Abstract
Varicella-zoster virus (VZV) open reading frame (ORF) 63 protein (ORF63p) is one of six VZV ORFs shown to be transcribed and translated in latently infected human dorsal root ganglia. ORF63p accumulates exclusively in the cytoplasm of latently infected sensory neurons, whereas it is both nuclear and cytoplasmic during lytic infection and following reactivation from latency. Here, we demonstrate that infection of primary guinea pig enteric neurons (EN) with an adenovirus expressing ORF63p results in the exclusive cytoplasmic localization of the protein reminiscent of its distribution during latent VZV infection in humans. We show that the addition of the simian virus 40 large-T-antigen nuclear localization signal (NLS) results in the nuclear import of ORF63p in EN and that the ORF63p endogenous NLSs are functional in EN when fused to a heterologous protein. These data suggest that the cytoplasmic localization of ORF63p in EN results from the masking of the NLSs, thus blocking nuclear import. However, the coexpression of ORF61p, a strictly lytic VZV protein, and ORF63p in EN results in the nuclear import of ORF63p in a proteasome-dependent manner, and both ORF63p NLSs are required for this event. We propose that the cytoplasmic localization of ORF63p in neurons results from NLS masking and that the expression of ORF61p removes this block, allowing nuclear import to proceed.
Varicella-zoster virus (VZV), a neurotropic alphaherpesvirus, is the etiological agent of chicken pox (varicella) and shingles (zoster). Upon primary infection of humans, VZV invades the dermis and epidermis, resulting in chicken pox. Following primary infection, the virus infects cranial nerve and dorsal root ganglia to form a latent infection. VZV can subsequently reactivate to cause shingles (28). During latency, only a subset of VZV genes is expressed. Transcripts (11-17, 19, 37, 49) and proteins (15, 29, 37, 46, 48) from open reading frames (ORFs) 4, 21, 29, 62, 63, and 66 have been detected in latently infected human ganglia. These latency-associated proteins (LAPs) appear to accumulate only in the cytoplasm of latently infected neurons. This is in contrast to their nuclear and cytoplasmic localizations during lytic and reactivated VZV infection (29, 46, 48). These data suggest a correlation between the nuclear import of LAPs and the outcome of VZV infection. Of all the transcripts detected in latently infected human ganglia, ORF63 is the most prevalent and abundant (13, 16). In addition, ORF63 transcripts and protein were detected in the ganglia of experimentally infected rodents (8, 21, 38). Therefore, ORF63 transcription and accumulation of the protein in the cytoplasm of latently infected neurons are some of the hallmarks of VZV latency.
VZV ORF63 encodes a 278-amino-acid protein (ORF63p) (20) that is expressed as an immediate-early protein and is present in the viral tegument (41). The protein is extensively phosphorylated during transient expression and lytic VZV infection in cell culture by both host and viral kinases (2, 5, 18, 30, 33, 39, 40, 61). Mutational analysis has identified two nuclear localization signals (NLSs) within the C terminus that are required for nuclear localization (2, 5). ORF63p is a transcriptional repressor and activator of cellular and viral promoters (5, 22, 24, 30, 31, 35, 42, 47, 68). It carries out these functions by interacting with ORF62p (the major VZV transactivator protein), RNA polymerase II, and the host transcriptional machinery (2, 24, 47, 58). Recently, ORF63p was shown to promote the survival of cultured primary human neurons by inhibiting apoptosis (34). In addition, by inhibiting the phosphorylation of the alpha subunit of eukaryotic initiation factor 2, ORF63p antagonizes the alpha interferon-induced antiviral response (1). These functions suggest possible roles of ORF63p in the establishment and maintenance of VZV latency in humans. In support of this, Cohen et al. previously suggested that ORF63p is critical for the establishment of latency in rodents (9, 10).
The subcellular localization of ORF63p appears to correlate with the fate of VZV infection. Therefore, an understanding of the mechanisms regulating the localization of ORF63p (and other LAPs) during VZV infection may expand our understanding of virus-cell interactions involved in the establishment and maintenance of VZV latency.
VZV infection of primary guinea pig enteric neuronal cultures provides a potential model for studying latency (8, 59). Following infection of enteric neurons (EN), VZV expresses a subset of LAPs, and these proteins localize exclusively in the cytoplasm. The localization of these proteins in the EN is reminiscent of their localization during latent VZV infection of human neurons. To further understand the mechanisms regulating VZV LAP localization in a neuronal system, we used cultures of EN to investigate ORF63p localization in the absence and presence of other viral proteins.
In this report, we analyze the differential subcellular localization of ORF63p in guinea pig epithelial (EP) cells and EN. Our results demonstrate that ORF63p accumulates in nuclear and cytoplasmic compartments in a cell-type-dependent manner and does not require other viral proteins for this process. We show that ORF63p is both nuclear and cytoplasmic in EP cells; however, it is exclusively cytoplasmic in EN. The localization of ORF63p in EP cells and EN is reminiscent of its distribution during lytic and latent VZV infection in humans. The nuclear import of ORF63p can occur in EN if the simian virus 40 (SV40) large-T-antigen NLS is fused to the protein, and ORF63p's NLSs are functional in EN when fused to a heterologous protein. These data suggest that the cytoplasmic localization of ORF63p in EN results from the masking of the NLSs, thus blocking nuclear import. However, the coexpression of ORF61p, a lytic VZV protein, and ORF63p in EN results in the nuclear import of ORF63p, and both NLSs are required for this event. Finally, we demonstrate that the ORF61p-dependent nuclear import of ORF63p in EN requires an active proteasome. We propose that the cytoplasmic localization of ORF63p in neurons results from NLS masking and blocking of nuclear import. However, the coexpression of ORF61p results in the release of this block in a proteasome-dependent manner, allowing the nuclear import of ORF63p. The experiments described in this report demonstrate that the localization of the most abundant latency-associated protein, ORF63, in neurons is regulated by a strictly lytic protein.
MATERIALS AND METHODS
Mammalian cells.
Transformed guinea pig EP cells (ATCC CCL-242) were maintained as monolayers in Dulbecco's modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, Carlsbad, CA) at 37°C in a 5% CO2 atmosphere. Twenty-four hours prior to infection, the cells were seeded into six-well tissue culture dishes. For indirect immunofluorescence experiments, glass coverslips were added to wells before seeding. During and after infection, cells were maintained in DMEM supplemented with 2% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Guinea pig EN were isolated and cultured as previously described (8). In brief, ganglia were seeded for culture in two-well chamber slides (Nalge Nunc International, Rochester, NY) coated with 10 μg/ml each of poly-d-lysine (Sigma, St. Louis, MO) and mouse laminin (Invitrogen, Carlsbad, CA) in DMEM-F12 (Gibco, Carlsbad, CA) supplemented with 2% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml gentamicin, and 5.25 μg/ml amphotericin B (Fungizone). Mitotic inhibitors (10 μM 5-fluoro-2′-deoxyuridine, 10 μM uridine, and 1 μM cytosine β-d-arabinofuranoside; Sigma, St. Louis, MO) were also added. Prior to infection, the mitotic inhibitors were removed, and the cells were subsequently cultured in their absence. For infection of guinea pig EN, the multiplicity of infection (MOI) per ganglion was calculated. For infection of guinea pig EP cells, the MOI per cell was calculated.
Adenoviruses.
Adenovirus expressing ORF63 (AdORF63), AdCompletePhos, AdORF63SV40NLS, AdORF63NLS1, AdORF63NLS2, AdORF63NLS1+2, AdLuc, AdLucSV40NLS, AdLucNLS1, AdLucSubNLS1, AdLucNLS2, and AdLucSubNLS2 were all constructed using a pDC516-derived expression plasmid where the insert was transcribed from a murine cytomegalovirus promoter and a pBHGfrtΔE1 E3FLP system (Microbix Biosystems, Toronto, Ontario, Canada) (53). AdEmpty, AdORF61, and AdORF61ires63 were constructed using empty pDC516, pCK-mflag-ORF61, and pCK-m61ires63, respectively. All adenoviruses were plaque purified, and their DNAs were sequenced.
Plasmid construction.
All PCRs for ORF63 and luciferase were carried out using Vent DNA polymerase (New England Biolabs, Inc., Beverly, MA) unless otherwise indicated. All ORF63 wild-type and mutant PCR products were amplified using primers containing EcoRI (forward primer) and SacI (reverse primer) restriction enzyme sites. Utilizing these unique restriction sites, each ORF63 PCR product was ligated into the plasmid pDC516 as an EcoRI/SacI restriction digestion fragment. Wild-type ORF63 was amplified from VZV genomic DNA (Jones strain) using specific forward (F) and reverse (R) primers (F primer 5′-GGGAATTCGCCACCATGTTTTGCACCTCACCGGCT-3′ and R primer 5′-GAGCTCATAAAGACTACACGCCATGGG-3′) and cloned to yield pDC516-ORF63. The CompletePhos mutant was amplified using the same primers and pLXIN-CompletePhos− as a template (2) to yield pDC516-CompletePhos. ORF63SV40NLS was amplified using F primer 5′-GGGAATTCGCCACCATGCCCAAGAAGAAGCGTAAGGTAATGTTTTGCACCTCACCGGCT-3′, which encodes the SV40 large-T-antigen NLS (PKKKRKV); R primer 5′-GAGCTCATAAAGACTACACGCCATGGG-3′; and pDC516-ORF63 as a template. The subsequent PCR product was cloned to create pDC516-ORF63SV40NLS. The mutant ORF63NLS1 PCR product was generated in a three-step method. First, a 687-bp PCR fragment was generated using pDC516-ORF63 as a template with F primer 5′-GGGAATTCGCCACCATGTTTTGCACCTCACCGGCT-3′ and R primer 5′-TGCTGCTGCTGCTGGGGACGGGGTGTTGCACC-3′. The last 12 nucleotides of the reverse primer result in the mutation of NLS1 within ORF63p from KRPQ to AAAA. Second, another PCR fragment of 158 bp was generated using pDC516-ORF63 as a template and specific primers (F primer 5′-GCAGCAGCAGCACGTGCCATCGAGCGATACGCG-3′ and R primer 5′-GAGCTCATAAAGACTACACGCCATGGG-3′). The first 12 nucleotides of the forward primer result in mutations of NLS1 within ORF63p from KRPQ to AAAA. These resulting PCR fragments generated from two independent reactions contain 12 bp of overlapping sequence homology. The overlapping PCR products were gel purified and used in a single PCR with F primer 5′-GGGAATTCGCCACCATGTTTTGCACCTCACCGGCT-3′ and R primer 5′-GAGCTCATAAAGACTACACGCCATGGG-3′ to generate full-length ORF63NLS1 and, subsequently, pDC516-ORF63NLS1. The ORF63NLS2 PCR product was generated using F primer 5′-GGGAATTCGCCACCATGTTTTGCACCTCACCGGCT-3′, R primer 5′-GGGAGCTCCTACACACCATGGGGGGGCGGTATATCATGCCGGCGCGGGGCTTCGTGTGCTGCTGCTGCCCAATCTACACCCCCCTC-3′, and pDC516-ORF63 as a template. The reverse primer is designed to mutate NLS2 of ORF63p from KRRR to AAAA. The subsequent cloning of the PCR product yielded pDC516-ORF63NLS2. The ORF63NLS1+2 PCR product was generated using the same primer set used for ORF63NLS2; however, pDC516-ORF63NLS1 was used as a template. The subsequent cloning of the PCR product created pDC516-ORF63NLS1+2.
All luciferase wild-type and mutant PCR products were amplified using primers containing HindIII (forward primer) and SacI (reverse primer) restriction enzyme sites, and each luciferase PCR product was cloned into pDC516 as a HindIII/SacI fragment. Luciferase PCR fragments were amplified from pcDNA-Firefly luciferase (a gift from Daniel Wolf, Columbia University, New York, NY) as a template and F primer 5′-GGAAGCTTGCCACCATGGAAGACGCCAAAAACATAAAG-3′ in conjunction with a specific reverse primer. These primers are Luc (R primer 5′-GGGAGCTCTTACAATTTGGACTTTCCGCCCTT-3′), LucSV40NLS (R primer 5′-GGGAGCTCTTATACCTTACGCTTCTTCTTAGGCAATTTGGACTTTCCGCCCTT-3′), LucNLS1 (R primer 5′-GGGAGCTCTTACTCGATGGCACGCTGGGGTCTCTTTGGGGACGGGGTCAATTTGGACTTTCCGCCC TT-3′), LucSubNLS1 (R primer 5′-GGGAGCTCTTACTCGATGGCACGT GCTGCTGCTGCTGGGGACGGGGTCAATTTGGACTTTCCGC CCTT-3′), LucNLS2 (R primer 5′-GGGAGCTCTTACGGGGCTTCGTGT CGACGTCGCTTCCAGTCTACACCCAATTTGGACTTTCCGCCCTT-3′), and LucSubNLS2 (R primer 5′-GGGAGCTCTTACGGTGCTTCGTGTGCTGCTGCTGCCCAGTCTACACCCAATTTGGACTTTCCGCCCTT-3′). Following PCR amplification, the fragments were cloned into pDC516 to create pDC516-Luc, pDC516-LucSV40NLS, pDC516-LucNLS1, pDC516-LucSubNLS1, pDC516-LucNLS2, and pDC516-LucSubNLS2, respectively.
The ORF61 gene was amplified from VZV genomic DNA (Jones strain) using Pfu polymerase (Stratagene, La Jolla, CA) and primers 5′-Eco-61 (5′-GGGAATTCTACCATGGATACCATATTAGC-3′) and 3′-Sal-61 (5′-CCGTCGACCCCAACAAACTAGGACTTCT-3′). The PCR product was cloned into pCR2.1 TOPO TA (Invitrogen, Carlsbad, CA) to yield pCK-ORF61. An EcoRI/SalI digestion fragment from pCK-ORF61 was cloned into EcoRI/SalI-digested pCMV-Tag2A to create pCK-flag-ORF61. pCK-mflag-ORF61 was constructed by cloning a NotI (Klenow-filled)/SalI digestion fragment from pCK-flag-ORF61 into EcoRI (Klenow-filled)/SalI-digested pDC516.
An ORF61-internal ribosome entry site (IRES)-ORF63 expression construct was made after first amplifying the three individual components using Pfu polymerase (Stratagene, La Jolla, CA) and primers Notflag61 (5′-CCGCGGCCGCCACCATGGATTAC-3′), IRES61 (5′-ACCCCCGGCTTTCAACTAGGACTTCTTCATCTTGTTTGGA-3′), 61IRES (5′-ATGAAGAAGTCCTAGTTGAAAGCCGGGGGTGGGAGAT-3′), 63IRES (5′-CGGTGAGGTGCAAAACATATTATCATCGTGTTTTTCAAAGGA-3′), IRES63 (5′-AAACACGATGATAATATGTTTTGCACCTCACCGGCTAC-3′), and Sac63 (5′-CCGAGCTCAGACTACACGCCATGGGGGGG-3′) and pCK-flagORF61, pE-C4 (32), and VZV genomic DNA (Jones strain) as templates, respectively. The three overlapping PCR products were gel purified and used in a single PCR with Pfu polymerase (Stratagene, La Jolla, CA) and primers Notflag61 and Sac63. The resulting PCR product was cloned into pCR-TOPO 2.1 TA (Invitrogen, Carlsbad, CA) to yield pCK-61ires63. pCK-m61ires63 was constructed by ligating a NotI (Klenow-filled)/SacI fragment from pCK-61ires63 into EcoRI (Klenow-filled)/SacI-digested pDC-516. All primers used were manufactured by Proligo LLC (Boulder, CO), and all vector inserts were verified by DNA sequencing.
Indirect immunofluorescence microscopy.
Cells grown on glass coverslips or two-well chamber slides were washed twice with phosphate-buffered saline (PBS), fixed for 20 min with 3.7% formaldehyde in PBS, washed two more times in PBS, and permeabilized with PBS plus 0.2% Triton (Sigma, St. Louis, MO) for 10 min at room temperature. Cells were washed twice more in PBS and blocked with 10% normal goat serum (Roche, Indianapolis, IN) in PBS plus 0.1% Tween 20 (PBST) (Sigma, St. Louis, MO) for 30 min. Cells were incubated with a 1:100 dilution of the appropriate primary antibody in 10% normal goat serum in PBST for 1 h and then washed three times for 5 min in PBST. Cells were then incubated for 1 h with an Alexa Fluor-conjugated secondary antibody diluted 1:1,000 in 10% normal goat serum in PBST and then washed twice with PBST. Once washed, cells were incubated for 30 min with PBST plus 0.5 μg/ml Hoechst stain (Sigma, St. Louis, MO). Following incubation, cells were washed twice for 5 min with PBST and then once with PBS. Coverslips were subsequently mounted with Gel/Mount (Biomeda, Foster City, CA).
All samples were visualized with a Zeiss Axiovert 200 M inverted microscope using a 63× objective (Carl Zeiss Microimaging Inc., Thornwood, NY), and images were acquired with a Hamamatsu C4742-80-12AG digital charge-coupled-device camera (Hamamatsu Photonics, Hamamatsu City, Japan) using Openlab 5 software (Improvision, Lexington, MA). Images were analyzed using Openlab 5 and assembled in Photoshop CS3 (Adobe Systems, San Jose, CA).
SDS-PAGE and Western blotting.
Infected or transfected cells were washed twice with cold PBS, scraped from tissue culture dishes, resuspended in radioimmunoprecipitation lysis buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM NaF) plus Complete protease inhibitor cocktail (Roche, Mannheim, Germany), and incubated on ice for 30 min. Halt phosphatase inhibitor cocktail (Pierce, Rockford, IL) was also added when phosphorylation levels of ORF63p were analyzed. Lysates were clarified by centrifugation at 22,500 × g for 10 min in a Tomy MX-160 high-speed refrigerated microcentrifuge. The total protein concentration was measured using the Bio-Rad (Hercules, CA) protein assay (7). SDS sample buffer (5×) (250 mM Tris HCl [pH 6.8], 500 mM dithiothreitol, 10% SDS, 0.5% bromophenol blue, 50% glycerol) was added to each sample before boiling for 10 min and SDS-polyacrylamide gel electrophoresis (PAGE) analysis (44). Proteins were transferred onto nitrocellulose membranes with a Bio-Rad Semi-Dry apparatus before Western blotting. After blocking membranes in 4% nonfat milk in PBST, immobilized proteins were reacted with ORF63p and c-Jun antibodies at a 1:1,000 dilution and tubulin or luciferase antibody at a 1:4,000 dilution in 4% nonfat milk in PBST. Membranes were washed three times for 5 min each with PBST, incubated with an anti-rabbit or anti-mouse antibody conjugated to horseradish peroxidase at a 1:5,000 dilution, and washed again three times for 5 min with PBST and twice with PBS, and antibodies were visualized after the addition of LumiGLO substrate (KPL, Gaithersburg, MD) by exposure to X-ray film.
Cell fractionation.
Cytoplasmic and nuclear fractions were prepared as previously described (45) except that Triton X-100 was used at a final concentration of 0.025% to lyse the plasma membrane.
Radioactive labeling of cells.
Proteins were radiolabeled after starving infected or control cell cultures of inorganic phosphate (Pi) or l-methionine (Met) and cysteine (Cys) by washing cells three times with PBS and incubation in serum-free DMEM or Met-free (MET−), Cys− DMEM (Gibco, Carlsbad, CA), respectively, for 30 min. Starvation medium was replaced with labeling medium (serum-free or Met−, Cys− DMEM supplemented with 1% dialyzed calf serum) and 500 μCi/ml of 32Pi (carrier-free H332PO4) or 500 μCi/ml trans 35S label (ICN, Irvine, CA). After 16 h of labeling, cells were washed twice with PBS, and lysates were prepared as described above. ORF63p was immunoprecipitated as described below. The bound material was subjected to SDS-PAGE, and the proteins were transferred onto nitrocellulose membranes with a Bio-Rad Semi-Dry apparatus. The membranes were subsequently exposed to X-ray film.
Immunoprecipitation.
Immunoprecipitations were performed as previously described (43).
Drug treatment.
Infected EN cultures were treated with either dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) or 20 μM MG132 (EMD Biosciences, La Jolla, CA) in growth medium as indicated in the figure legends.
Antibodies.
Rabbit polyclonal antibody against amino acids 1 to 265 of ORF63p was described previously (46).
A portion of ORF61 encoding amino acids 136 to 248 was cloned into pALEX (54). The glutathione S-transferase fusion protein was overexpressed in Escherichia coli strain BL21(DE3) purified to apparent homogeneity by affinity chromatography (57) and used to immunize rabbits. ORF61p-specific antibodies were purified by affinity chromatography as described previously (43).
Mouse monoclonal anti-Flag M5 antibody was purchased from Sigma (St. Louis, MO). Mouse monoclonal antibody to human tubulin and rabbit polyclonal antibody to human c-Jun were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibody to firefly luciferase was purchased from Abcam (Cambridge, MA). Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 546-conjugated anti-rabbit antibodies were obtained from Invitrogen (Carlsbad, CA). Goat anti-rabbit and anti-mouse antibodies conjugated to horseradish peroxidase for immunoblotting were obtained from KPL (Gaithersburg, MD).
RESULTS
ORF63p localizes exclusively to the cytoplasm of guinea pig EN.
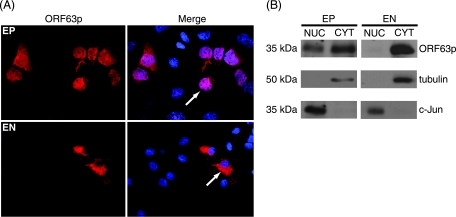
To further understand LAP localization in neurons, we used cultures of EN to investigate the localization of ORF63p in the absence and presence of other VZV proteins. For these studies, we constructed a replication-deficient adenovirus expressing ORF63p (AdORF63). Guinea pig EP cells and EN were infected with AdORF63, and the localization of ORF63p was assayed at 48 h postinfection by indirect immunofluorescence microscopy (Fig. 1A). As observed during lytic VZV infection, ORF63p was detected in the nucleus and cytoplasm of EP cells. However, ORF63p was exclusively cytoplasmic in EN, reminiscent of its localization during latency. Therefore, the intracellular localization of ORF63p is cell type dependent and occurs in the absence of other VZV proteins. The subcellular localization of ORF63p in EP and EN was examined after separation into nuclear and cytoplasmic fractions. Proteins in these fractions were subjected to Western blot analysis (Fig. 1B). These blots confirmed that ORF63p is present in both compartments in EP cells; however, it is exclusively cytoplasmic in EN. Costaining for c-Jun, a nuclear marker, and tubulin, a cytoplasmic marker, validated the fractionation procedure. This biochemical analysis supports the microscopy data and demonstrates that ORF63p localization in EN cells mimics that found during latency in human neurons. Thus, this system provides a model for studying the contribution of host and viral proteins to the differential localization of VZV LAPs during lytic and latent infections.
FIG. 1.
Subcellular localization of ORF63p. Guinea pig EP cells and primary guinea pig EN were infected with AdORF63 at MOIs of 0.2 and 100, respectively. (A) The subcellular localization of ORF63p was examined 48 h postinfection by indirect immunofluorescence microscopy using ORF63p-specific antisera. Nuclei were counterstained with Hoechst stain. (B) The subcellular localization of ORF63p was examined 48 h postinfection after biochemical separation into nuclear (NUC) and cytoplasmic (CYT) fractions. Equal amounts of total protein for each fraction (20 μg and 5 μg for EN and EP, respectively) were analyzed by Western blotting using antisera specific for ORF63p, c-Jun (nuclear marker), and tubulin (cytoplasmic marker).
ORF63p is phosphorylated in guinea pig EN.
ORF63p expressed from VZV or plasmid-derived vectors is phosphorylated by both viral and cellular kinases (2, 5, 18, 30, 33, 39, 40, 61). Phosphorylation of ORF63p is known to affect its cellular localization (5, 30). Therefore, we asked if ORF63p is phosphorylated in EN and, if so, if the mutation of specific serine and threonine phosphorylation sites alters its localization. Cultures of EN were mock infected or infected with AdORF63, and at 28 h postinfection, cells were washed, starved for 30 min in labeling medium, and labeled with either trans 35S label or 32Pi. After labeling for 20 h, whole-cell extracts were prepared, and ORF63p was captured by ORF63p-specific antisera bound to Sepharose beads. Bound proteins were subjected to SDS-PAGE and visualized by autoradiography (Fig. 2B). A specific protein (∼35 kDa in mass) was detected in infected cells labeled with 35S. This protein, which corresponds to the molecular weight of ORF63p, was absent from mock-infected lysates. These data demonstrate that ORF63p was expressed, labeled, and successfully identified from lysates of EN infected with AdORF63. In AdORF63-infected EN labeled with 32P, a ∼35-kDa protein was also immunoprecipitated, and it was specific for AdORF63-infected lysates, demonstrating that ORF63p is phosphorylated in cultures of EN.
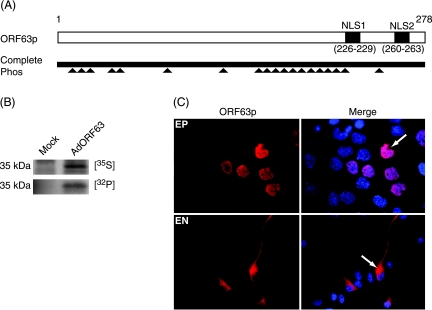
FIG. 2.
Phosphorylation of ORF63p in primary guinea pig EN. (A) Schematic of wild-type ORF63p and the CompletePhos mutant. ORF63p is a 278-amino-acid phosphoprotein that contains two NLSs (NLS1 and NLS2) located between amino acids 226 and 229 and 260 and 263. Mutations of putative phosphorylation sites (▴) were made by alanine substitution of 19 serine (S) and threonine (T) residues (S12, S13, S15, T41, S42, S82, S129, S165, S171, S173, S181, S185, S186, S197, T201, S203, T222, S224, and T244) that are predicted targets of cellular S/T kinases (CompletePhos). (B) EN were mock infected or infected with AdORF63 at an MOI of 100. At 28 h postinfection, cells were labeled with trans 35S label or 32Pi for 20 h. At the end of the labeling period, whole-cell extracts were prepared, and equal amounts of total protein were immunoprecipitated, as described in Materials and Methods, with antisera directed against ORF63p. Immunoprecipitated ORF63p was resolved by SDS-PAGE and visualized by autoradiography. (C) Subcellular localization of the CompletePhos mutant. EP cells and EN were infected with AdCompletePhos at MOIs of 0.2 and 100, respectively. The subcellular localization of CompletePhos was examined 48 h postinfection by indirect immunofluorescence microscopy using ORF63p-specific antisera. Nuclei were counterstained with Hoechst stain.
A mutant ORF63 gene with alanine substitutions at 19 putative phosphorylation sites (CompletePhos) (Fig. 2A) (2) was used to determine what effect the lack of phosphorylation of ORF63p has on localization in EN. The protein encoded by this mutant has a reduced phosphorylation level compared to that of wild-type ORF63p (2). Its subcellular localization was examined in EP cells and EN infected with a replication-deficient adenovirus expressing the mutant protein (AdCompletePhos). The subcellular localization of this protein was assayed by indirect immunofluorescence microscopy (Fig. 2C). The CompletePhos mutant showed increased nuclear localization in EP cells compared to those of the wild-type protein (Fig. 1A). This result confirmed a previous report showing that the inhibition of phosphorylation at serine/threonine residues results in the increased nuclear localization of ORF63p in Vero cells (30). However, the CompletePhos protein was exclusively cytoplasmic in EN and localized like wild-type ORF63p. Moreover, the treatment of EN expressing wild-type ORF63p with an inhibitor (Roscovitine) or activator (phorbol-12-myristate-13-acetate) of phosphorylation had no effect on ORF63p localization (data not shown). Therefore, we conclude that the nuclear exclusion of ORF63p in EN is probably not regulated by phosphorylation.
The SV40 large-T-antigen NLS directs nuclear import of ORF63p in EN.
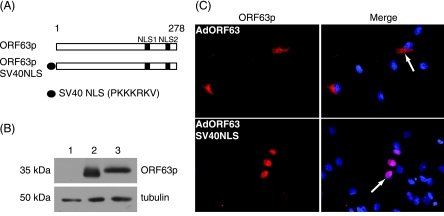
To gain further insight into why ORF63p accumulation is restricted to the cytoplasm in EN, we asked if the addition of a strong heterologous NLS would drive the nuclear import of ORF63p. Accordingly, the NLS from SV40's large T antigen (36) was fused to the N terminus of ORF63p, and a replication-deficient adenovirus expressing this protein (AdORF63SV40NLS) was generated (Fig. 3A). Western blot analysis revealed that both wild-type and SV40-ORF63p fusion proteins accumulated to equivalent levels (Fig. 3B). EN were infected with AdORF63 or AdORF63SV40NLS, and ORF63p localization was assayed by indirect immunofluorescence microscopy (Fig. 3C). Wild-type ORF63p localized exclusively to the cytoplasm of EN; however, the addition of the SV40 T-antigen NLS resulted in its accumulation in the nucleus. Thus, the localization of ORF63p in EN can be changed from cytoplasmic to nuclear by the addition of a functional heterologous NLS.
FIG. 3.
Subcellular localization and accumulation of an ORF63p-SV40 NLS fusion protein in primary guinea pig EN. (A) Schematic diagram of ORF63p SV40 NLS fusion protein. The SV40 large-T-antigen NLS amino acid sequence (PKKKRKV) ( ) was fused in frame to the N terminus of ORF63p (ORF63pSV40NLS). (B) Western blot analysis of ORF63p and ORF63pSV40NLS fusion proteins. EP were mock infected (lane 1) or infected with AdORF63 (lane 2) or AdORF63SV40NLS (lane 3) at MOIs of 0.2, and 48 h postinfection, whole-cell lysates were harvested. Equal amounts of total protein (20 μg) for each sample were analyzed by Western blotting using antisera specific for ORF63p and tubulin (loading control). (C) EN were infected with AdORF63 or AdORF63SV40NLS at an MOI of 100. The subcellular localization of ORF63p was examined 48 h postinfection by indirect immunofluorescence microscopy using ORF63p-specific antisera. Nuclei were counterstained with Hoechst stain.
) was fused in frame to the N terminus of ORF63p (ORF63pSV40NLS). (B) Western blot analysis of ORF63p and ORF63pSV40NLS fusion proteins. EP were mock infected (lane 1) or infected with AdORF63 (lane 2) or AdORF63SV40NLS (lane 3) at MOIs of 0.2, and 48 h postinfection, whole-cell lysates were harvested. Equal amounts of total protein (20 μg) for each sample were analyzed by Western blotting using antisera specific for ORF63p and tubulin (loading control). (C) EN were infected with AdORF63 or AdORF63SV40NLS at an MOI of 100. The subcellular localization of ORF63p was examined 48 h postinfection by indirect immunofluorescence microscopy using ORF63p-specific antisera. Nuclei were counterstained with Hoechst stain.
The endogenous NLSs of ORF63p are functional in EN when fused to firefly luciferase.
The addition of the SV40 T-antigen NLS to ORF63p demonstrated that it could be driven into the nucleus of EN by the addition of a functional heterologous NLS. Therefore, the cytoplasmic localization of ORF63p in EN may result from masking the endogenous NLSs of ORF63p or their being nonfunctional in EN. Two NLSs (NLS1 and NLS2) in the C-terminal domain of ORF63p (Fig. 2A) have been described (2, 5, 61). To determine if these NLSs function in EN, we fused each one to the C terminus of the cytoplasmic protein firefly luciferase to mimic their location in ORF63p. For each fusion protein, 12 amino acids of the ORF63p sequence spanning each NLS were fused to the C terminus of luciferase to generate LucNLS1 and LucNLS2 (Table 1). Subsequently, using alanine substitution mutagenesis, the four core amino acids known to be essential for NLS function (2, 5) were altered. The resulting mutant fusion proteins LucSubNLS1 and LucSubNLS2 (Table 1) were used to validate the specificity of each ORF63p NLS sequence. As a positive control, the SV40 NLS was also fused to luciferase (LucSV40NLS) (Table 1). Replication-deficient adenoviruses were constructed to express each fusion protein.
TABLE 1.
Subcellular localization of luciferase NLS fusion proteins in guinea pig EP cells and ENa
| Adenovirus | Amino acid sequence of NLS fused to C terminus of firefly luciferase | ORF63 amino acids | Subcellular localization of luciferase NLS fusion protein
|
|
|---|---|---|---|---|
| EP | EN | |||
| AdLuc | C | C | ||
| AdLucSV40NLS | PKKKRKV | N+ | N+ | |
| AdLucNLS1 | TPSPKRPQRAIE | 222-233 | N | N |
| AdLucSubNLS1 | TPSPAAAARAIE | 222-233 | C | C |
| AdLucNLS2 | GVDWKRRRHEAP | 256-267 | N+ | N+ |
| AdLucSubNLS2 | GVDWAAAAHEAP | 256-267 | C | C |
C, cytoplasmic staining; N, nuclear staining; N+, major nuclear staining.
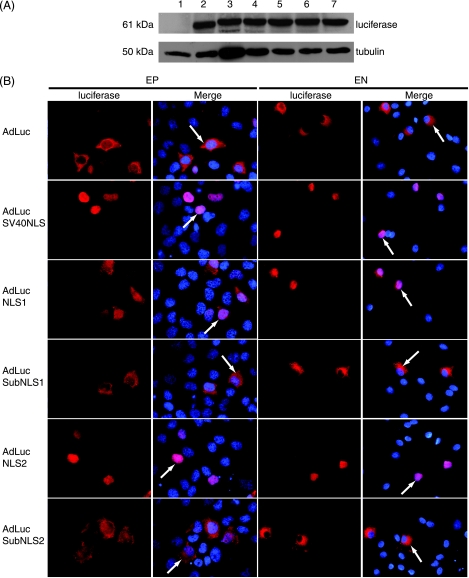
The expression of the fusion proteins and their subcellular localization in EP cells were examined following infection with each adenovirus (Table 1). Infected cells were processed for Western blot analysis or indirect immunofluorescence microscopy. Western blot analysis revealed that each fusion protein was expressed at equivalent levels. Therefore, the fusion of NLS sequences to luciferase did not affect protein stability (Fig. 4A). As expected, luciferase was predominantly cytoplasmic in EP cells, whereas the addition of the SV40 NLS resulted in nuclear localization (Fig. 4B). The addition of NLS1 also resulted in the nuclear accumulation of luciferase, whereas the substitution of the core KRPQ sequence with alanines abolished nuclear import. ORF63p NLS2 also directed the nuclear import of luciferase, and the alanine-substituted core KRRR sequence abolished import. These data reveal that in EP cells, both NLS1 and NLS2 can direct the nuclear import of a heterologous protein and function in the same manner as when they are part of ORF63p.
FIG. 4.
Subcellular localization of ORF63p NLS-luciferase fusion proteins in guinea pig EP cells and primary guinea pig EN. (A) Western blot analysis of ORF63p NLS-luciferase fusion proteins. EP cells were mock infected (lane 1) or infected with AdLuc (lane 2), AdLucSV40NLS (lane 3), AdLucNLS1 (lane 4), AdLucSubNLS1 (lane 5), AdLucNLS2 (lane 6), or AdLucSubNLS2 (lane 7) at an MOI of 0.2, and 48 h postinfection, whole-cell lysates were harvested. Equal amounts of total protein (20 μg) for each sample were analyzed by Western blotting using antisera specific for luciferase and tubulin (loading control). (B) Subcellular localization of ORF63p NLS-luciferase fusion proteins in EP and EN. EP and EN were infected with each adenovirus at MOIs of 0.2 and 100, respectively. The subcellular localization of the ORF63p NLS-luciferase fusion proteins was examined 48 h postinfection by indirect immunofluorescence microscopy using luciferase-specific antisera. Nuclei were counterstained with Hoechst stain.
We then determined the fate of these fusion proteins in EN. EN were infected with each adenovirus, and the subcellular localization of luciferase fusion proteins was assayed as described above. As expected, luciferase was predominantly cytoplasmic in EN, and the addition of the SV40 NLS resulted in its nuclear localization (Fig. 4B). Given that ORF63p is cytoplasmic in EN, we were surprised to observe that each NLS enabled nuclear import of luciferase in EN. Importantly, alanine substitution of the core sequences in both NLSs abrogated nuclear import. Thus, both NLS1 and NLS2 can function in EN. However, within the context of ORF63p, we hypothesize that they are inaccessible in EN and accordingly unavailable to direct nuclear import of ORF63p.
Coexpression of ORF61p and ORF63p results in nuclear import of ORF63p in EN.
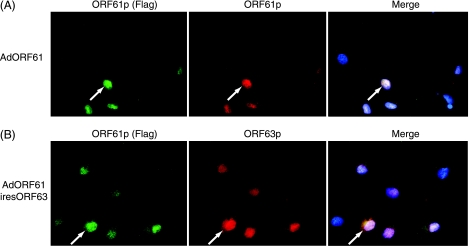
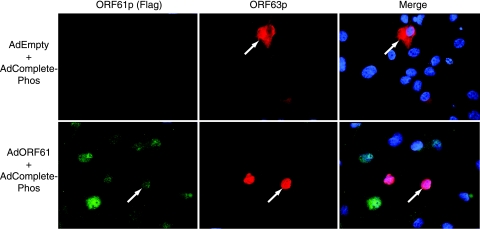
If there is an inherent block to nuclear import of ORF63p in EN, one might hypothesize that a similar block exists in human sensory neurons. If so, how does VZV overcome this block upon reactivation from latency? Stallings et al. previously demonstrated that latent VZV infection of EN can be reactivated after infection with an adenovirus expressing ORF61p. Upon the reactivation of VZV, LAPs such as ORF29p and ORF62p accumulate in the nucleus (59). Accordingly, we tested whether the expression of ORF61p was sufficient to cause the nuclear import of ORF63p in EN. An adenovirus expressing ORF61p and ORF63p (AdORF61iresORF63) was constructed. This virus expresses both proteins in the same cell. A Flag epitope was fused to the N terminus of ORF61p to facilitate its detection in dual-labeling analyses. An adenovirus expressing only Flag-ORF61p (AdORF61) was also constructed to determine ORF61p localization. EN were first infected with AdORF61 to examine the subcellular localization of the protein in this cell type. Indirect immunofluorescence analysis using antisera specific to either the Flag epitope or ORF61p (Fig. 5A) revealed that the protein localized in the nucleus of EN as previously described for other cell types (60). Staining patterns with Flag antibody and ORF61p-specific antisera were identical. To determine the effect of ORF61p expression on ORF63p localization in EN, cells were infected with AdORF61ires63, and protein localization was studied (Fig. 5B). For the first time, we observed the nuclear localization of ORF63p in EN. Thus, the presence of ORF61p abrogates the block to ORF63p nuclear import in EN. Furthermore, coinfection with individual adenoviruses expressing ORF61p and ORF63p also resulted in the nuclear import of ORF63p (data not shown). As expected, ORF63p remained cytoplasmic in EN when cells were coinfected with AdORF63 and a control empty adenovirus vector (AdEmpty) (data not shown). Interestingly, ORF61p expression caused the nuclear import of ORF63p CompletePhos in EN (Fig. 6), confirming our hypothesis that regulation of ORF63p localization in EN is not controlled by phosphorylation.
FIG. 5.
Subcellular localization of ORF61p and ORF63p following coexpression in primary guinea pig EN. (A) ORF61p subcellular localization in EN. EN were infected with AdORF61 at an MOI of 100. The subcellular localization of ORF61p was examined 48 h postinfection by indirect immunofluorescence microscopy using antisera specific for the Flag epitope and ORF61p. (B) ORF61p and ORF63p subcellular localization in EN following coexpression. EN were infected with AdORF61iresORF63 at an MOI of 100. The subcellular localization of ORF61p and ORF63p was examined 48 h postinfection by indirect immunofluorescence microscopy using antisera specific for the Flag epitope and ORF63p. Nuclei were counterstained with Hoechst stain.
FIG. 6.
Subcellular localization of ORF61p and ORF63p CompletePhos following coexpression in primary guinea pig EN. EN were coinfected with AdCompletePhos at an MOI of 100 with either AdEmpty or AdORF61 at an MOI of 10,000. The subcellular localizations of ORF61p and ORF63p CompletePhos were examined 48 h postinfection by indirect immunofluorescence microscopy using Flag epitope- and ORF63p-specific antisera, respectively. Nuclei were counterstained with Hoechst stain.
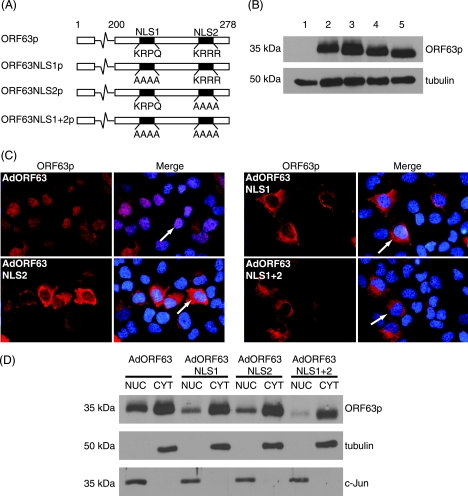
Both NLSs of ORF63p are required for ORF61p-dependent nuclear import in EN.
To further elucidate the requirements of the ORF61p-dependent nuclear import of ORF63p in EN, ORF63p NLS mutants containing alanine substitutions in one or both NLSs were constructed and cloned into an adenovirus background (ORF63NLS1p, ORF63NLS2p, and ORF63NLS1+2p) (Fig. 7A). EP were infected with each mutant adenovirus, and cells were processed for Western blot analysis, indirect immunofluorescence microscopy, or biochemical fractionation into nuclear and cytoplasmic fractions as described above. Western blots revealed the expression of each NLS mutant with the expected molecular mass at levels comparable to those of wild-type ORF63p (Fig. 7B). The variance in migration of the mutant proteins is a reflection of substitution of alanine residues for the indigenous amino acids within the NLSs. As described above, immunofluorescence analysis demonstrated that wild-type ORF63p localized to both the nucleus and cytoplasm of EP cells (Fig. 7C). However, the mutation of either NLS1 or NLS2 resulted in an increased abundance of cytoplasmic ORF63p. The abolition of both NLSs resulted in greater cytoplasmic accumulation of ORF63p, with an almost complete absence of protein in the nucleus. The subcellular distribution of ORF63p NLS mutants was confirmed by Western blot analysis of fractionated cell extracts (Fig. 7D). The distribution of ORF63p in the nuclear and cytoplasmic fractions confirmed the immunofluorescence data. Compared to wild-type ORF63p, the mutation of either NLS1 or NLS2 results in decreased levels of nuclear ORF63p, with the mutation of both NLSs enhancing this effect. Costaining for nuclear (c-Jun) and cytoplasmic (tubulin) markers validated the fractionation procedure.
FIG. 7.
Subcellular localization of ORF63p NLS mutants in guinea pig EP cells. (A) Schematic diagram of ORF63p NLS substitution mutants. The four core amino acids of each ORF63p NLS were substituted with alanines to disrupt NLS function. (B) Western blot analysis of ORF63p NLS mutants. EP cells were mock infected (lane 1) or infected with AdORF63 (lane 2), AdORF63NLS1 (lane 3), AdORF63NLS2 (lane 4), or AdORF63NLS1+2 (lane 5) at an MOI of 0.2, and 48 h postinfection, whole-cell lysates were prepared. Equal amounts of total protein (20 μg) for each sample were analyzed by Western blotting using antisera specific for ORF63p and tubulin (loading control). (C) Subcellular localization of ORF63p NLS mutants. EP cells were infected with AdORF63, AdORF63NLS1, AdORF63NLS2, or AdORF63NLS1+2 at an MOI of 0.2, and localization was examined 48 h postinfection by indirect immunofluorescence microscopy using ORF63p-specific antisera. Nuclei were counterstained with Hoechst stain. (D) The subcellular localization of ORF63p NLS mutants was examined 48 h postinfection after biochemical separation of nuclear (NUC) and cytoplasmic (CYT) fractions. Equal amounts of total protein for nuclear and cytoplasmic fractions (20 μg) were analyzed by Western blotting using antisera specific for ORF63p, c-Jun, and tubulin.
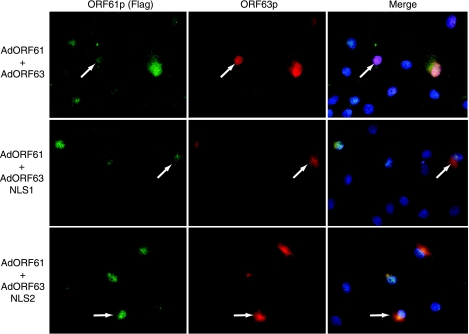
Because we discovered that coexpression of ORF61p targeted ORF63p to the nucleus, we questioned whether either of the NLSs within ORF63p were required for nuclear import in EN. Accordingly, EN were coinfected with AdORF61 and either AdORF63, AdORF63NLS1, or AdORF63NLS2. The localization of ORF61p and ORF63p was examined by indirect immunofluorescence microscopy. As expected, ORF63p was nuclear in cells coexpressing ORF61p (Fig. 8). However, both ORF63NLS1p and ORF63NLS2p remained cytoplasmic in cells coexpressing ORF61p. Therefore, both ORF63p NLSs are required for ORF61p-dependent nuclear import in EN.
FIG. 8.
Subcellular localization of ORF63p NLS mutants in primary guinea pig EN during coexpression of ORF61p. EN were coinfected with AdORF61 at an MOI of 10,000 with either AdORF63, AdORF63NLS1, or AdORF63NLS2 at an MOI of 100. The subcellular localizations of ORF61p and the ORF63p NLS mutants were examined after 48 h by indirect immunofluorescence microscopy using Flag epitope- and ORF63p-specific antisera, respectively. Nuclei were counterstained with Hoechst stain.
MG132 blocks ORF61p-dependent nuclear import of ORF63p in EN.
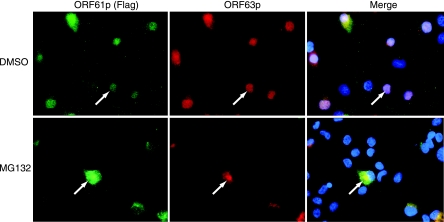
While ORF61p expression is sufficient to drive ORF63p nuclear import in EN, the mechanism behind this phenomenon remains unclear. ORF61p is a C3HC4 RING finger domain-containing protein and an orthologue of herpes simplex virus type 1 ICP0 (51, 55). The E3 ubiquitin ligase activity of ICP0 allows it to influence cell protein abundance via a ubiquitin-dependent proteasome degradation pathway (6, 23, 25, 26, 66). To test if proteasome activity is required for ORF61p-dependent nuclear import of ORF63p in EN, we treated EN infected with AdORF61iresORF63 with DMSO or MG132 for 6 h. Following drug treatment, the subcellular localizations of ORF61p and ORF63p were observed. MG132 treatment blocked nuclear import of ORF63p (Fig. 9). Thus, the ORF61p-dependent nuclear import of ORF63p in EN requires proteasome activity. Of note, we also observed that following MG132 treatment, there was an apparent increase in levels of cytoplasmic ORF61p. Cytoplasmic staining of ORF61p overlapped with that of ORF63p, suggesting that they colocalized and/or form part of a complex. Furthermore, MG132 treatment of EN expressing ORF61p alone also resulted in increased cytoplasmic staining compared to that with the DMSO-treated control (data not shown). However, the increase in levels of cytoplasmic ORF61p was far less than that observed in the presence of ORF63p. MG132 treatment of EN expressing ORF63p alone had no effect on its localization (data not shown). These experiments reveal roles for ORF61p and the proteasome in regulation of ORF63p localization in EN.
FIG. 9.
Subcellular localization of ORF61p and ORF63p in primary guinea pig EN treated with proteasome inhibitors. EN were infected with AdORF61iresORF63 at an MOI of 100. At 42 h postinfection, cells were treated with either DMSO or 20 μM MG132 for 6 h prior to analysis. The subcellular localization of ORF61p and ORF63p was examined by indirect immunofluorescence microscopy using Flag epitope- and ORF63p-specific antisera, respectively. Nuclei were counterstained with Hoechst stain.
DISCUSSION
The establishment and maintenance of a latent infection are critical for lifelong persistence by herpesviruses. During latency, VZV is novel among the neurotropic human alphaherpesvirus family in that multiple viral genes are transcribed (11-17, 19, 37, 49) and translated (15, 29, 37, 46, 48). The expressed LAPs accumulate exclusively in the cytoplasm; however, they are nuclear and cytoplasmic following reactivation (29, 46, 48). An understanding of the virus and host contributions to regulation of LAP localization in neurons may further our understanding of what regulates the fate of VZV infection.
In this report, we analyze the subcellular localization of the VZV LAP ORF63p in guinea pig EP cells and primary guinea pig EN. Our studies demonstrate that ORF63p subcellular localization is cell type dependent and occurs in the absence of other viral proteins. The localization of ORF63p in EP and EN cells mimics the protein's distribution during lytic and latent VZV infection, thus validating the use of guinea pig cells to study LAP localization (Fig. 1). We have also assayed the contribution of each of the ORF63p NLSs in maintaining the cytoplasmic localization of ORF63p in EN and discuss hypotheses to explain our observations. Finally, we show that nuclear import of ORF63p in EN is induced by the coexpression of ORF61p (a lytic protein) and that this occurs in a proteasome-dependent manner.
In eukaryotic cells, separation of the nuclear and cytoplasmic contents into discrete compartments requires controlled nuclear entry and exit. Molecules of >40 kDa are excluded from passive diffusion across the nuclear pore complex and must possess a specific targeting signal (NLS), or they will be excluded from the nucleus. NLS-containing molecules are actively transported between the cytoplasmic and nuclear compartments in an energy-dependent manner. The classical nuclear import cycle is based on a carefully timed series of interactions that are controlled spatially and temporally in a precise manner. In brief, cytoplasmic cargo proteins that contain a classical or nonclassical NLS form an import complex with an importin-α:β heterodimer (or directly with importin-β) that facilitates movement through the nuclear pore complex. Once an import complex reaches the nucleus, it is dissociated by RanGTP. The binding of RanGTP to importin-β causes a conformational change that results in release of the importin-α-cargo complex. Importin-β bound to RanGTP is then recycled to the cytoplasm, where conversion to RanGDP releases it to enable binding to new cargoes (56, 62, 63). Because of the multicomponent hierarchal nature of nuclear import, transport and, hence, localization of cargo proteins are regulated at several levels.
To gain further insight into why ORF63p localization is restricted to the cytoplasm in EN, we asked if the addition of a heterologous NLS could drive the nuclear import of ORF63p in these cells. Accordingly, we fused the NLS from the SV40 large T antigen to ORF63p and found that the resulting fusion protein localized to the nucleus (Fig. 3C). Thus, within EN, ORF63p can be moved into the nucleus by addition of a functional heterologous NLS. This finding suggested that nuclear exclusion of ORF63p might result from the NLSs being nonfunctional or masked in EN. One explanation for these findings is the differential expression of importin receptor families between EP cells and EN. The expression of the transport receptor that recognizes ORF63p for nuclear import may be absent in EN. Indeed, several studies have documented that transport can be regulated by changes in transport receptor or adaptor expression levels. For example, Drosophila heat shock factor is recognized by importin-α3 for nuclear transport. Importin-α3 is not expressed during early embryonic development; therefore, Drosophila heat shock factor nuclear entry is prohibited until later in development, when importin-α3 is expressed (27).
To determine if both ORF63p NLSs can function in EN, we fused each of them to the C terminus of the cytoplasmic protein luciferase and studied their intracellular localizations. Our results demonstrated that both NLS1 and NLS2 of ORF63p can function in EN and direct the nuclear import of luciferase; thus, the import receptors for ORF63p are present in EN (Fig. 4B). Therefore, our results support the conclusion that within the context of ORF63p, the NLSs are inaccessible to the nuclear import machinery in EN, resulting in cytoplasmic retention.
The masking of NLSs from the transport machinery may occur by multiple mechanisms including intermolecular or intramolecular interactions and/or posttranslational modifications. The modulation of nuclear import by phosphorylation is a common posttranslational modification. For example, the nuclear import of SV40 T antigen is enhanced by the phosphorylation of sites flanking its NLS (67). In contrast, phosphorylation of the NLSs of nuclear factor of activated T cells 2 (NF-AT2) results in masking and prevents recognition by importin, thus blocking nuclear import. The subsequent dephosphorylation unmasks the NLSs, permitting the nuclear import of NF-AT2 (3). Along these lines, previous studies showed that the phosphorylation state of ORF63p at specific residues can affect its localization (5, 30). Here, we show that ORF63p is phosphorylated in cultures of EN (Fig. 2B). To investigate what role phosphorylation of ORF63p had on its subcellular localization in EN, a mutant ORF63p with alanine substitutions at 19 putative serine and threonine phosphorylation sites (CompletePhos) (Fig. 2A) (2) was used to ask what effect the lack of phosphorylation had on its subcellular localization. The CompletePhos mutant showed increased nuclear localization in EP cells compared to the wild-type protein (compare Fig. 1A and 2C), a result that is consistent with a previous report showing that inhibition of phosphorylation results in increased nuclear localization of ORF63p in Vero cells (30). However, we show for the first time that this mutant remained exclusively cytoplasmic in EN behaving like the wild-type ORF63 protein. We also assayed the localization of three other phosphorylation mutants (5′Phos, CenterPhos, and 3′Phos), each containing six or seven alanine mutations present in the CompletePhos mutant (2). All of these mutant proteins were cytoplasmic like wild-type ORF63p in EN (data not shown). In addition to ORF63 mutagenesis, we also investigated the effect of inhibitors and inducers of phosphorylation on ORF63p localization. The treatment of EN expressing ORF63p with an inhibitor (Roscovitine) or activator (phorbol-12-myristate-13-acetate) of phosphorylation had no effect on localization (data not shown). Taken together, these genetic and pharmacological assays indicate that ORF63p localization in EN is not regulated by its phosphorylation state. This is in contrast to EP cells, where enhanced nuclear localization of ORF63p was observed upon dephosphorylation. However, our results do not exclude the possibility that modifications other than phosphorylation contribute to regulation of ORF63p localization in EN.
Here, we demonstrate for the first time that coexpression of ORF61p is sufficient to result in nuclear import of ORF63p in EN (Fig. 5). Thus, ORF61p expression overcomes the block to ORF63p nuclear import in EN. ORF61p is a C3HC4 RING finger domain-containing protein and functions as a transcriptional repressor and activator (50-52). Based on its position in the genome and homology of the RING finger-containing domain, ORF61p is an orthologue of herpes simplex virus ICP0 (51, 55). ICP0 has E3 ubiquitin ligase activity that influences cell protein abundance via the ubiquitin-dependent proteasome degradation pathway (6, 23, 25, 26, 66). At present, it is unknown if ORF61p contains E3 ubiquitin ligase activity. However, our finding that proteasome inhibition with MG132 prevents ORF61p-dependent nuclear import of ORF63p in EN supports a role for ubiquitination and protein degradation in controlling ORF63p nuclear import (Fig. 9). Based on these results, we hypothesize that ORF63p localization in EN is regulated by intermolecular masking of the NLSs in a fashion analogous to that of NF-κB. The nuclear import of the NF-κB heterodimer p65-p50 is regulated by the binding of IκBα, which occludes the NLS of p65-p50, resulting in its cytoplasmic localization. In the presence of proinflammatory stimuli, IκBα is phosphorylated and targeted for proteasome degradation, resulting in unmasking of the NF-κB NLS and subsequent nuclear import (4, 64). Therefore, we postulate that in EN, a neuron-specific protein binds to ORF63p, masking its NLSs and blocking nuclear import. This putative protein is targeted for proteasomal degradation in response to ORF61p, resulting in unmasking of the ORF63p NLSs and nuclear import.
Alternatively, ORF61p may compete with a neuronal protein for binding to ORF63p. The subsequent ORF61p-ORF63p interaction results in release of the inhibitory neuronal protein (which in turn may lead to its degradation) and unmasking of ORF63p's NLSs. Following MG132 treatment of EN, there was an increase in cytoplasmic staining of ORF61p that overlapped with ORF63p, suggesting that they colocalize and/or form part of a complex. It is plausible that treatment with MG132 stabilized formation of a transient ORF61p-ORF63p interaction with the inhibitory neuronal protein in the cytoplasm. Furthermore, MG132 treatment of EN expressing ORF61p alone also resulted in increased cytoplasmic staining; however, the increase in cytoplasmic ORF61p levels was far less than that observed when it was expressed with ORF63p. Although we recently detected a weak interaction between ORF61p and ORF63p in vitro, it has not been possible to reliably detect this interaction in vivo. This may reflect a weak and transient association between these proteins.
Besides NLS masking, the block may occur at the intramolecular level, involving the direct posttranslational modification of ORF63p. It is plausible that ORF61p may directly modify ORF63p to initiate nuclear import. If ORF61p possesses E3 ubiquitin ligase activity, it may direct ubiquitination of ORF63p to drive import in a mechanism similar to that described for PTEN. The phosphatase and tumor suppressor protein PTEN is efficiently imported after monoubiquitination of specific lysine residues (65). At present, we are investigating ORF61p for E3 ubiquitin ligase activity.
We further characterized the requirements of the ORF61p-dependent nuclear import of ORF63p in EN and found that both ORF63p NLSs were required (Fig. 8). Indeed, the loss-of-function mutation of either NLS1 or NLS2 by alanine substitution prevented ORF61p-dependent nuclear import. This was surprising, because in EP cells, one intact ORF63p NLS is sufficient, although less efficient, for nuclear import (Fig. 7C and D). The finding that both ORF63p NLSs are required for ORF61p-dependent nuclear import in EN demonstrates that even if a physical interaction occurs between ORF61p and ORF63p, it is not sufficient for nuclear import. Therefore, it is highly unlikely that ORF61p-dependent import of ORF63p in EN occurs by ORF63p hijacking the NLS of ORF61p via physical interaction. Together, these data highlight the differential regulation of ORF63p localization in neurons versus nonneuronal cells. We also show that ORF61p expression results in nuclear localization of the ORF63p CompletePhos mutant, confirming our belief that localization in EN is not regulated by phosphorylation (Fig. 6).
The findings in this report coupled with previously published data (59) show that localization of VZV LAPs is regulated in EN by expression of the VZV lytic protein ORF61p. Stallings et al. demonstrated that ORF61p expression caused nuclear accumulation of autonomously expressed ORF29p in EN and U373MG cells. Although ORF61p can modulate both ORF29p and ORF63p localization in EN, it appears that regulation of localization of these proteins is unique and distinct. These data suggest that ORF61p is a multifunctional protein playing a major role in modulating the cell environment to allow for redistribution of VZV LAPs. These results lead us to conclude that ORF61p is a central player in VZV reactivation.
In summary, we propose that cytoplasmic localization of ORF63p in neurons results from NLS masking, which prevents nuclear import. The coexpression of ORF61p results in release of this block in a proteasome-dependent manner and allows nuclear import of ORF63p. Similar to ORF29p, the experiments described here demonstrate that localization of the major latency-associated protein ORF63p in neurons is regulated by a strictly lytic protein, suggesting that lytic gene expression precedes nuclear translocation of LAPs during reactivation. These findings provide insight into an understanding of the localization of LAPs during VZV latency and the subsequent processes that occur upon reactivation.
Acknowledgments
These studies were supported by a grant from the Public Health Service, grant AI-024021, to S.S.
We thank Anne and Michael D. Gershon for valuable advice and stimulating discussions. We thank Michelle Staudt, Daniel Wolf, and members of the Silverstein laboratory for critical comments on the manuscript. We also thank the laboratory of Ann Arvin for providing reagents.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Ambagala, A. P., and J. I. Cohen. 2007. Varicella-zoster virus IE63, a major viral latency protein, is required to inhibit the alpha interferon-induced antiviral response. J. Virol. 817844-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baiker, A., C. Bagowski, H. Ito, M. Sommer, L. Zerboni, K. Fabel, J. Hay, W. Ruyechan, and A. M. Arvin. 2004. The immediate-early 63 protein of varicella-zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo. J. Virol. 781181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beals, C. R., N. A. Clipstone, S. N. Ho, and G. R. Crabtree. 1997. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 11824-834. [DOI] [PubMed] [Google Scholar]
- 4.Beg, A. A., S. M. Ruben, R. I. Scheinman, S. Haskill, C. A. Rosen, and A. S. Baldwin, Jr. 1992. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 61899-1913. [DOI] [PubMed] [Google Scholar]
- 5.Bontems, S., E. Di Valentin, L. Baudoux, B. Rentier, C. Sadzot-Delvaux, and J. Piette. 2002. Phosphorylation of varicella-zoster virus IE63 protein by casein kinases influences its cellular localization and gene regulation activity. J. Biol. Chem. 27721050-21060. [DOI] [PubMed] [Google Scholar]
- 6.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. J., A. A. Gershon, Z. S. Li, O. Lungu, and M. D. Gershon. 2003. Latent and lytic infection of isolated guinea pig enteric ganglia by varicella zoster virus. J. Med. Virol. 70(Suppl. 1)S71-S78. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., E. Cox, L. Pesnicak, S. Srinivas, and T. Krogmann. 2004. The varicella-zoster virus open reading frame 63 latency-associated protein is critical for establishment of latency. J. Virol. 7811833-11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, J. I., T. Krogmann, S. Bontems, C. Sadzot-Delvaux, and L. Pesnicak. 2005. Regions of the varicella-zoster virus open reading frame 63 latency-associated protein important for replication in vitro are also critical for efficient establishment of latency. J. Virol. 795069-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 702789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohrs, R. J., M. B. Barbour, R. Mahalingam, M. Wellish, and D. H. Gilden. 1995. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J. Virol. 692674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohrs, R. J., and D. H. Gilden. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 812950-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohrs, R. J., and D. H. Gilden. 2003. Varicella zoster virus transcription in latently-infected human ganglia. Anticancer Res. 232063-2069. [PubMed] [Google Scholar]
- 15.Cohrs, R. J., D. H. Gilden, P. R. Kinchington, E. Grinfeld, and P. G. Kennedy. 2003. Varicella-zoster virus gene 66 transcription and translation in latently infected human ganglia. J. Virol. 776660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohrs, R. J., J. Randall, J. Smith, D. H. Gilden, C. Dabrowski, H. van Der Keyl, and R. Tal-Singer. 2000. Analysis of individual human trigeminal ganglia for latent herpes simplex virus type 1 and varicella-zoster virus nucleic acids using real-time PCR. J. Virol. 7411464-11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohrs, R. J., K. Srock, M. B. Barbour, G. Owens, R. Mahalingam, M. E. Devlin, M. Wellish, and D. H. Gilden. 1994. Varicella-zoster virus (VZV) transcription during latency in human ganglia: construction of a cDNA library from latently infected human trigeminal ganglia and detection of a VZV transcript. J. Virol. 687900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohrs, R. J., J. Wischer, C. Essman, and D. H. Gilden. 2002. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 767228-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croen, K. D., J. M. Ostrove, L. J. Dragovic, and S. E. Straus. 1988. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc. Natl. Acad. Sci. USA 859773-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 671759-1816. [DOI] [PubMed] [Google Scholar]
- 21.Debrus, S., C. Sadzot-Delvaux, A. F. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 693240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desloges, N., M. Rahaus, and M. H. Wolff. 2005. The varicella-zoster virus-mediated delayed host shutoff: open reading frame 17 has no major function, whereas immediate-early 63 protein represses heterologous gene expression. Microbes Infect. 71519-1529. [DOI] [PubMed] [Google Scholar]
- 23.Diao, L., B. Zhang, J. Fan, X. Gao, S. Sun, K. Yang, D. Xin, N. Jin, Y. Geng, and C. Wang. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-kappaB by catalyzing IkappaBalpha ubiquitination. Cell. Signal. 17217-229. [DOI] [PubMed] [Google Scholar]
- 24.Di Valentin, E., S. Bontems, L. Habran, O. Jolois, N. Markine-Goriaynoff, A. Vanderplasschen, C. Sadzot-Delvaux, and J. Piette. 2005. Varicella-zoster virus IE63 protein represses the basal transcription machinery by disorganizing the pre-initiation complex. Biol. Chem. 386255-267. [DOI] [PubMed] [Google Scholar]
- 25.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 726581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 177161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang, X., T. Chen, K. Tran, and C. S. Parker. 2001. Developmental regulation of the heat shock response by nuclear transport factor karyopherin-alpha3. Development 1283349-3358. [DOI] [PubMed] [Google Scholar]
- 28.Gilden, D. H., R. J. Cohrs, and R. Mahalingam. 2003. Clinical and molecular pathogenesis of varicella virus infection. Viral Immunol. 16243-258. [DOI] [PubMed] [Google Scholar]
- 29.Grinfeld, E., and P. G. Kennedy. 2004. Translation of varicella-zoster virus genes during human ganglionic latency. Virus Genes 29317-319. [DOI] [PubMed] [Google Scholar]
- 30.Habran, L., S. Bontems, E. Di Valentin, C. Sadzot-Delvaux, and J. Piette. 2005. Varicella-zoster virus IE63 protein phosphorylation by roscovitine-sensitive cyclin-dependent kinases modulates its cellular localization and activity. J. Biol. Chem. 28029135-29143. [DOI] [PubMed] [Google Scholar]
- 31.Habran, L., N. El Mjiyad, E. Di Valentin, C. Sadzot-Delvaux, S. Bontems, and J. Piette. 2007. The varicella-zoster virus immediate-early 63 protein affects chromatin-controlled gene transcription in a cell-type dependent manner. BMC Mol. Biol. 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn, H., and A. C. Palmenberg. 1995. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J. Virol. 692697-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heineman, T. C., and J. I. Cohen. 1995. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J. Virol. 697367-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood, C., A. L. Cunningham, B. Slobedman, A. M. Arvin, M. H. Sommer, P. R. Kinchington, and A. Abendroth. 2006. Varicella-zoster virus ORF63 inhibits apoptosis of primary human neurons. J. Virol. 801025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoover, S. E., R. J. Cohrs, Z. G. Rangel, D. H. Gilden, P. Munson, and J. I. Cohen. 2006. Downregulation of varicella-zoster virus (VZV) immediate-early ORF62 transcription by VZV ORF63 correlates with virus replication in vitro and with latency. J. Virol. 803459-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39499-509. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy, P. G., E. Grinfeld, and J. E. Bell. 2000. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 7411893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy, P. G., E. Grinfeld, S. Bontems, and C. Sadzot-Delvaux. 2001. Varicella-zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289218-223. [DOI] [PubMed] [Google Scholar]
- 39.Kenyon, T. K., E. Homan, J. Storlie, M. Ikoma, and C. Grose. 2003. Comparison of varicella-zoster virus ORF47 protein kinase and casein kinase II and their substrates. J. Med. Virol. 70(Suppl. 1)S95-S102. [DOI] [PubMed] [Google Scholar]
- 40.Kenyon, T. K., J. Lynch, J. Hay, W. Ruyechan, and C. Grose. 2001. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J. Virol. 758854-8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J. Virol. 694274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kost, R. G., H. Kupinsky, and S. E. Straus. 1995. Varicella-zoster virus gene 63: transcript mapping and regulatory activity. Virology 209218-224. [DOI] [PubMed] [Google Scholar]
- 43.Kyratsous, C. A., and S. J. Silverstein. 2007. BAG3, a host cochaperone, facilitates varicella-zoster virus replication. J. Virol. 817491-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 45.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 3021972-1975. [DOI] [PubMed] [Google Scholar]
- 46.Lungu, O., C. A. Panagiotidis, P. W. Annunziato, A. A. Gershon, and S. J. Silverstein. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. USA 957080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 30271-82. [DOI] [PubMed] [Google Scholar]
- 48.Mahalingam, R., M. Wellish, R. Cohrs, S. Debrus, J. Piette, B. Rentier, and D. H. Gilden. 1996. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. USA 932122-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier, J. L., R. P. Holman, K. D. Croen, J. E. Smialek, and S. E. Straus. 1993. Varicella-zoster virus transcription in human trigeminal ganglia. Virology 193193-200. [DOI] [PubMed] [Google Scholar]
- 50.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1994. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology 205238-246. [DOI] [PubMed] [Google Scholar]
- 51.Moriuchi, H., M. Moriuchi, H. A. Smith, S. E. Straus, and J. I. Cohen. 1992. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J. Virol. 667303-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus (VZV) open reading frame 61 protein transactivates VZV gene promoters and enhances the infectivity of VZV DNA. J. Virol. 674290-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng, P., D. T. Cummings, C. M. Evelegh, and F. L. Graham. 2000. Yeast recombinase FLP functions effectively in human cells for construction of adenovirus vectors. BioTechniques 29524-526, 528. [DOI] [PubMed] [Google Scholar]
- 54.Panagiotidis, C. A., and S. J. Silverstein. 1995. pALEX, a dual-tag prokaryotic expression vector for the purification of full-length proteins. Gene 16445-47. [DOI] [PubMed] [Google Scholar]
- 55.Perry, L. J., F. J. Rixon, R. D. Everett, M. C. Frame, and D. J. McGeoch. 1986. Characterization of the IE110 gene of herpes simplex virus type 1. J. Gen. Virol. 672365-2380. [DOI] [PubMed] [Google Scholar]
- 56.Pouton, C. W., K. M. Wagstaff, D. M. Roth, G. W. Moseley, and D. A. Jans. 2007. Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 59698-717. [DOI] [PubMed] [Google Scholar]
- 57.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 6731-40. [DOI] [PubMed] [Google Scholar]
- 58.Sommer, M. H., E. Zagha, O. K. Serrano, C. C. Ku, L. Zerboni, A. Baiker, R. Santos, M. Spengler, J. Lynch, C. Grose, W. Ruyechan, J. Hay, and A. M. Arvin. 2001. Mutational analysis of the repeated open reading frames, ORFs 63 and 70 and ORFs 64 and 69, of varicella-zoster virus. J. Virol. 758224-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stallings, C. L., G. J. Duigou, A. A. Gershon, M. D. Gershon, and S. J. Silverstein. 2006. The cellular localization pattern of varicella-zoster virus ORF29p is influenced by proteasome-mediated degradation. J. Virol. 801497-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevenson, D., K. L. Colman, and A. J. Davison. 1992. Characterization of the varicella-zoster virus gene 61 protein. J. Gen. Virol. 73521-530. [DOI] [PubMed] [Google Scholar]
- 61.Stevenson, D., M. Xue, J. Hay, and W. T. Ruyechan. 1996. Phosphorylation and nuclear localization of the varicella-zoster virus gene 63 protein. J. Virol. 70658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8195-208. [DOI] [PubMed] [Google Scholar]
- 63.Terry, L. J., E. B. Shows, and S. R. Wente. 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 3181412-1416. [DOI] [PubMed] [Google Scholar]
- 64.Traenckner, E. B., S. Wilk, and P. A. Baeuerle. 1994. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J. 135433-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trotman, L. C., X. Wang, A. Alimonti, Z. Chen, J. Teruya-Feldstein, H. Yang, N. P. Pavletich, B. S. Carver, C. Cordon-Cardo, H. Erdjument- Bromage, P. Tempst, S. G. Chi, H. J. Kim, T. Misteli, X. Jiang, and P. P. Pandolfi. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 988815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao, C. Y., S. Hubner, and D. A. Jans. 1997. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J. Biol. Chem. 27222191-22198. [DOI] [PubMed] [Google Scholar]
- 68.Zuranski, T., H. Nawar, D. Czechowski, J. M. Lynch, A. Arvin, J. Hay, and W. T. Ruyechan. 2005. Cell-type-dependent activation of the cellular EF-1alpha promoter by the varicella-zoster virus IE63 protein. Virology 33835-42. [DOI] [PubMed] [Google Scholar]