4). Viral RNAs were cloned by reverse transcription-PCR and sequenced.
In-line structural probing.
In-line probing was performed essentially as previously described (39). Briefly, RNA fragments synthesized using T7 RNA polymerase were 3′ end labeled using T4 RNA ligase (Ambion) and [α-32P]pCp or 5′ end labeled using [γ-32P]ATP and polynucleotide kinase. Fragments were purified by polyacrylamide gel electrophoresis, heated to 75°C, and allowed to slow cool over 2 h to room temperature. RNAs were then incubated at room temperature for 14 h in 50 mM Tris-HCl (pH 8.5) and 20 mM MgCl2. Reactions were ethanol precipitated, heated at 95°C for 5 min, and subjected to electrophoresis through 8% and 20% sequencing-length denaturing polyacrylamide gels. RNA cleavage ladders were generated by incubating end-labeled RNA in a reaction mix containing 1 μg of yeast tRNA, 50 mM NaHCO3/Na2CO3 (pH 9.2), and 1 mM EDTA for 5 min at 95°C. RNase T1 digests were carried out by incubating 5 pmol of partially denatured labeled RNA in 1 μg of yeast tRNA, 20 mM sodium citrate (pH 5), 1 mM EDTA, 7 M urea, and 1 U RNase T1 for 15 min at room temperature.
RNA 3-D structure prediction and analysis.
RNA2D3D uses constraints obtained from standard A-form helices to obtain a 3-D structure by embedding the 3-D nucleotides into a planar secondary structure and ultimately winding the structure. The sequence and secondary structure of the TCV 3′ UTR including Ψ2 and Ψ3 were presented to RNA2D3D to produce initial 3-D computational models. Each of the models was subjected to extensive molecular modeling protocols including manual adjustments and refinements of the structural models, followed by mechanics minimization and molecular dynamics (MD) equilibrations (18, 33, 40). The manual adjustments included rotation and repositioning of the helical segments, strands, and individual bases. The H5 helix GAAA tetraloop with its closing base pair was spliced in from the Protein Data Bank entry 1F9L's equivalent fragment. This splicing, as well as the alignment of motifs with known RNA structural motifs, was performed with DSViewer Pro. All simulations were performed using the ff99 Cornell force field for RNA (38), which is a reliable and refined force field for nucleic acids, and using the MD software Amber 9.0 (5). MD simulations were performed using explicit water molecules and ions. Particle-mesh Ewald summation was used to calculate the electrostatic interactions (9). The nonbonded interactions were truncated at 9 Å. For a 100-base TCV molecule subjected to MD simulation, 99 Na+ counterions and 63 Na+/Cl− pairs were used with 28,075 TIP3P water molecules to solvate the RNA molecule to a 0.1-mol/liter relative salt concentration. The final box dimensions were 106 Å by 112 Å by 88 Å, and the total system consisted of 87,666 atoms. The system was minimized by constraining the solute and then the solvent and was then heated to 300 K, constraining the RNA. Equilibration was performed in multiple stages by slowly releasing the constraints. SHAKE was applied to all hydrogen bonds in the system. Pressure was maintained at 1.0 Pa using the Berendsen algorithm (3), and a periodic boundary condition was imposed. A production simulation was performed for 50 ns with a 2-fs time step. SGI Altix and SGI Origin computers utilizing 8 to 12 processors were used to perform all simulations. Analysis of the MD results, excluding the equilibration stage, was performed using the PTRAJ module of the Amber 9.0 package.
RESULTS
The 194-nt 3′-terminal region of TCV contains five hairpins and three pseudoknots.
MPGAfold, a massively parallel genetic algorithm that predicts secondary and limited tertiary RNA structures (H-type pseudoknots) (29, 32) was used to identity potential structural elements within the 3′-terminal region of TCV (positions 3860 to 4054) that might contribute to replication and/or translational enhancement (Fig. 1B). MPGAfold predicted the presence of previously identified hairpins H4, H5, and Pr, along with two additional juxtaposed hairpins (H4a and H4b) located just upstream of H5. In addition to these hairpins, MPGAfold predicted a possible H-type pseudoknot between 5 nt in the loop of H4a and the upstream flanking sequence, labeled Ψ3 (Fig. 1B). Sequences that form an important satC pseudoknot (Ψ2) connecting the H4b loop sequence (5′-UGGA) with sequences flanking the 3′ side of H5 (3′-GCCU) (45) are also conserved in TCV.
A compensatory mutagenesis approach was used to assess the presence of H4a, H4b, Ψ2, and Ψ3. For H4a, two putative adjacent base pairs in the stem, A·U/G·C, were converted to two sets of mismatched pairs (C-U/C-U [m53]; G-A/G-A [m54]) and the putative compensatory exchanges U·A/C·G (m53 plus m54) (Fig. 1B). m53 and m54 accumulated to 20% and 10% of the wt activity, respectively, while the compensatory m53 plus m54 regained 71% of the wt activity (Fig. 1C). H4b was subjected to a similar analysis, with two consecutive base pairs near the base of the stem (G·C/C·G) converted to G-G/C-C (m51), C-C/G-G (m52), and the putative compensatory C·G/G·C (m51+m52). Mutations in the H4b stem were less detrimental than those in the H4a stem, with single-side alterations accumulating to approximately 30% of the wt levels. The alterations together (m51 plus m52) were compensatory, with virus accumulating to 76% of the wt levels (Fig. 1C). These results confirm the presence of H4a and H4b in TCV and also suggest that these hairpins are necessary but may not be critical for TCV accumulation.
Two sets of single compensatory alterations were used to assess the presence of Ψ3 (Fig. 1B). TCV containing G3913C (m26) upstream of H4a or C3922G (m27) in the H4a loop accumulated to barely detectable levels in Arabidopsis protoplasts, whereas virus containing both mutations (m26+m27) predicted to reform Ψ3 accumulated to 55% of the wt levels (Fig. 1C). The second pair of mutations, G3912U (m40) and U3923G (m41), individually reduced TCV levels to 11% and 16% of the wt levels, respectively, while TCV containing both mutations (m40+m41) accumulated slightly more than the single mutants (22% of the wt levels). These results support the presence of Ψ3 in TCV and also suggest that efficient biological function is sequence dependent.
Two sets of compensatory mutations were generated to determine if Ψ2 exists in TCV (Fig. 1B). Converting a G to a C at position 3947 in the H4b loop (G3947C; m46) or a C to G in its putative partner at the base of H5 (C4007G; m47) reduced TCV accumulation in protoplasts to 31 and 34% of that of the wt, respectively (Fig. 1C). Together, the mutations (m46+m47) had a compensatory effect, with mutant TCV accumulating to 80% of that of the wt (Fig. 1C). Alteration of H4b loop position G3946C (m44) was less deleterious to TCV accumulation (50% of that of the wt), possibly due to a three-base-pair Ψ2 interaction that can exist if pairing with the sequence at the base of H5 is shifted 1 nt downstream. Altering the putative partner residue at the base of H5 (C4008G; m45) reduced accumulation fivefold, while both alterations improved TCV accumulation to 58% of that of the wt. These results support the existence of Ψ2 in TCV.
At least four hairpins and two pseudoknots coexist in a 3′-terminal fragment of TCV.
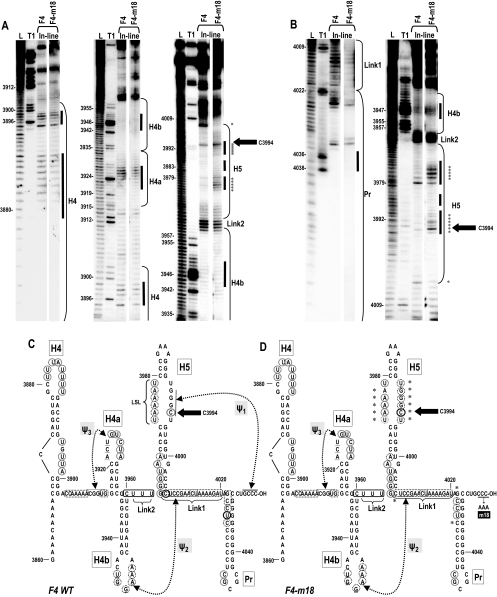
In-line probing (39) was used to analyze which computer-predicted and genetically confirmed hairpins and pseudoknots coexist in an in vitro-synthesized fragment (F4) containing the 3′-terminal region (positions 3860 to 4054). In-line probing reports on the spontaneous cleavage of the RNA backbone mediated by 2′ hydroxyl that are geometrically in-line with oxyanion leaving groups on backbone phosphates. Such in-line geometry occurs primarily in nonstructured regions of RNA, where nucleotides are not torsionally constrained by hydrogen bond pairings (39).
Bases susceptible to in-line cleavage in fragment F4 were located predominantly in proposed internal and terminal loop regions in the MPGAfold-predicted structure (Fig. 2). Most of the residues in the linker between H4 and H4a and H5 and Pr were susceptible to various amounts of cleavage, suggesting they are not substantially participating in a higher-order structure within the fragment. Residues more strongly susceptible to in-line cleavage were located in the terminal and internal loops of H4, the short linker sequence between H4b and H5 (Link2), the linker between H5 and Pr (Link1), the cytidylate at position 3994 in the LSL of H5, and the cytidylate at the 3′ base of H5 (position 4005). Most residues predicted by MPGAfold to participate in Ψ3 were protected from cleavage, as were the stems of H4a and H4b, with the exception of C3960 at the base of H4b. Most of the residues in the terminal loop of H4b were slightly susceptible to cleavage, including residues that participate in Ψ2. The Ψ2 partner residues downstream of H5 also displayed low-level cleavages; thus, Ψ2 cannot be confirmed as occurring in this fragment. The H5 LSL was consistent with a structure that participates in Ψ1. However, six residues (positions 3964 to 3969) located in the 5′ side of the lower stem of H5 were reproducibly susceptible to moderate levels of cleavage, suggesting that the entire phylogenetically conserved H5 structure (42) may not exist in this fragment. In the Pr region, five residues at the 5′ base of the stem were not protected from cleavage, suggesting that the lower portion of the stem is also not paired in this conformation. Interestingly, enzymatic and base modification probing of the biologically relevant preactive structure of full-length satC similarly indicated that the phylogenetically conserved structure of H5 was not present and that residues at the 5′ base of the Pr stem were unpaired (43, 46).
FIG. 2.
In-line probing of the 3′ UTR region of TCV. (A) In-line probing of fragments containing wt TCV and TCV with a 3′-terminal CCC-to-AAA alteration (TCV-m18). F4 fragments (positions 3860 to 4054 [H4→3′ end]) were 5′ end labeled (A) or 3′ end labeled (B) and incubated at 23°C for 14 h before gel electrophoresis. Left panels, 20% polyacrylamide gel electrophoresis; center and right panels, 8% polyacrylamide gel electrophoresis. L, ladder generated by nonspecific partial cleavages; T1, ladder generated by partial RNase T1 digests using partially denatured RNA; F4, F4 fragment; F4-m18, F4-m18 fragment. Locations of the hairpins, Link1, and Link2 are indicated. Bars indicate predicted terminal and internal loops within the hairpins. Positions of selected guanylates are given. Asterisks denote cleavage differences between wt F4 and F4-m18. The large black arrow denotes the location of C3994, which is strongly susceptible to in-line cleavage. This susceptibility to in-line cleavage was also predicted by MD simulations (see text and Fig. 5). (C) Position of residues cleaved by in-line probing in wt F4. Circled bases were susceptible to cleavage. Darker circles indicate a higher level of spontaneous cleavages relative to other cleavages. Hairpin and pseudoknot designations are as shown in Fig. 1B. (D) Position of residues cleaved by in-line probing in F4-m18. The altered bases that comprise the m18 mutation are indicated. Asterisks denote cleavage differences between wt F4 and F4-m18. (C and D) The location of C3994 is denoted by large black arrows.
The three guanylates that participate in Ψ1 within the H5 LSL were protected from cleavage, suggesting that Ψ1 is present. To gain support for the existence of this pseudoknot in the fragment, F4 containing a three-base alteration at the 3′ terminus (CCC to AAA; m18) that should disrupt Ψ1 was also subjected to in-line probing. The cleavage patterns of mutant F4-m18 and wt F4 were identical in the regions upstream and downstream of H5 (Fig. 2). The structure of H5, however, differed significantly as follows: (i) most residues on the 5′ side of the LSL were more susceptible to cleavage, (ii) residues on the 3′ side of the LSL that normally participate in Ψ1 were newly susceptible to cleavage, and (iii) the strong cleavage site at C4005 (the 3′ base of H5) was absent. Since elimination of Ψ1 uniquely affected the structure of H5, particularly within the LSL region, Ψ1 is likely present in F4. Taken together with our previous findings, the structural probing data and/or genetic data support the existence of five hairpins and three pseudoknots in the 3′ UTR of TCV, with the conformation of the F4 fragment likely containing H4, H4a, H4b, portions of H5 and Pr, Ψ3, and Ψ1.
CCFV 3′ UTR contains sequences capable of forming TCV-like hairpins and pseudoknots.
CCFV is the most closely related carmovirus to TCV, with 66% pairwise similarity across the genome (65% in the 3′ UTR). Using a combination of mFold and manual building of structural elements based on the TCV model (Fig. 1B), the existence of similar hairpins and pseudoknots in the CCFV 3′ region is proposed (Fig. 1B). The CCFV Pr is similar to that of TCV, with 8 consecutive guanylates (compared with 10 for TCV), 10 possible paired bases (1 less than that of TCV), and an identical sequence in the terminal loop. The CCFV H5 differs from that of TCV by incorporating a different stable tetraloop, a smaller LSL, and an additional non-Watson-Crick pair or bases in the lower symmetrical loop. As with other carmoviruses, CCFV 3′-terminal bases are complementary to the sequence in the 3′ side of the H5 LSL, which is necessary for formation of Ψ1. The existence of H4b is strongly suggested, as multiple covariant bases that maintain the structure are found in a second CCFV isolate (Fig. 1B). CCFV H4b shares five identical residues in its terminal loop with TCV H4b (UGGAA), whereas the CCFV H4b stem is proposed to contain a single bulged base. The putative CCFV Ψ2 differs from TCV Ψ2 by having only three possible Watson-Crick base pairs beginning 1 nt downstream of H5. Ψ3 base pairing predicted between the loop of H4a and the upstream sequences also appears less stable than the comparable pseudoknot in TCV, with one less Watson-Crick pair. A portion of the H4a loop sequence, but not the stem, is conserved between CCFV and TCV (UCCGUCC and ACUGUCUA, respectively).
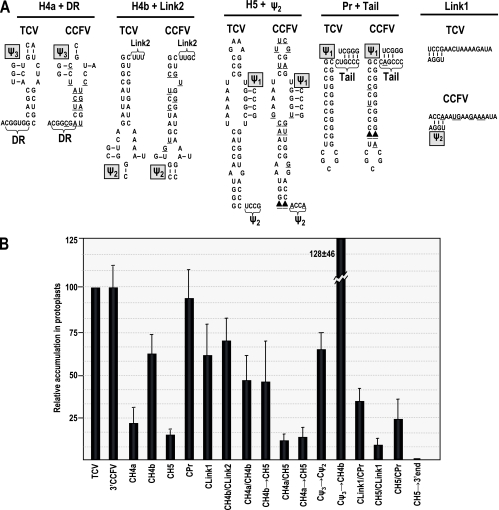
The 3′-terminal 142 nt of CCFV and various encompassed regions form cooperative modules.
To determine if the structurally similar 3′ end of CCFV can substitute for the analogous region in TCV, the elements from Ψ3 to the 3′ end (positions 3909 to 4054) in TCV were replaced with the equivalent region from CCFV (positions 3930 to 4072). TCV with the 3′ end of CCFV (3′CCFV) accumulated to TCV-TSNL5 levels (the parental construct for generation of the chimeric viruses) in protoplasts (Fig. 3B), indicating that this CCFV fragment is compatible with the remaining TCV cis- or trans-acting factors required for replication and/or translation. To determine if substitution of individual elements allows for efficient viral accumulation, TCV hairpins H4a, H4b, H5, and Pr or the linker between H5 and Pr (Link1) were replaced with the analogous hairpins and the linker region from CCFV. TCV with the H4a of CCFV (CH4a), which maintains four of five Ψ3 base pairings (Fig. 3A), accumulated to only 21% of the TCV-TSNL5 levels (Fig. 3B), similar to the level of virus accumulating with a disrupted H4a (m53+m54, Fig. 1). TCV with CCFV H4b (CH4b), which would maintain a TCV-like Ψ2, accumulated to 62% of that of the wt TCV, indicating that the sequence differences in the stem and asymmetric bulge in CCFV H4b had a moderate effect on its activity. To determine if H4b requires association with its 3′ flanking linker sequence (Link2) for a more efficient function, CH4b was altered to also contain Link2. The addition of Link2 (CH4b/CLink2) had no significant effect on the accumulation of CH4b (Fig. 3B), suggesting that other factors decrease the efficiency of TCV accumulation when associated with CCFV H4b.
FIG. 3.
Stepwise replacements of TCV elements with analogous regions from CCFV reveal functional domains. (A) Comparison of sequences comprising elements of TCV and CCFV. Underlined bases in the CCFV sequences denote differences from TCV. Putative Ψ3, Ψ2, and Ψ1 that could form when CCFV sequences are partnered with TCV background sequences are shown. The wt TCV pseudoknot pairings are also shown. Filled triangles denote missing bases. Locations of the hairpins, pseudoknots, and linker (Link) sequences are shown in Fig. 1B. The DR sequence was previously identified in satC as being a sequence-specific element required for the conformational switch between preactive and active structures (44). (B) Relative accumulation of chimeric TCV RNAs in protoplasts at 40 hpi compared with that of parental TCV-TSNLS (TCV) as assayed by RNA gel blots using a TCV-specific probe. The composition of the replacement elements is given, preceded by a “C” for CCFV. Results are the average of three experiments. Standard deviation bars are shown. 3′CCFV, 142 nt at the 3′ terminus of CCFV.
Replacement of TCV H5 with CCFV H5 (CH5) could putatively retain the important Ψ1 interaction, though it may contain an additional A·U pair and substitute a weaker G·U pair for a G·C pair (Fig. 3A). The presence of CCFV H5 reduced TCV accumulation to 14% of that of the wt (Fig. 3B), indicating that the differences between the hairpins significantly reduced TCV viability in the absence of additional surrounding sequences. In contrast, substitution of the highly related CCFV Pr for the TCV Pr (CPr) had no significant effect on chimeric TCV accumulation in protoplasts (Fig. 3B). This latter result is in contrast to an earlier evaluation of this construct, which suggested a more substantial effect on TCV accumulation (46). The reason for the discrepancy between these two sets of data is not known. However, support for the more recent data was obtained when CPr was independently retested in triplicate and found to accumulate at 76% of the wt levels (data not shown). Replacement of the TCV Link1 sequence with the comparable region of CCFV (CLink1) reduced virus levels by 39%, which may reflect the altered location and sequence forming Ψ2 (Fig. 3). All together, these results indicate that (i) the complete CCFV 3′ end is fully functional when replacing the analogous TCV segment and (ii) individual elements are not as viable when associated with surrounding TCV sequences.
To determine if substitution of multiple CCFV elements can enhance the function of individual elements, combinations of hairpins and linking sequences were substituted for the analogous regions in TCV. Recent studies replacing H4a and H4b of satC with those of CCFV revealed that satC accumulated to higher levels when both hairpins originated from the same virus (45). To determine if a similar compensatory effect exists in TCV, H4a and H4b from CCFV were substituted for the analogous hairpins in TCV. CH4a/CH4b accumulated to 46% of TCV levels, a 2.2-fold improvement in comparison to the substitution of H4a alone (Fig. 3B). This suggests that a relationship between H4a and H4b also exists for TCV. Substitution of the CCFV sequence from H4b through H5 (CH4b→CH5) improved chimeric virus accumulation by 3.2-fold in comparison to the substitution of H5 alone, also indicating a possible connection between H5 and adjacent upstream sequences.
TCV accumulated to only 11 or 13% of that of the wt, respectively, when the substitutions included H4a and H5 (CH4a/CH5, with TCV H4b and Link2 sequences unchanged) or the entire region from H4a through H5 (CH4a→CH5) (Fig. 3B). However, when three single-base substitutions just upstream and downstream of H4a and H5 were included in the latter construct (CΨ3→CΨ2), accumulation was enhanced to 65% of the wt levels. The single-nucleotide substitution near the base of H4a is located just downstream of the Ψ3-interacting sequence and within the DR element (CGGUGG; the underlined residue was replaced with the CCFV cytidylate), which is required for the conformational shift from the preactive to active structure in satC (43). The two substitutions downstream of H5 are in the sequence that forms Ψ2 (Fig. 3A). When the CCFV DR residue was combined with substitutions of H4a and H4b (CΨ3→CH4b), chimeric TCV accumulation was also enhanced over the substitution of just H4a and H4b, reaching greater levels than those of parental TCV (Fig. 3B). These results suggest that a 100-nt region encompassing Ψ3→Ψ2 forms a functional domain, and subsets of the included elements, especially Ψ3→H4b, also perform more efficiently when associated with cognate sequences.
CCFV H5 and Pr do not form a fully functional domain in TCV.
A recent report suggested that TCV H5 could replace H5 of HCRSV RNA if the substitution included the TCV Pr (37). HCRSV, however, does not form discernible H4a or H4b, and thus, exchanges that are tolerated by HCRSV may not be similarly tolerated by TCV. To examine the possible association of elements in the region from H5 to the 3′ end, TCV constructs were generated containing CCFV Link1 and either H5 or Pr (CH5/CLink1; CLink1/CPr), H5 and Pr (CH5/CPr), and the sequence from H5 to the 3′ end (CH5→3′ end). CH5/CLink1 and CLink1/CPr accumulated less than the replacement of the hairpins alone (8% and 34% of that of the wt), indicating that hairpin function was not improved by association with flanking cognate linker sequences. CH5/CPr accumulation was low (23% of that of the wt), but was nearly twofold higher than the substitution of just H5, suggesting that a weak association between H5 and Pr may exist. The addition of Link1 and the 3′ tail sequence to this combination (CH5→3′ end), however, did not permit the virus to accumulate to detectable levels. These results suggest that, in contrast with the results of Wang and Wong (37), where substitution of TCV H5 and Pr resulted in wt levels of chimeric HCRSV, the H5 and Pr of CCFV do not lead to robust accumulation in a TCV background in the presence or absence of Link1 and the single-stranded tail. Since 3′CCFV was as viable as that of wt TCV, these results also indicate that the function of the CCFV H5→3′ end requires the cognate Ψ3→H4b. However, since the combination of CCFV Ψ3→H4b and TCV H5→3′ end was fully functional, the presence of the TCV H5→3′ end region eliminates sensitivity to the origin of the upstream Ψ3→H4b unit. The reason for the different outcomes of the reciprocal exchanges of the Ψ3→H4b and H5→3′ end regions is not known.
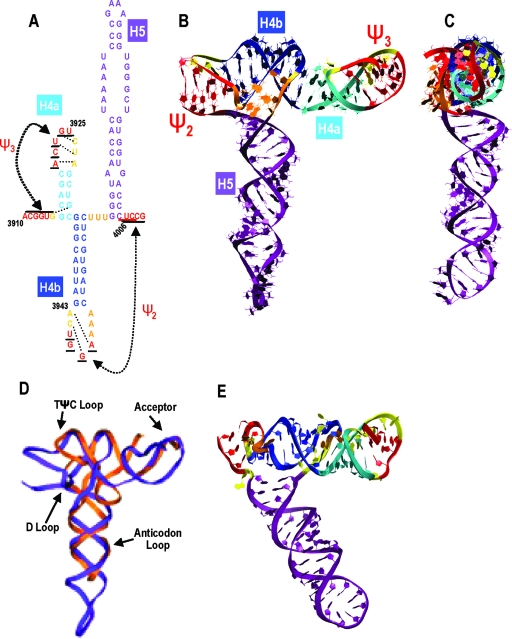
The region encompassed by Ψ2 and Ψ3 is predicted to form a stable structure that bears some resemblance to a tRNA.
To determine if elements comprising the Ψ3→Ψ2 domain are capable of simultaneous existence, a 3-D modeling protocol was employed involving initial prediction by RNA2D3D, followed by molecular modeling and MD simulation (18, 33, 40). Several starting structures for the TCV model were used, with different positions of the bases not participating in the secondary structure base pairing. The Ψ3→Ψ2 region was predicted to stably fold into a single structural domain that is T-shaped (Fig. 4). The two pseudoknots with H4a and H4b were predicted to coaxially stack, with the H5 helix folding perpendicularly to the stacked structure. Other tertiary contacts involved triple base-pair formations in both Ψ2 and Ψ3, which were disrupted over the course of the MD trajectory. Superimposing the predicted TCV structure onto a canonical tRNA revealed some tRNA-like topology in the H4b/H4a helix and in the lower portion of the H5 stem (Fig. 4D). The upper stem-loop of H5, however, extends beyond the position of a tRNA anticodon loop. Using the TCV structure as a model, the CCFV analogous region was predicted by RNA2D3D to form a bent T-shaped structure (Fig. 4E). The molecular modeling predictions thus support the substitution analyses that indicate that the region from Ψ3→Ψ2 is capable of forming a structural domain.
FIG. 4.
T-shaped structure predicted by RNA2D3D and molecular modeling. (A) Ψ3→Ψ2 region of TCV subjected to RNA2D3D and molecular modeling. The secondary structure of the TCV modeled region with predicted tertiary interactions is indicated by dotted lines. (B) Predicted tertiary structure of TCV colored according to the secondary structure shown in panel A. Ψ2 and Ψ3 are red, and unpaired bases are yellow. (C) Side view of the predicted TCV structure. (D) Superimposed structures of the TCV region between Ψ2 and Ψ3 (purple) and Phe-tRNA (orange). tRNA sites are indicated. (E) Predicted structure of CCFV region from Ψ2 through Ψ3.
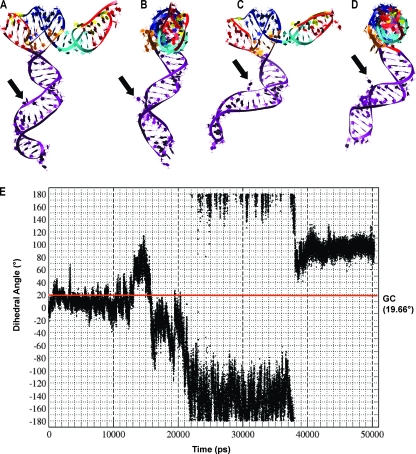
MD simulation performed on the 3-D TCV model shown in Fig. 4B and C (the Ψ3→Ψ2 region) indicated several intervals of differing stability. During the approximately first 12 to 13 ns, helix H5 moves as a relatively rigid structure. However, even early on, nucleotide C3994 begins to intermittently rotate out into the major groove, which can be seen in the graph of its dihedral angle relative to the preceding base G3993 (Fig. 5E). Past the 13-ns point, rearrangements in the interactions in the central internal loop region of helix H5 lead to its distortions away from the helical form, and, past the 17.5-ns point, to the bending of the entire helix toward the 3′ end of the modeled structure. Figure 5A and B illustrate such an early bent state, while Fig. 5C and D show a much deeper bend in H5 at 37.0 ns in the MD trajectory. For short periods of time between the states illustrated in Fig. 5, bending toward the 3′ end is even more pronounced (not shown). Finally, past the 38.0-ns point, H5 begins to return to a form similar to that shown in Fig. 5A. Exceptional rotational freedom of C3994 is captured in a graph of the dihedral angles over the MD simulation time, measured between nucleotides G3993 and C3994 (atoms G[N1], G[C3′], C[C3′], and C[N3]). Nucleotide G3993 is relatively, though not completely, rotationally stable, and thus, most of the change in the measured angle is due to C3994 movement, first at an angle to the major groove and then to the minor groove, and then through the solvent back into the major groove. The observed distortions of the H5 internal loop region and the resulting bending of the entire H5 helix follow the movements of this nucleotide. Stabilization of the angle past the approximately 38.0-ns point is due to C3994 forming multiple interactions with upstream nucleotides G3992 and G3993, which also marks the beginning of the return to the less-distorted and less-bent conformation of the H5 helix (Fig. 5E). This dynamic behavior of C3994 corresponds with the results of in-line probing of fragment F4 in the presence and absence of Ψ1 (Fig. 2), which showed that C3994 had enhanced susceptibility to cleavage compared with other residues in the H5 LSL. Additionally, the stability of the non-Watson-Crick base pair A3968-G4001, illustrated as a small symmetric internal loop in the TCV secondary structure drawings, showed it was a strong pair that maintained the helicity of H5 in the MD simulations.
FIG. 5.
MD simulation of the Ψ3→Ψ2 region. The 3-D structures are colored according to the code shown in Fig. 4. The selected structures illustrate rearrangements of interactions in the central internal loop region of helix H5, the full 360° rotational mobility of C3994 (denoted by the black arrow), and deformations of the helix H5, which bends toward the 3′ end of the modeled fragment. (A) Tertiary structure from the 21.6-ns point in the MD simulation. (B) Side view of the structure shown in panel A. (C) Tertiary structure from the 37.0-ns point. (D) Side view of the structure shown in panel C. (E) Graph of the dihedral angles over the MD simulation time, measured between the neighboring nucleotides G3993 and C3994: G(N1), G(C3′), C(C3′), and C(N3). Nucleotide G3993 is relatively stable, and most of the change in the angle is due to C3994 movement. The red line is the reference angle measured for the same atoms in an ideal RNA A-type helix (19.66°). Points above the line correspond (approximately, due to some G3993 movement) to the nucleotide C3994 pointing out into the major groove; points below the line indicate movement into the minor groove. Stabilization of the angle past the approximately 38.0-ns point is due to the interactions of C3994 with G3992 and G3993.
JINRV does not contain similar functional domains in its 3′ UTR.
Japanese iris necrotic ring virus (JINRV) shares 55% pairwise sequence similarity across the genome with TCV, with 50% sequence similarity in the 3′ UTR. One possible structure for the 3′ end of JINRV, based on the known structure of TCV and predictions by mFold, is shown in Fig. 6A. Compared with TCV elements, the JINRV Pr is more divergent than that of CCFV, with 12 possible paired bases and a dissimilar terminal loop, and Link1 is increased in length with little sequence similarity. The H5 LSL and upper stem-loop are nearly identical in JINRV and TCV, whereas the lower portion of the hairpins has little sequence similarity, and the lower internal loop in JINRV H5 is larger and asymmetric (Fig. 6B). The sequence upstream of H5 is able to fold into a putative H4a and H4b, with Link2 being larger in comparison with TCV or CCFV (Fig. 6A). The putative H4b in JINRV contains a shorter stem and loop than the comparable hairpin in TCV, and the conserved loop sequence is limited to “UGG.” The H4b loop could possibly form a 4-nt Ψ2 with the sequence located one base downstream of H5. The proposed JINRV H4a has no sequence similarity with TCV, and only two Watson-Crick base pairs are possible for Ψ3 formation.
FIG. 6.
The JINRV 3′ end does not contain similar functional domains. (A) Possible structure of the JINRV 3′ end based on the TCV 3′-end model and mFold structural predictions. The designation of elements is according to the known elements from TCV. (B) Comparison of sequences between TCV and JINRV. Underlined bases in the JINRV sequences denote differences with TCV. Putative Ψ3, Ψ2, and Ψ1 that could form when JIRNV sequences are partnered with TCV background sequences are shown. The wt TCV pseudoknot pairings are also shown. Filled triangles denote absent bases. (C) Relative accumulation of chimeric TCV RNAs in protoplasts at 40 hpi compared with that of TCV-TSNLS (TCV), as assayed by RNA gel blots using a TCV-specific probe. The identity of the replacement elements is given. Results are the average of three experiments. Standard deviation bars are shown.
While substitution of the entire CCFV 3′ end was fully functional, substituting the JINRV 3′ end for that of TCV did not produce detectable virus (data not shown). To determine if single JINRV elements can substitute for the comparable elements in TCV, putative H4a, H4b, H5, Link1, and Pr from JINRV were substituted for their counterparts in TCV and the chimeric viral RNAs were assayed for accumulation in protoplasts. JINRV H4a, which can maintain four of five Ψ3 base pairings, was poor at replacing the TCV sequence, with accumulation of JH4a at 16% of the wt TCV levels (Fig. 6C). Replacement of the TCV Link1 sequence with the comparable region of JINRV reduced virus levels by 64%, which may reflect the altered location and sequence that would comprise Ψ2. H4b and Pr were unable to substitute for the TCV sequences, with no detected accumulation of JH4b or JPr. JINRV H5 was a poor substitute, with JH5 accumulating to only 11% of that of the wt (Fig. 6C). Substitutions of multiple elements, including JINRV versions of all the combinations described in Fig. 3 for CCFV, were uniformly detrimental with none accumulating to detectable levels in protoplasts (data not shown). This suggests that, unlike CCFV, JINRV elements either do not exist or are not forming similar functional domains, or the domains are not compatible with other cis- or trans-acting TCV factors necessary for replication and/or translation.
Symmetry and position of the H5 small internal loop are important for TCV accumulation.
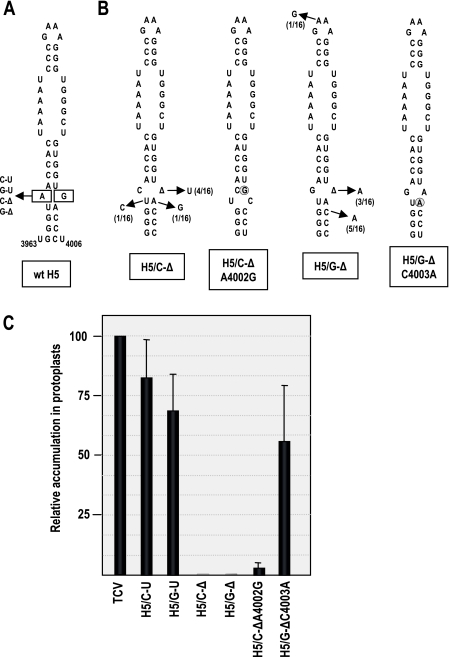
JINRV H5 substituted poorly for TCV H5 though the JINRV LSL and upper stem-loop are nearly identical with those of TCV (Fig. 6B). The H5 lower stem, however, differs substantially between TCV and JINRV, with the JINRV H5 lower stem having a larger and asymmetric internal loop. To determine the importance of specific residues or symmetry of the lower internal loop for TCV H5 function, mutations were engineered into this region in wt TCV and viral accumulation levels were measured in protoplasts. Replacing the A-G loop with C-U (H5/C-U) reduced TCV accumulation by only 17% (Fig. 7C), suggesting that the identities of non-Watson Crick residues are not critical in this portion of the hairpin. Closing the lower internal loop by replacing the A-G with G-U (H5/G-U) was more detrimental, reducing accumulation in protoplasts by 31%. To create asymmetry in the lower internal loop, the A-G was replaced with either C-Δ or G-Δ, forming H5/C-Δ and H5/G-Δ, respectively (Fig. 7A). Accumulation of H5/C-Δ and H5/G-Δ in protoplasts was below the level of detection (Fig. 7C), suggesting that symmetry in the lower stem is of critical importance for H5 function in TCV.
FIG. 7.
Importance of the TCV H5 small symmetrical loop. (A) Boxed nucleotides denote residues comprising the small symmetrical loop in TCV H5. Mutations generated in the loop are indicated. Triangles denote deleted bases. (B) Two mutants (H5/C-Δ and H5/G-Δ) assayed in planta produced progeny with single second-site alterations. Second-site changes within H5 and the number of clones containing the alterations at 20 dpi out of 16 clones sequenced are shown. Possible H5 structures of H5/C-Δ containing an adenylate-to-guanylate alteration at position 4002 (H5/C-ΔA4002G) and H5/G-Δ containing a cytidylate-to-adenylate alteration at position 4003 (H5/G-ΔC4003A) are shown. The second-site mutations are circled. (C) Relative accumulation levels of mutants in comparison to TCV-TSNLS (TCV) in protoplasts at 40 hpi. Results are the average of three experiments. Standard deviation bars are shown.
To determine if second-site alterations that improve accumulation could be recovered in the region, transcripts of H5/C-U, H5/G-U, H5/C-Δ, and H5/G-Δ were inoculated onto turnip seedlings, and progeny at 20 days postinoculation were cloned and sequenced. Progeny of H5/C-U and H5/G-U maintained the alterations with no second-site changes within H5. In contrast, six different second-site mutations were identified in H5 for H5/C-Δ and H5/G-Δ (Fig. 7B). Four of the 16 H5/C-Δ progeny clones sequenced contained an insertion of a uridylate opposite the bulged cytidylate, which would result in the well-tolerated C-U symmetrical bulge at this location (Fig. 7C). An additional clone contained an adenylate-to-guanylate transition at position 4002 (H5/C-ΔA4002G). This new guanylate could pair with the bulged cytidylate, with a symmetrical loop restored by C4003 forming an interior loop with U3967 and C4004/C4005/U4006 pairing with the three guanylates in the H5 lower stem (Fig. 7B). The third second-site alteration (U3967C) would disrupt an additional base pair adjacent to the bulged cytidylate.
Of the three second-site alterations associated with H5/G-Δ, two are predicted to reform a symmetrical lower loop. Three of 16 clones contained an adenylate inserted opposite the bulged guanylate, whereas 5 of 16 clones converted the cytidylate at 4003 to an adenylate. The latter alteration is predicted to generate a G-A pairing in the lower stem, with C4004/C4005/U4006 forming canonical base pairs with G3964/G3965/G3966 (Fig. 7B). The third alteration converted the GAAA tetraloop to GGAA. To assess if some of the second-site alterations were enhancing TCV accumulation, two mutants containing second-site alterations predicted to reform a symmetrical lower loop were generated and tested for accumulation in protoplasts. H5/C-ΔA4002G accumulated to low but detectable levels (3% of the wt levels), whereas H5/G-ΔC4003A accumulated to 56% of the wt TCV levels. The greater compensatory effect of C4003A compared with that of A4002G suggests that placement of the loop within the lower stem of H5 may be an additional important feature of TCV H5.
DISCUSSION
The 3′ UTRs of plant and animal RNA viruses were once thought to be exclusively dedicated to promoting complementary strand synthesis. However, an increasing number of studies report the presence of 3′ elements unrelated to poly(A) tails that are required for or enhance the translation of viruses with and without 5′ methylguanosine nucleotide caps (7, 8). Antagonistic processes such as translation and replication can be controlled by the dynamic nature of RNA, which allows for conformational transitions producing different structural folds to promote particular activities (27). Genetic assays that employ mutagenic approaches to disrupt and reform putative structures can confirm the existence of particular elements but do not reveal which elements functionally coexist. Biochemical structure probing provides a “snapshot” of the structure of an in vitro-generated RNA. However, since RNA can assume multiple intermediate states with similar stabilities when transitioning between alternative, biologically relevant conformations (16, 27, 47), it is difficult to determine if an RNA fragment is assuming a functional, productive conformation or one that is not valid due to the absence of interactive elements or the trapping of intermediates during folding. The use of RNA fragments to assess the efficiency of transcription in vitro can also provide misleading information regarding the importance of particular sequences, especially if the loss of interacting sequences causes a conformational switch from a nonactive to an active form (43).
Addressing these concerns requires additional approaches to identify elements in RNA viruses that coexist and interact to form functional domains. Our approach consisted of first mapping additional secondary and tertiary structures in the 3′ UTR region of TCV, followed by exchanges of structures and intervening sequences with those of related carmoviruses. The following four elements in TCV were identified or verified in this report: the juxtaposed hairpins H4a and H4b; Ψ3, which links the terminal loop of H4a with upstream flanking sequences; and Ψ2, which bridges the terminal loop of H4b and sequence flanking the 3′ side of H5. We recently found that Ψ3 does not form or has no importance in untranslated satC (R. Guo, J. Zhang, and A. E. Simon, unpublished data). However, the region flanking H4a in satC (the DR) is important for accumulation in vivo, a result that correlates with a role for the DR in the conformational switch activating satC templates in vitro (43).
In-line probing of fragment F4 produced cleavages on the 5′ side of the lower stem of H5 (Fig. 2). The absence of cleavages in all but one of the residues on the 3′ side of the phylogenetically conserved lower stem suggests that the 3′ side bases are not paired with the 5′ side in the F4 fragment. Elimination of Ψ1 (F4-m18) abolished the strong cleavage site at position C4005, possibly due to the relief of torsional stress. However, absence of the pseudoknot did not affect the remaining cleavages in the lower H5 stem, showing that their presence is unrelated to Ψ1. The absence of Ψ1 enhanced cleavages in the 5′ side of the LSL in F4-m18 (Fig. 2), suggesting that these residues participate in Ψ1, possibly through triple base-pair interactions. The phylogenetically conserved H5 structure (42) is also absent from the satC preactive structure (43), though genetic selection (SELEX) of the complete satC lower H5 stem indicated that pairing in this region is important (44). Therefore, complete formation of H5 in TCV is likely occurring in another conformer that is necessary for viral replication.
Of the known carmoviruses, only CCFV and JINRV have 3′ UTR regions that have the capacity to form all or nearly all of the TCV-like hairpins and pseudoknots. Substitution of comparable single elements from CCFV into TCV revealed that only TCV containing CCFV H4b (CH4b) and Pr (CPr) maintained at least 50% of its normal accumulation in protoplasts (Fig. 3B). While substitutions of some combinations of elements reduced TCV levels further than any of the single exchanges (e.g., CH4a/H5 and CH5/Link1), several combinations enhanced accumulation over single-element replacements, suggesting the existence of a modular, cointeracting function (Fig. 3). For example, substitution of both H4a and H4b (CH4a/CH4b) enhanced accumulation by 2.2-fold compared with substitution of H4a alone (CH4a), similar to the finding for substitutions of CCFV H4a and H4b in satC (45). Since the H4a/H4b module is important for both TCV and untranslated satC, an unknown interaction between these two hairpins, or between their sequences in an alternative conformation, may be important for replication. Surprisingly, altering one additional residue (position 3914) from the TCV uridylate to the CCFV cytidylate (CΨ3→CH4b) enhanced the accumulation of CH4a/H4b by threefold (Fig. 3B). This residue is located at the edge of Ψ3, eliminating a possible A·U pair, and converted the DR to the satC-equivalent sequence (CGGCGG). The reason that CΨ3→CH4b accumulates efficiently is unknown, and we are currently determining if the residue at position 3914 influences the function of H4a or H4b individually.
A larger functional module appears to form from elements encompassed by Ψ3→Ψ2. Substitution of the region between H4a and H5 (CH4a→CH5) was poorly tolerated, with accumulation of the chimeric TCV reaching only 13% of that of the wt, similar to the accumulation level of virus containing only CCFV H5 (14% of the wt level) (Fig. 3B). When three CCFV residues just upstream and downstream of the region were included in the construct, accumulation reached 65% of that of the wt, suggesting a more functional H5 was now present. Compared with CH4a→CH5, inclusion of these additional residues would not discernibly alter the stability of Ψ3, but would alter Ψ2 by eliminating one A·U pair and converting a G·U pair to an A·U pair. Which of the three residues (or if all three) enhance the interactive function of the Ψ3→Ψ2 region is currently unknown.
Subjecting the Ψ3→Ψ2 region in TCV to the RNA2D3D modeling protocol revealed the possible formation of a highly stable, T-shaped structural domain with two flanking pseudoknots. A similar structure with a bent H5 was also predicted for the analogous region of CCFV, supporting our substitution results that this region forms a functional module. The complete 3′ end of CCFV (3′-CCFV) was also fully functional when substituted, in contrast to substituting CCFV H5→3′ end, which did not lead to detectable accumulation. These results suggest a critical interaction is required between elements in the two regions, such as connecting H5 with 5′ and 3′ elements that together form the T-shaped structure.
C3994 was particularly susceptible to cleavage in F4 and F4-m18. This suggested that in the presence or absence of Ψ1, C3994 is more dynamic than surrounding nucleotides, allowing it to more readily assume the in-line conformation required for cleavage. This result was predicted by the MD simulation of the predicted 100-nt T-shaped structure, which revealed unique 360° flexibility in C3994 that correlated with movement of the H5 helix toward the 3′ end of the modeled TCV fragment. The bending of the H5 helix toward the 3′ end did not require Ψ1 but may facilitate its formation in the full 3′-terminal region of TCV (Fig. 1B). Previous mutagenesis of C3994 (to a guanylate) (46) revealed a 14-fold decrease in TCV accumulation, compared with only a threefold decrease in TCV levels when its presumptive Ψ1 partner residue (G4001) was altered to a cytidylate. TCV with the combined mutations achieved only 50% of the wt levels, suggesting a requirement for particular nucleotides at one of both locations. satC did not accumulate to detectable levels with the identical single and compensatory mutations (42), which may reflect different requirements in the usage of H5, since satC is not predicted to form a T-shaped structure in this region (Y. Yingling, B. Shapiro and A. Simon, unpublished data).
TCV with JINRV H5 (JH5) was poorly functional (11% of that of the wt), despite an identical sequence in the LSL and the presence of a similar GNRA tetraloop (Fig. 7C). The major differences between TCV H5 and JINRV H5 are in the lower stem, which contains an asymmetric internal loop in JINRV H5 and a symmetrical loop in TCV and CCFV H5. The poor viability of TCV containing an asymmetric lower loop in its H5, and the enhanced accumulation of TCV that reestablishes symmetry through second site alterations, suggest that a symmetrical lower loop is critical for H5 function in TCV. This difference between TCV and JINRV H5 may reflect the participation of TCV H5 in the Ψ3→Ψ2 domain, which does not appear to be present in JINRV.
A recent report suggested that TCV H5 and Pr were fully functional in HCRSV if substituted together (37). The same substitutions of CCFV hairpins into TCV allowed accumulation to only 23% of that of the wt. While this value was greater than substitution of H5 alone (14% of that of the wt), the effect was far less than the substitutions into HCRSV. We also previously demonstrated that CCFV H5 and Pr do not form a functional unit in satC (45). The HCRSV sequence upstream of H5 is very pyrimidine rich, with no possibility of forming H4a or H4b. Therefore, TCV H5 and Pr may be capable of efficiently replacing the HCRSV hairpins when added in tandem because H5 is not part of a comparable functional domain with upstream elements in HCRSV.
Recently, Li and Wong (17) reported that TCV mutants containing deletions in the 3′ UTR accumulated at or near wt levels in Arabidopsis protoplasts, including extensive deletions that removed nearly all of the 3′ UTR (except the Pr) or substantially altered the structure of the Pr. These results contradict our current results, as well as three previous reports not cited by Li and Wong (4, 19, 46). We therefore have no explanation for the variant results reported by Li and Wong (17).
In conclusion, we have identified a novel 100-nt structural domain composed of three hairpins and two pseudoknots that is predicted to fold into a T-shaped structure. Interestingly, this region was recently found to be part of the TCV 3′ translational enhancer and binds to 60S ribosomal subunits (V. Stupina, A. Meskauskas, J. McCormack, J. Dinman, B. Shapiro, and A. Simon, submitted for publication). We are continuing to examine how this element interacts with surrounding sequences and how viruses in the same genus perform similar functions in the apparent absence of comparable contiguous elements.
Acknowledgments
We are very grateful to Ping Yu for providing us with T7 RNA polymerase.
This work was supported by grants from the U.S. Public Health Service (GM 061515-05A2/G120CD and GM 61515-01) and the National Science Foundation (MCB-0086952) to A.E.S. J.C.M. was supported by NIH Institutional Training Grant no. T32-AI51967-01. This research was supported in part by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. This publication has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400. The computational support was provided in part by the National Cancer Institute's Advanced Biomedical Computing Center.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W.-M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino, R., N. Böddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 776-82. [DOI] [PubMed] [Google Scholar]
- 3.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 813684-3690. [Google Scholar]
- 4.Carpenter, C. D., J.-W. Oh, C. Zhang, and A. E. Simon. 1995. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J. Mol. Biol. 245608-622. [DOI] [PubMed] [Google Scholar]
- 5.Case, D. A., T. E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K. M. Merz, Jr., A. Onufriev, C. Simmerling, B. Wang, and R. J. Woods. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 261668-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, L., C. U. Rang, L. E. Wong. 2002. Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage ϕ6. J. Virol. 763276-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreher, T. W., and W. A. Miller. 2006. Translational control in positive strand RNA plant viruses. Virology 344185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgil, D., and E. Harris. 2006. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 11943-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essmann, U., L. Perera, M. L. Berkowitz, T. A. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 1038577-8593. [Google Scholar]
- 10.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker, D. L., I. T. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 1861-8. [DOI] [PubMed] [Google Scholar]
- 12.Kasprzak, W., and B. A. Shapiro. 1999. Stem Trace: an interactive visual tool for comparative RNA structure analysis. Bioinformatics 1516-31. [DOI] [PubMed] [Google Scholar]
- 13.Koev, G., S. Liu, R. Beckett, and W. A. Miller. 2002. The 3′-terminal structure required for replication of Barley yellow dwarf virus RNA contains an embedded 3′ end. Virology 292114-126. [DOI] [PubMed] [Google Scholar]
- 14.Koh, D. C., D. X. Liu, and S. M. Wong. 2002. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 761144-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PloS Biol. 5e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leulliot, N., and G. Varani. 2001. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 407947-7956. [DOI] [PubMed] [Google Scholar]
- 17.Li, W., and S.-M. Wong. 2007. Host-dependent effects of the 3′ untranslated region of turnip crinkle virus RNA on accumulation in Hibiscus and Arabidopsis. J. Gen. Virol. 88680-687. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, H. M., J. V. Maizel, and B. A. Shapiro. 2008. RNA2D3D: a program for generating, viewing and comparing 3-dimensional models of RNA. J. Biomol. Struct. Dyn. 25669-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormack, J. C., and A. E. Simon. 2004. Biased hypermutagenesis associated with mutations in an untranslated hairpin of an RNA virus. J. Virol. 787813-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Na, H., and K. A. White. 2006. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology 345305-316. [DOI] [PubMed] [Google Scholar]
- 21.Olsthoorn, R. C., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 184856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 7910608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 225602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu, F., and T. J. Morris. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 741085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanfaçon, H. 2005. Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can. J. Bot. 831529-1549. [Google Scholar]
- 27.Schroeder, R., A. Barta, and K. Semrad. 2004. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 5908-919. [DOI] [PubMed] [Google Scholar]
- 28.Schuppli, D., J. Georgijevic, and H. Weber. 2000. Synergism of mutations in bacteriophage Qbeta RNA affecting host factor dependence of Qbeta replicase. J. Mol. Biol. 295149-154. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro, B. A., D. Bengali, W. Kasprzak, and J. C. Wu. 2001. RNA folding pathway functional intermediates: their prediction and analysis. J. Mol. Biol. 31227-44. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro, B. A., and W. Kasprzak. 1996. STRUCTURELAB: a heterogeneous bioinformatics system for RNA structure analysis. J. Mol. Graph. 14194-205. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro, B. A., W. Kasprzak, C. Grunewald, and J. Aman. 2006. Graphical exploratory data analysis of RNA secondary structure dynamics predicted by the massively parallel genetic algorithm. J. Mol. Graph. Model. 25514-531. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro, B. A., J. C. Wu, D. Bengali, and M. J. Potts. 2001. The massively parallel genetic algorithm for RNA folding: MIMD implementation and population variation. Bioinformatics 17137-148. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro, B. A., Y. G. Yingling, W. Kasprzak, and E. Bindewald. 2007. Bridging the gap in RNA structure prediction. Curr. Opin. Struct. Biol. 17157-165. [DOI] [PubMed] [Google Scholar]
- 34.Simon, A. E., and S. H. Howell. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′ end of the helper virus genome. EMBO J. 53423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 2546-14. [DOI] [PubMed] [Google Scholar]
- 36.Sun, X., and A. E. Simon. 2006. A cis-replication element functions in both orientations to enhance replication of Turnip crinkle virus. Virology 35239-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, H. H., and S. M. Wong. 2004. Significance of the 3′-terminal region in minus-strand RNA synthesis of Hibiscus chlorotic ringspot virus. J. Gen. Virol. 851763-1776. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J. M., P. Cieplak, and P. A. Kollman. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 211049-1074. [Google Scholar]
- 39.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 9915908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yingling, Y. G., and B. A. Shapiro. 2006. The prediction of the wild-type telomerase RNA pseudoknot structure and the pivotal role of the bulge in its formation. J. Mol. Graph. Model. 25261-274. [DOI] [PubMed] [Google Scholar]
- 41.Yoshii, M., M. Nishikiori, K. Tomita, N. Yoshioka, R. Kozuka, S. Naito, and M. Ishikawa. 2004. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 786102-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, G., J. Zhang, and A. E. Simon. 2004. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 787619-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, G., J. Zhang, A. T. George, T. Baumstark, and A. E. Simon. 2006. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA 12147-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, J., and A. E. Simon. 2005. Importance of sequence and structural elements within a viral replication repressor. Virology 333301-315. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, J., G. Zhang, R. Guo, B. A. Shapiro, and A. E. Simon. 2006. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J. Virol. 809181-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J., G. Zhang, J. C. McCormack, and A. E. Simon. 2006. Evolution of virus-derived sequences for high-level replication of a subviral RNA. Virology 351476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Q., A. C. Stelzer, C. K. Fisher, and H. M. Al-Hashimi. 2007. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 4501263-1267. [DOI] [PubMed] [Google Scholar]