Abstract
The horse-adapted virulent Bucyrus (VB) strain of equine arteritis virus (EAV) established persistent infection in high-passage-number human cervix cells (HeLa-H cells; passages 170 to 221) but not in low-passage-number human cervix cells (HeLa-L cells; passages 95 to 115) or in several other cell lines that were evaluated. However, virus recovered from the 80th passage of the persistently infected HeLa-H cells (HeLa-H-EAVP80) readily established persistent infection in HeLa-L cells. Comparative sequence analysis of the entire genomes of the VB and HeLa-H-EAVP80 viruses identified 16 amino acid substitutions, including 4 in the replicase (nsp1, nsp2, nsp7, and nsp9) and 12 in the structural proteins (E, GP2, GP3, GP4, and GP5). Reverse genetic studies clearly showed that substitutions in the structural proteins but not the replicase were responsible for the establishment of persistent infection in HeLa-L cells by the HeLa-H-EAVP80 virus. It was further demonstrated that recombinant viruses with substitutions in the minor structural proteins E and GP2 or GP3 and GP4 were unable to establish persistent infection in HeLa-L cells but that recombinant viruses with combined substitutions in the E (Ser53→Cys and Val55→Ala), GP2 (Leu15→Ser, Trp31→Arg, Val87→Leu, and Ala112→Thr), GP3 (Ser115→Gly and Leu135→Pro), and GP4 (Tyr4→His and Ile109→Phe) proteins or with a single point mutation in the GP5 protein (Pro98→Leu) were able to establish persistent infection in HeLa-L cells. In summary, an in vitro model of EAV persistence in cell culture was established for the first time. This system can provide a valuable model for studying virus-host cell interactions, especially virus-receptor interactions.
Equine arteritis virus (EAV) is the prototype member of the family Arteriviridae in the order Nidovirales (12). The EAV genome is a single-stranded, positive-sense RNA molecule of 12.7 kb that includes 5′ and 3′ untranslated regions (UTRs) and nine functional open reading frames (ORFs) (60, 61). The first two ORFs (1a and 1b) are located in the 5′-terminal three-quarters of the genome and encode two replicase polyproteins (pp1a and pp1ab) that are posttranslationally processed by three ORF 1a-encoded proteinases (nsp1, nsp2, and nsp4) into at least 13 nonstructural proteins (nsp1 to nsp12, including nsp7α and nsp7β) required for viral replication and transcription (63, 67, 72). The other seven ORFs (2a, 2b, and 3 to 7) are located in the 3′ one-quarter of the genome and encode structural proteins E, GP2, GP3, GP4, GP5, M, and N, respectively (24, 61).
EAV is the causative agent of equine viral arteritis, a globally distributed infectious disease of equids (65). Some 10 to 70% of stallions acutely infected with EAV can subsequently become carriers and constantly shed the virus in semen (65). Persistently infected stallions are the principal reservoir of EAV and are responsible for the perpetuation and dissemination of EAV in equine populations (64-66). Carrier stallions are also a significant natural source of genetic and phenotypic diversity of EAV (4, 5, 33). There is convincing evidence that the establishment and maintenance of the carrier state in the stallion is testosterone dependent, with the underlying mechanism as yet unknown (38, 44, 47). Other than testosterone, the host and viral factors that contribute to the establishment and maintenance of persistent EAV infection in the reproductive tracts of stallions remain to be determined.
The persistent infection of cell cultures has been documented previously for many viruses, including certain nidoviruses, e.g., human coronaviruses OC43 and 229E (2, 3), severe acute respiratory syndrome-associated coronavirus (13, 49, 53, 69), murine hepatitis virus (8, 16, 41, 42, 58), bovine coronavirus (35-37), and lactate dehydrogenase-elevating virus (62). Persistently infected cell cultures can provide a model system for elucidating mechanisms of viral persistence in vivo. In vitro persistence has also proven to be a useful tool for investigating virus and cell evolution (1, 15, 16, 18, 22, 23, 36, 40). Genetic and phenotypic characterization of virus and host cell mutants during persistent infection has facilitated the study of virus-receptor interactions (10, 11, 18, 22, 30, 31, 54-57). Such studies have also served to define genetic determinants of virulence (19, 20, 26, 27) and to identify viral and host cell determinants involved in the establishment of persistent viral infection (9-11, 14, 25, 29, 30, 52, 57).
This study was undertaken in an attempt to develop a model system of persistent EAV infection in cell culture. Various cell lines originating from different species and tissues were evaluated; these included equine pulmonary artery endothelial cells (EECs) and equine dermis (ED; NBL-6), baby hamster kidney (BHK-21), rabbit kidney (RK-13), mouse muscle (C2C12), mouse connective tissue (L-M), human cervix (HeLa), and human epidermoid larynx (Hep-2) cells. In the course of this study, HeLa cells were found to become more susceptible to EAV infection after extended serial passage. The respective HeLa cell lines were identified as HeLa-L (passage 95 [P95] to P115) and HeLa-H (P170 to P221). The horse-adapted virulent Bucyrus (VB) strain of EAV successfully established persistent infection in HeLa-H cells but not in any of the other cell lines tested. This is the first report of an in vitro model of EAV persistence. Genetic and phenotypic variations of the virus during the course of persistent infection were investigated.
MATERIALS AND METHODS
Cells and viruses.
The cell lines used in the study included ED cells (NBL-6 [ATCC CCL-57]; P18 to P25), EECs (P9 to P20) (34, 50), baby hamster kidney cells (BHK-21 [ATCC CCL-10]; P61 to P114), rabbit kidney cells (RK-13 [ATCC CCL-37]; P193 to P204), mouse muscle cells (C2C12 [ATCC CRL-1772]; P4 to P14), mouse connective tissue cells (L-M [ATCC CCL-1.2]; P300 to P393), human cervix cells (HeLa [ATCC CCL-2]; P95 to P221), and human epidermoid larynx cells (Hep-2 [ATCC CCL-23]; P451 to P531). Cells were grown at 37°C in Eagle's minimum essential medium with 10% ferritin-supplemented bovine calf serum, penicillin, streptomycin, amphotericin B, and sodium bicarbonate. The horse-adapted highly VB (horse P15) strain of EAV (48) was propagated once in BHK-21 cells to produce the virus stock that was used in this study.
Chromosomal analysis.
Karyotypic analysis of HeLa-L (P98) and HeLa-H (P202) cells was performed using the G-banding technique (32, 70) in the Department of Cytogenetics, University of Kentucky. Twenty Giemsa-banded metaphase cells from each of the HeLa-L and HeLa-H cell lines were analyzed.
Indirect immunofluorescence assay (IFA).
Mock- or EAV-infected or EAV RNA-transfected cells grown in eight-well chamber slides were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) and washed with PBS containing 10 mM glycine. After permeabilization with 0.2% Trixton X-100 in PBS, slides were incubated with monoclonal antibody (MAb) 3E2 against the EAV nucleocapsid (N) protein (45) or MAb 12A4 against the EAV nsp1 protein (68), followed by fluorescein-conjugated goat anti-mouse immunoglobulin (Pierce). The cells were counterstained with Evans blue. The percentage of EAV fluorescence-positive cells was determined based on counting of 500 cells.
Growth characteristics of EAV in various cell lines.
Subconfluent monolayers of each cell line (EECs and ED, BHK-21, RK-13, C2C12, HeLa-L, HeLa-H, Hep-2, and L-M cells) grown in six-well plates were inoculated with the VB strain of EAV at a multiplicity of infection of 3 and held at 37°C for 1 h. The inoculum was aspirated off, and the cell sheets were washed three times with PBS (pH 7.4) to remove unbound virus and then overlaid with 4 ml of Eagle's minimum essential medium. This time point was designated time zero with respect to infection. At 0, 12, 24, 36, 48, 60, and 72 h postinfection, supernatants were harvested and virus titers were determined by plaque assays of RK-13 cells as described previously (46).
Attempts to establish persistent EAV infection in various cell lines.
Subconfluent monolayers of each cell line grown in T-25 flasks were inoculated with EAV at a multiplicity of infection of 3. Following 1 h of adsorption at 37°C, cell monolayers were washed three times with PBS and 10 ml of fresh culture medium was added. Inoculated cultures were incubated at 37°C, microscopically monitored daily for the development of a cytopathic effect (CPE), and subcultured once every 4 days. Tissue culture supernatants were harvested and titrated by plaque assays of RK-13 cells.
Transfection of cells with IVT EAV RNA.
Viral RNA transcripts were in vitro transcribed (IVT) from the XhoI-linearized full-length infectious cDNA clones, and cells were transfected with RNA transcripts by means of electroporation according to previously described protocols (7).
Isolation of viral RNA, RT-PCR amplification, and sequencing.
Viral RNAs of the VB virus and the virus recovered from P80 of persistently infected HeLa-H cells (HeLa-H-EAVP80 virus) were extracted using the QIAamp viral RNA mini kit (Qiagen). The full-length genomes of these two viruses were reverse transcription (RT)-PCR amplified using SuperScript III (Invitrogen) and the high-fidelity proofreading Pfu Turbo DNA polymerase (Stratagene) according to previously described procedures (71). Both sense and antisense strands were sequenced using an ABI 377 automatic sequencer (Applied Biosystems). Sequence data were analyzed using Aligner version 1.5.2 (CodonCode) and Vector NTI Advance 10 (Invitrogen).
Reverse genetics.
The recombinant chimeric infectious cDNA clone pEAVrVBS/P80S was constructed by replacing the structural-protein genes (ORFs 2a to 7) of the infectious cDNA clone pEAVrVBS, which was derived from the VB strain of EAV (6), with the corresponding structural-protein genes of the HeLa-H-EAVP80 virus. The recombinant infectious cDNA clone pEAVrVBS/P80NS4m was constructed by introducing the following four site-specific nucleotide changes that were present in HeLa-H-EAVP80 virus into the pEAVrVBS clone: C658→T (nsp1 Ala145→Val), A1954→G (nsp2 Asp577→Gly), A4900→G (nsp7 Lys1559→Arg), and C6020→T (nsp9 Pro1933→Ser). A similar approach was used to construct the recombinant infectious cDNA clones pEAVrVBS/P80ORFs2ab (9867T→C [GP2 Leu15→Ser], 9907A→T [E Ser53→Cys], 9914T→C [E Val55→Ala and GP2 Trp31→Arg], 10082G→C [GP2 Val87→Leu], 10157G→A [GP2 Ala112→Thr], and 10189T→C), pEAVrVBS/P80ORFs34 (10648A→G [GP3 Ser115→Gly], 10709T→C [GP3 Leu135→Pro and GP4 Tyr4→His], and 11024A→T [GP4 Ile109→Phe]), pEAVrVBS/P80ORFs234 (9867T→C, 9907A→T, 9914T→C, 10082G→C, 10157G→A, 10189T→C, 10648A→G, 10709T→C, and 11024A→T), and pEAVrVBS/GP5P98→L (11438C→T). BHK-21 cells were transfected with IVT RNAs by following previously described protocols (7) to generate the recombinant viruses rVBS/P80S, rVBS/P80NS4m, rVBS/P80ORFs2ab, rVBS/P80ORFs34, rVBS/P80ORFs234, and rVBS/GP5P98→L.
Nucleotide sequence accession numbers.
The nucleotide sequences of the VB and HeLa-H-EAVP80 viruses were deposited in GenBank under accession numbers DQ846750 and EU252114, respectively.
RESULTS
Permissiveness of various cell lines to EAV infection.
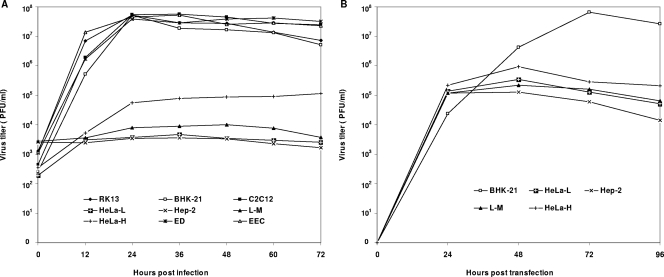
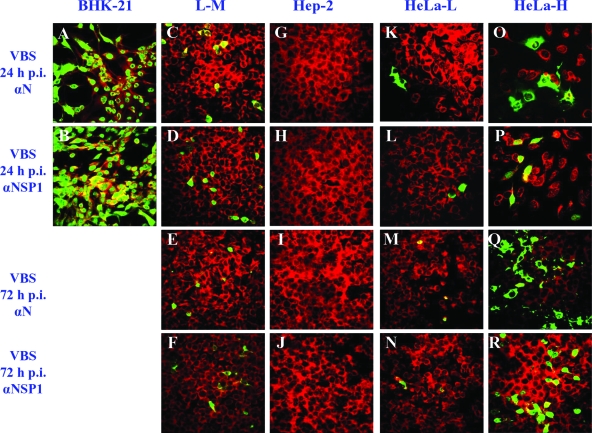
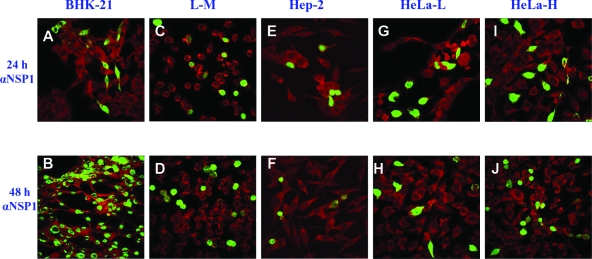
A variety of cell lines from different species and tissues were evaluated for their permissiveness of infection with the VB strain of EAV. The EECs and the ED, BHK-21, RK-13, and C2C12 cell lines were all highly permissive and produced high titers of infectious virus and fully lytic infections by 48 h postinfection (Fig. 1A). In contrast, CPE on either the Hep-2 or L-M cell line was not detected even at 72 h postinfection and virus titers did not increase substantially (Fig. 1A). These findings were confirmed by IFAs using MAbs to the N and nsp1 proteins, as there was no staining of Hep-2 cells after infection and as only approximately 6% of L-M cells were positive at 24 h postinfection and there was little subsequent spread to adjacent uninfected cells even after 72 h (Fig. 2). The susceptibilities of HeLa cells of different passage levels (P95 to P221) to the VB strain of EAV were compared. Infected HeLa-H cells (P170 to P221) produced significantly higher virus titers than HeLa-L cells (P95 to P115) (Fig. 1A), and approximately 12% of HeLa-H cells were positive by IFA staining at 24 h, whereas less than 2% of HeLa-L cells were positive (Fig. 2). Furthermore, in contrast to the pattern among infected HeLa-L cells, there was spreading of IFA positivity in the infected HeLa-H monolayers (∼12 and ∼20% of cells were positive at 24 and 72 h postinfection, respectively) (Fig. 2). These data indicate that HeLa cells become increasingly susceptible to EAV infection after extended serial passage.
FIG. 1.
Growth characteristics of the VB strain of EAV in various cell lines. (A) Infection of EECs and ED, BHK-21, RK-13, C2C12, HeLa-H, HeLa-L, Hep-2, and L-M cell lines with the VB strain of EAV. Three separate experiments were carried out, and representative data are shown. (B) Transfection of BHK-21, HeLa-L, HeLa-H, L-M, and Hep-2 cell lines with IVT viral RNA from the infectious cDNA clone pEAVrVBS, which was derived from the VB strain of EAV. Data representative of results from two separate experiments are shown.
FIG. 2.
Results from IFAs of cells infected with the VB strain (VBS) of EAV. At 24 and 72 h postinfection (h p.i.), EAV-infected cells were examined by IFA using MAb 3E2 against EAV N protein (αN) and MAb 12A4 against EAV nsp1 protein (αNSP1). The cells were counterstained with Evans blue, shown as red coloring. EAV fluorescence-positive cells are shown in green.
Productive infection of BHK-21, L-M, Hep-2, HeLa-L, and HeLa-H cells occurred following the transfection of these cells with RNA transcripts from the infectious cDNA clone pEAVrVBS, which was derived from the VB strain of EAV (6). Approximately 20% of the monolayer of each of these cell lines was positive by IFA at 24 h posttransfection (Fig. 3). Virus yields from these transfected cells were similar (104 to 105 PFU/ml) at 24 h post transfection (Fig. 1B). However, virus spread from transfected cells to untransfected cells occurred efficiently only among BHK-21 cells and did not occur among transfected L-M, Hep-2, and HeLa-L cells (Fig. 3). In the transfected HeLa-H cells, spread of infection occurred but was not substantial (∼20 and ∼24% of cells were infected at 24 and 48 h posttransfection, respectively) (Fig. 3).
FIG. 3.
Results from IFAs of cells transfected with IVT viral RNA from the infectious cDNA clone pEAVrVBS. At 24 and 48 h posttransfection, viral RNA-transfected cells were examined by IFA using MAb 12A4 against the nsp1 protein of EAV (αNSP1). The cells were counterstained with Evans blue, shown as red coloring. EAV fluorescence-positive cells are shown in green.
These data collectively confirm that EECs and the ED, BHK-21, RK-13, and C2C12 cell lines are all highly susceptible to EAV infection, whereas HeLa-L and L-M cell lines have only limited susceptibilities and the Hep-2 cell line is apparently resistant to infection. The HeLa-H cells are clearly more susceptible to EAV infection than L-M, HeLa-L, and Hep-2 cells, but they are less susceptible than EECs and ED, BHK-21, RK-13, and C2C12 cells. The L-M, Hep-2, HeLa-L, and HeLa-H cells support EAV replication upon transfection with viral RNA, indicating a lack of any intracellular block during the virus replication cycle. Thus, EAV infection of these cell lines appears to be restricted at the attachment and/or entry step.
Ability of the VB strain of EAV to establish persistent infection in HeLa-H cells.
The EECs and ED, BHK-21, RK-13, C2C12, L-M, Hep-2, HeLa-L, and HeLa-H cells were infected once with the VB strain of EAV and subjected to continuous subculture to determine whether persistent EAV infection could be established. The EECs and ED, BHK-21, RK-13, and C2C12 cells infected with EAV all developed 100% CPE at 48 h postinfection without the establishment of persistent infection. After two serial passages, infectious virus was no longer detected in HeLa-L, Hep-2, and L-M cell lines inoculated with EAV (Fig. 4B), indicating that persistent infection was not established in these cells. In contrast, the HeLa-H cell line maintained EAV infection during serial cell culture passage, indicating the establishment of persistent infection. The persistently infected HeLa-H cell line was subjected to 94 serial passages without the loss of production of infectious progeny virus (Fig. 4A). In addition, virus was readily recovered from these cells after storage in liquid nitrogen for 5 years, with virus titers similar to those obtained prior to the freezing of the cells.
FIG. 4.
Attempt to establish persistent infection in various cell lines with the VB strain of EAV or its viral RNA. (A) Persistent infection was readily established in the HeLa-H cell line with the VB strain of EAV. This finding was confirmed in three separate experiments, and representative data are shown. h p.i., hours postinfection. (B) The VB strain of EAV was unable to establish persistent infection in the HeLa-L, L-M, and Hep-2 cell lines. Tissue culture supernatants from serial subcultures up to the 20th passage were harvested and titrated. (C) Attempt to establish persistent infection by serially subculturing the HeLa-H, HeLa-L, Hep-2, and L-M cell lines which were transfected with RNA transcripts from the infectious cDNA clone pEAVrVBS. Tissue culture supernatants from serial subcultures up to the 10th passage were harvested and titrated. The results presented in panels B and C were confirmed in two separate experiments, and representative data are shown.
An attempt to establish persistent infection by serially subculturing L-M, Hep-2, HeLa-L, and HeLa-H cells that were transfected with RNA transcripts derived from the infectious cDNA clone pEAVrVBS was also performed. As shown in Fig. 4C, although all transfected cell lines initially produced substantial titers of virus, persistent EAV infection eventually was established only in the HeLa-H cell line but not in the L-M, Hep-2, and HeLa-L cell lines, implying that the establishment and maintenance of persistent EAV infection may require horizontal virus spread in the cell population.
Ability of HeLa-H-EAVP80 virus to establish persistent infection in both HeLa-L and HeLa-H cell lines.
The tissue culture fluid harvested from the 80th passage of the persistently infected HeLa-H cell line (yielding HeLa-H-EAVP80 virus) was used to evaluate whether virus recovered from persistently infected cells had selectively acquired features that enabled it to establish persistent infection. Five cell lines (BHK-21, HeLa-L, HeLa-H, Hep-2, and L-M) were infected with the HeLa-H-EAVP80 virus. Infection caused 100% CPE in BHK-21 cells at 48 h postinfection without the establishment of persistent infection. The HeLa-H-EAVP80 virus also did not establish persistent infection in the Hep-2 and L-M cell lines, in which virus production ceased after two or three serial passages (Fig. 5). Remarkably, the HeLa-H-EAVP80 virus did establish persistent infections in both the HeLa-H and HeLa-L cell lines (Fig. 5).
FIG. 5.
Attempt to establish persistent infection in various cell lines with the HeLa-H-EAVP80 virus. Tissue culture supernatants from serial subcultures up to the 10th passage were harvested and titrated. Data representative of results from two separate experiments are shown. h p.i., hours postinfection.
Genetic basis of persistent EAV infection in HeLa cells.
To investigate the genetic evolution of EAV during persistent infection in HeLa-H cells, the genomes of the VB and HeLa-H-EAVP80 viruses were sequenced. Comparative analyses of the nucleotide and deduced amino acid sequences of these two viruses are summarized in Table 1. The entire genomes of the VB strain and HeLa-H-EAVP80 virus were each 12,704 nucleotides in length, with a total of 33 nucleotide differences. All of these nucleotide changes occurred in ORFs 1a, 1b, 2a, 2b, and 3 to 5; no nucleotide changes occurred in the 5′ UTR, ORFs 6 and 7, or the 3′ UTR. Compared to the VB strain, the HeLa-H-EAVP80 virus had 4 amino acid changes in the replicase and 12 amino acid changes in the structural proteins (Table 1).
TABLE 1.
Nucleotide and amino acid differences between the VB strain and HeLa-H-EAVP80 virus
| Protein(s) and region(s) (length in amino acids) | Genome region or ORF (nucleotide position)a | Nucleotide ina:
|
Amino acid inb:
|
||||
|---|---|---|---|---|---|---|---|
| Position | VB strain | HeLa-H- EAVP80 | Position | VB strain | HeLa-H- EAVP80 | ||
| None | 5′ UTR (1-224) | −c | − | − | |||
| Nonstructural (nsp) proteins | ORF 1ab (225-9751) | ||||||
| pp1ab polyprotein (3,175) | |||||||
| nsp1, Met1-Gly260 (260) | 658 | C | T | 145 | Ala | Val | |
| 695 | C | T | |||||
| nsp2, Gly261-Gly831 (571) | 1280 | C | T | ||||
| 1889 | T | C | |||||
| 1954 | A | G | 577 | Asp | Gly | ||
| 1979 | C | T | |||||
| 2057 | C | T | |||||
| 2483 | A | T | |||||
| nsp3, Gly832-Glu1064 (233) | 2898 | C | T | ||||
| 3011 | G | A | |||||
| nsp4, Gly1065-Glu1268 (204) | 3707 | Yd | C | ||||
| 3797 | T | C | |||||
| nsp5, Ser1269-Glu1430 (162) | 4349 | G | A | ||||
| nsp6, Gly1431-Glu1452 (22) | − | − | − | − | − | − | |
| nsp7, Ser1453-Glu1677 (225) | 4900 | A | G | 1559 | Lys | Arg | |
| nsp8, Gly1678-Asn1727 (50) | — | — | — | — | — | — | |
| nsp9, Gly1678-Glu2370 (693) | 6020 | C | T | 1933 | Pro | Ser | |
| 6752 | T | C | |||||
| 7021 | Rd | A | |||||
| nsp10, Ser2371-Gln2837 (467) | 8260 | Y | C | ||||
| 8290 | Y | C | |||||
| 8398 | T | C | |||||
| nsp11, Ser2838-Glu3056 (219) | − | − | − | − | − | − | |
| nsp12, Gly3057-Val3175 (119) | − | − | − | − | − | − | |
| Structural proteins | |||||||
| E (67) | ORF 2a (9751-9954) | 9867 | T | C | |||
| 9907 | A | T | 53 | Ser | Cys | ||
| 9914 | T | C | 55 | Val | Ala | ||
| GP2 (227) | ORF 2b (9824-10507) | 9867 | T | C | 15 | Leu | Ser |
| 9907 | A | T | |||||
| 9914 | T | C | 31 | Trp | Arg | ||
| 10082 | G | C | 87 | Val | Leu | ||
| 10157 | G | A | 112 | Ala | Thr | ||
| 10189 | T | C | |||||
| GP3 (163) | ORF 3 (10306-10797) | 10648 | A | G | 115 | Ser | Gly |
| 10709 | T | C | 135 | Leu | Pro | ||
| GP4 (152) | ORF 4 (10700-11158) | 10709 | T | C | 4 | Tyr | His |
| 11024 | A | T | 109 | Ile | Phe | ||
| GP5 (255) | ORF 5 (11146-11913) | 11171 | T | C | 9 | Phe | Ser |
| 11438 | C | T | 98 | Pro | Leu | ||
| 11475 | T | C | |||||
| 11704 | T | C | |||||
| M (162) | ORF 6 (11901-12389) | − | − | − | − | − | − |
| N (110) | ORF 7 (12313-12645) | − | − | − | − | − | − |
| None | 3′ UTR (12646-12704) | − | − | − | |||
Nucleotides are numbered according to the published sequence of EAV030 virus (GenBank accession no. NC_002532).
Only nonsynonymous mutations are shown, and silent mutations are not shown. Amino acids of nonstructural proteins are numbered according to their locations in the replicase polyprotein pp1ab. Amino acids of structural proteins are numbered according to their locations in the individual structural proteins.
−, no nucleotide or amino acid change occurred.
Y, T or C; R, A or G.
Reverse genetic studies using the infectious cDNA clone pEAVrVBS were carried out to explore the genetic basis of the persistent infection of the HeLa-L cell line with the HeLa-H-EAVP80 virus. The recombinant virus rVBS/P80NS4m, which had a sequence identical to that of the recombinant VB strain (rVBS) virus generated from the clone pEAVrVBS, with the exception of four amino acid substitutions in the replicase polyproteins (nsp1 Ala145→Val, nsp2 Asp577→Gly, nsp7 Lys1559→Arg, and nsp9 Pro1933→Ser), was unable to establish persistent infection in the HeLa-L cell line (Fig. 6). In contrast, the recombinant virus rVBS/P80S, which carried the replicase gene of the parental rVBS and the structural-protein genes of the HeLa-H-EAVP80 virus, did establish persistent infection in the HeLa-L cell line (Fig. 6). This finding clearly indicates that it is the changes in the structural proteins (E, GP2, GP3, GP4, and GP5) and not the replicase that were responsible for the establishment of persistent infection in the HeLa-L cell line by the HeLa-H-EAVP80 virus.
FIG. 6.
Attempt to establish persistent infection in the HeLa-L cell line with the recombinant viruses rVBS/P80NS4m, rVBS/P80S, rVBS/P80ORFs2ab, rVBS/P80ORFs34, rVBS/P80ORFs234, and rVBS/GP5P98→L. Tissue culture supernatants from serial subcultures up to the 10th passage were harvested and titrated. Data representative of results from two separate experiments are shown.
Additional mutants (rVBS/P80ORFs2ab, rVBS/P80ORFs34, rVBS/P80ORFs234, and rVBS/GP5P98→L) were constructed to further identify specific structural-protein genes responsible for the persistent infection of HeLa-L cells with the HeLa-H-EAVP80 virus. The recombinant virus rVBS/P80ORFs2ab, which had a nucleotide sequence identical to that of the rVBS virus with the exception that ORFs 2a and 2b were exchanged to include the corresponding regions of the HeLa-H-EAVP80 virus, was unable to establish persistent infection in the HeLa-L cell line (Fig. 6). Similarly, the recombinant virus rVBS/P80ORFs34, which included ORFs 3 and 4 from the HeLa-H-EAVP80 virus, was unable to establish persistent infection in the HeLa-L cell line (Fig. 6). In contrast, the recombinant virus rVBS/P80ORFs234, which carried ORFs 2a, 2b, 3, and 4 of the HeLa-H-EAVP80 virus, did establish persistent infection in the HeLa-L cell line, although virus titers were considerably lower than those in cells persistently infected with the rVBS/P80S virus (Fig. 6). Interestingly, the recombinant virus rVBS/GP5P98→L, which had only a single mutation in GP5 (nucleotide change 11438C→T and GP5 amino acid change Pro98→Leu compared to the rVBS virus), was also able to establish persistent infection in the HeLa-L cell line; however, virus titers were also lower than those in cells persistently infected with the rVBS/P80S virus (Fig. 6). These data indicate that the establishment and maintenance of persistent infection in HeLa-L cells by the HeLa-H-EAVP80 virus involves complex interactions of several structural proteins (E, GP2, GP3, GP4, and GP5).
DISCUSSION
This study confirms that a wide variety of cell lines from different species and tissues are susceptible to EAV infection. Specifically, we have demonstrated that EECs and ED, BHK-21, RK-13, C2C12, HeLa-H, L-M, HeLa-L, and Hep-2 cells have various degrees of susceptibility to EAV infection. The infection of L-M, Hep-2, HeLa-L, and HeLa-H cells appears to be restricted at the attachment or entry step. To date, neither the viral components nor the cellular receptors involved in the early events (e.g., attachment, penetration, and uncoating) of the EAV replication cycle have been fully characterized. These cell lines with various levels of susceptibility to EAV infection potentially could be utilized to identify the cellular receptor(s) involved in EAV attachment and entry. The VB strain of EAV established persistent infection in HeLa-H but not in HeLa-L cells, indicating that the HeLa cells had evolved during long-term serial passage. Karyotypic analysis revealed that both HeLa-L and HeLa-H cell lines contained typical HeLa marker chromosomes, confirming that the HeLa-H cell line was not contaminated with any other cell type. Changes between the HeLa-L and HeLa-H cell lines at the chromosomal level were observed (data not shown). However, the molecular changes that are responsible for the differences in susceptibility to EAV infection of the HeLa-L and HeLa-H cell lines have not yet been determined. The evolution of cells and the emergence of cell diversity in laboratory cell cultures are not unusual events (39, 59). The extent of cellular heterogeneity in tumors is greatly increased compared to that in other tissues, and tumor cell heterogeneity appears to result from the enhanced genetic instability of transformed cells (17, 21, 43, 51). HeLa cells were originally derived from a cervical adenocarcinoma and perhaps retain the potential to evolve if they undergo extended serial passage.
Viral determinants also are critical in mediating the persistent EAV infection of HeLa cells, as the HeLa-H-EAVP80 virus established persistent infection in HeLa-L cells whereas the VB strain of EAV was unable to do so. Comparative analysis of the entire genomes of the VB strain and the HeLa-H-EAVP80 virus identified amino acid residues in the replicase and structural proteins that had mutated during the process of adaptation of the VB virus to persistence in HeLa-H cells. Reverse genetic studies using the infectious cDNA clone pEAVrVBS clearly demonstrated that amino acid substitutions in the structural proteins (E, GP2, GP3, GP4, and GP5) but not the replicase were responsible for the altered tropism and persistence of HeLa-H-EAVP80 virus in HeLa-L cells. Further studies indicated that the recombinant viruses with substitutions in the minor structural proteins E and GP2 or GP3 and GP4 were unable to establish persistent infection in HeLa-L cells and that the recombinant viruses with combined substitutions in the E (Ser53→Cys and Val55→Ala), GP2 (Leu15→Ser, Trp31→Arg, Val87→Leu, and Ala112→Thr), GP3 (Ser115→Gly and Leu135→Pro), and GP4 (Tyr4→His and Ile109→Phe) proteins or with a single point mutation in the GP5 protein (Pro98→Leu) were able to establish persistent infection in HeLa-L cells. Cells that were persistently infected with these viruses, however, all produced lower virus titers than similar cells persistently infected with the rVBS/P80S virus. This finding strongly suggests that complex interactions of several structural proteins (E, GP2, GP3, GP4, and GP5) are required to alter the tropism of the HeLa-H-EAVP80 virus toward HeLa-L cells. It also implicates the structural proteins E, GP2, GP3, GP4, and GP5 of EAV in virus-receptor interactions during the infection of HeLa cells. A previous study showed that the ectodomain (amino acids 1 to 114) of the GP5 protein is not the main determinant of EAV tropism toward BHK-21 or RK-13 cells (28), whereas the present data suggest that the amino acid residue at position 98 in the EAV GP5 protein is involved in viral tropism toward HeLa cells. However, the replacement of the amino acid residue at this position did not alter tropism toward BHK-21 or RK-13 cells (data not shown). It is likely, therefore, that the receptors and viral proteins involved in attachment to and entry into individual cell lines are different.
Previous studies have shown that genetic variants emerge during the course of long-term persistent infection in the stallion, with significant variation in ORFs 2a to 5, which encode the E, GP2, GP3, GP4, and GP5 proteins, respectively (5, 33). The HeLa-H-EAVP80 virus likely is a variant selected from the quasispecies population during the persistent infection of HeLa-H cells, and it replicates more efficiently than the VB virus in HeLa cells. However, the amino acid substitutions that occurred during the persistent EAV infection of HeLa cells generally were located at different positions than those which accumulate during the persistent EAV infection of stallions. Nevertheless, persistently infected HeLa cells provide a convenient in vitro model system for studying persistent EAV infection.
In summary, we have established an in vitro cell culture system for EAV persistence. The persistently infected HeLa cells provide a convenient model system with which to study viral and host cell factors involved in EAV persistence and could potentially assist in defining the mechanisms of EAV persistence in carrier stallions.
Acknowledgments
We are grateful to Eric Snijder for helpful comments; Carole Moncman and the late George Allen for providing cell lines BHK-21, L-M, and C2C12; and Anjana Pettigrew and Pamela White for carrying out the karyotypic analysis of the HeLa cells. We thank Teri Lear for assistance with fluorescence microscopy and Kathleen Shuck for making cell culture medium.
This study was supported by the Frederick Van Lennep chair endowment fund, and J. Zhang was supported by the Geoffrey C. Hughes Foundation graduate fellowship program.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Ahmed, R., W. M. Canning, R. S. Kauffman, A. H. Sharpe, J. V. Hallum, and B. N. Fields. 1981. Role of the host cell in persistent viral infection: coevolution of L cells and reovirus during persistent infection. Cell 25325-332. [DOI] [PubMed] [Google Scholar]
- 2.Arbour, N., G. Cote, C. Lachance, M. Tardieu, N. R. Cashman, and P. J. Talbot. 1999. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J. Virol. 733338-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbour, N., S. Ekande, G. Cote, C. Lachance, F. Chagnon, M. Tardieu, N. R. Cashman, and P. J. Talbot. 1999. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J. Virol. 733326-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., J. F. Hedges, S. A. Nadler, W. H. McCollum, P. J. Timoney, and N. J. MacLachlan. 1999. Genetic stability of equine arteritis virus during horizontal and vertical transmission in an outbreak of equine viral arteritis. J. Gen. Virol. 801949-1958. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya, U. B., J. F. Hedges, V. L. Smalley, A. Navarrette, W. H. McCollum, P. J. Timoney, E. J. Snijder, and N. J. MacLachlan. 2004. Genetic characterization of equine arteritis virus during persistent infection of stallions. J. Gen. Virol. 85379-390. [DOI] [PubMed] [Google Scholar]
- 6.Balasuriya, U. B., E. J. Snijder, H. W. Heidner, J. Zhang, J. C. Zevenhoven-Dobbe, J. D. Boone, W. H. McCollum, P. J. Timoney, and N. J. MacLachlan. 2007. Development and characterization of an infectious cDNA clone of the virulent Bucyrus strain of Equine arteritis virus. J. Gen. Virol. 88918-924. [DOI] [PubMed] [Google Scholar]
- 7.Balasuriya, U. B., E. J. Snijder, L. C. van Dinten, H. W. Heidner, W. D. Wilson, J. F. Hedges, P. J. Hullinger, and N. J. MacLachlan. 1999. Equine arteritis virus derived from an infectious cDNA clone is attenuated and genetically stable in infected stallions. Virology 260201-208. [DOI] [PubMed] [Google Scholar]
- 8.Baybutt, H. N., H. Wege, M. J. Carter, and V. ter Meulen. 1984. Adaptation of coronavirus JHM to persistent infection of murine sac− cells. J. Gen. Virol. 65915-924. [DOI] [PubMed] [Google Scholar]
- 9.Benton, P. A., D. J. Barrett, R. L. Matts, and R. E. Lloyd. 1996. The outcome of poliovirus infections in K562 cells is cytolytic rather than persistent after hemin-induced differentiation. J. Virol. 705525-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borzakian, S., I. Pelletier, V. Calvez, and F. Colbere-Garapin. 1993. Precise missense and silent point mutations are fixed in the genomes of poliovirus mutants from persistently infected cells. J. Virol. 672914-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvez, V., I. Pelletier, S. Borzakian, and F. Colbere-Garapin. 1993. Identification of a region of the poliovirus genome involved in persistent infection of HEp-2 cells. J. Virol. 674432-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142629-633. [PubMed] [Google Scholar]
- 13.Chan, P. K., K. F. To, A. W. Lo, J. L. Cheung, I. Chu, F. W. Au, J. H. Tong, J. S. Tam, J. J. Sung, and H. K. Ng. 2004. Persistent infection of SARS coronavirus in colonic cells in vitro. J. Med. Virol. 741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, L. K., C. L. Liao, C. G. Lin, S. C. Lai, C. I. Liu, S. H. Ma, Y. Y. Huang, and Y. L. Lin. 1996. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology 217220-229.8599206 [Google Scholar]
- 15.Chen, W., and R. S. Baric. 1995. Function of a 5′-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J. Virol. 697529-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, W., and R. S. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 703947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cifone, M. A., and I. J. Fidler. 1981. Increasing metastatic potential is associated with increasing genetic instability of clones isolated from murine neoplasms. Proc. Natl. Acad. Sci. USA 786949-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colbere-Garapin, F., C. Christodoulou, R. Crainic, and I. Pelletier. 1989. Persistent poliovirus infection of human neuroblastoma cells. Proc. Natl. Acad. Sci. USA 867590-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couderc, T., N. Guedo, V. Calvez, I. Pelletier, J. Hogle, F. Colbere-Garapin, and B. Blondel. 1994. Substitutions in the capsids of poliovirus mutants selected in human neuroblastoma cells confer on the Mahoney type 1 strain a phenotype neurovirulent in mice. J. Virol. 688386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couderc, T., J. Hogle, H. Le Blay, F. Horaud, and B. Blondel. 1993. Molecular characterization of mouse-virulent poliovirus type 1 Mahoney mutants: involvement of residues of polypeptides VP1 and VP2 located on the inner surface of the capsid protein shell. J. Virol. 673808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crouch, E. C., K. R. Stone, M. Bloch, and R. W. McDivitt. 1987. Heterogeneity in the production of collagens and fibronectin by morphologically distinct clones of a human tumor cell line: evidence for intratumoral diversity in matrix protein biosynthesis. Cancer Res. 476086-6092. [PubMed] [Google Scholar]
- 22.de la Torre, J. C., E. Martinez-Salas, J. Diez, and E. Domingo. 1989. Extensive cell heterogeneity during persistent infection with foot-and-mouth disease virus. J. Virol. 6359-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Torre, J. C., E. Martinez-Salas, J. Diez, A. Villaverde, F. Gebauer, E. Rocha, M. Davila, and E. Domingo. 1988. Coevolution of cells and viruses in a persistent infection of foot-and-mouth disease virus in cell culture. J. Virol. 622050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. de Vries, M. C. Horzinek, and W. J. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 652910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desforges, M., J. Charron, S. Berard, S. Beausoleil, D. F. Stojdl, G. Despars, B. Laverdiere, J. C. Bell, P. J. Talbot, C. P. Stanners, and L. Poliquin. 2001. Different host-cell shutoff strategies related to the matrix protein lead to persistence of vesicular stomatitis virus mutants on fibroblast cells. Virus Res. 7687-102. [DOI] [PubMed] [Google Scholar]
- 26.Diez, J., M. Davila, C. Escarmis, M. G. Mateu, J. Dominguez, J. J. Perez, E. Giralt, J. A. Melero, and E. Domingo. 1990. Unique amino acid substitutions in the capsid proteins of foot-and-mouth disease virus from a persistent infection in cell culture. J. Virol. 645519-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diez, J., M. Hofner, E. Domingo, and A. I. Donaldson. 1990. Foot-and-mouth disease virus strains isolated from persistently infected cell cultures are attenuated for mice and cattle. Virus Res. 183-7. [DOI] [PubMed] [Google Scholar]
- 28.Dobbe, J. C., Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288283-294. [DOI] [PubMed] [Google Scholar]
- 29.Dryga, S. A., O. A. Dryga, and S. Schlesinger. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology 22874-83. [DOI] [PubMed] [Google Scholar]
- 30.Duncan, G., I. Pelletier, and F. Colbere-Garapin. 1998. Two amino acid substitutions in the type 3 poliovirus capsid contribute to the establishment of persistent infection in HEp-2c cells by modifying virus-receptor interactions. Virology 24114-29. [DOI] [PubMed] [Google Scholar]
- 31.Gosselin, A. S., Y. Simonin, F. Guivel-Benhassine, V. Rincheval, J. L. Vayssiere, B. Mignotte, F. Colbere-Garapin, T. Couderc, and B. Blondel. 2003. Poliovirus-induced apoptosis is reduced in cells expressing a mutant CD155 selected during persistent poliovirus infection in neuroblastoma cells. J. Virol. 77790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harnden, D. G., and H. P. Klinger (ed.) 1985. Birth defects: original article series, vol. 21, no. 1. ISCN (1985): an international system for human cytogenetics nomenclature. March of Dimes Birth Defects Foundation, New York, NY.
- 33.Hedges, J. F., U. B. Balasuriya, P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. 1999. Genetic divergence with emergence of novel phenotypic variants of equine arteritis virus during persistent infection of stallions. J. Virol. 733672-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedges, J. F., C. D. Demaula, B. D. Moore, B. E. McLaughlin, S. I. Simon, and N. J. MacLachlan. 2001. Characterization of equine E-selectin. Immunology 103498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann, M. A., R. Y. Chang, S. Ku, and D. A. Brian. 1993. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology 196163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann, M. A., S. D. Senanayake, and D. A. Brian. 1993. A translation-attenuating intraleader open reading frame is selected on coronavirus mRNAs during persistent infection. Proc. Natl. Acad. Sci. USA 9011733-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmann, M. A., P. B. Sethna, and D. A. Brian. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 644108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holyoak, G. R., T. V. Little, W. H. McCollam, and P. J. Timoney. 1993. Relationship between onset of puberty and establishment of persistent infection with equine arteritis virus in the experimentally infected colt. J. Comp. Pathol. 10929-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingram, V. M., M. P. Ogren, C. L. Chatot, J. M. Gossels, and B. B. Owens. 1985. Diversity among Purkinje cells in the monkey cerebellum. Proc. Natl. Acad. Sci. USA 827131-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan, G., A. Levy, and V. R. Racaniello. 1989. Isolation and characterization of HeLa cell lines blocked at different steps in the poliovirus life cycle. J. Virol. 6343-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamontagne, L. M., and J. M. Dupuy. 1984. Persistent infection with mouse hepatitis virus 3 in mouse lymphoid cell lines. Infect. Immun. 44716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavi, E., A. Suzumura, M. Hirayama, M. K. Highkin, D. M. Dambach, D. H. Silberberg, and S. R. Weiss. 1987. Coronavirus mouse hepatitis virus (MHV)-A59 causes a persistent, productive infection in primary glial cell cultures. Microb. Pathog. 379-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichtner, R. B., P. S. Moskwa, and G. L. Nicolson. 1987. Heterogeneous expression of cytokeratins in metastatic mammary adenocarcinoma cells in vitro and in vivo. Invasion Metastasis 7367-383. [PubMed] [Google Scholar]
- 44.Little, T. V., G. R. Holyoak, W. H. McCollum, and P. J. Timoney. 1991. Output of equine arteritis virus from persistently infected stallion is testosterone dependent, p. 225-229. In W. Plowright, P. D. Rossdale, and J. F. Wade (ed.), Proceedings of the 6th International Conference of Equine Infectious Diseases, Cambridge, United Kingdom.
- 45.MacLachlan, N. J., U. B. Balasuriya, J. F. Hedges, T. M. Schweidler, W. H. McCollum, P. J. Timoney, P. J. Hullinger, and J. F. Patton. 1998. Serologic response of horses to the structural proteins of equine arteritis virus. J. Vet. Diagn. Investig. 10229-236. [DOI] [PubMed] [Google Scholar]
- 46.McCollum, W. H., E. R. Doll, J. C. Wilson, and J. Cheatham. 1962. Isolation and propagation of equine arteritis virus in monolayer cell cultures of rabbit kidney. Cornell Vet. 52452-458. [PubMed] [Google Scholar]
- 47.McCollum, W. H., T. V. Little, P. J. Timoney, and T. W. Swerczek. 1994. Resistance of castrated male horses to attempted establishment of the carrier state with equine arteritis virus. J. Comp. Pathol. 111383-388. [DOI] [PubMed] [Google Scholar]
- 48.McCollum, W. H., and P. J. Timoney. 1999. Experimental observation on the virulence of isolates of equine arteritis virus, p. 558-559. In U. Wernery, J. F. Wade, J. A. Mumford, and O. R. Kaaden (ed.), Proceedings of the 8th International Conference on Equine Infectious Diseases, Dubai, United Arab Emirates, 1998.
- 49.Mizutani, T., S. Fukushi, K. Ishii, Y. Sasaki, T. Kenri, M. Saijo, Y. Kanaji, K. Shirota, I. Kurane, and S. Morikawa. 2006. Mechanisms of establishment of persistent SARS-CoV-infected cells. Biochem. Biophys. Res. Commun. 347261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore, B. D., U. B. Balasuriya, J. F. Hedges, and N. J. MacLachlan. 2002. Growth characteristics of a highly virulent, a moderately virulent, and an avirulent strain of equine arteritis virus in primary equine endothelial cells are predictive of their virulence to horses. Virology 29839-44. [DOI] [PubMed] [Google Scholar]
- 51.Nicolson, G. L. 1987. Tumor cell instability, diversification, and progression to the metastatic phenotype: from oncogene to oncofetal expression. Cancer Res. 471473-1487. [PubMed] [Google Scholar]
- 52.Okada, Y., G. Toda, H. Oka, A. Nomoto, and H. Yoshikura. 1987. Poliovirus infection of established human blood cell lines: relationship between the differentiation stage and susceptibility of cell killing. Virology 156238-245. [DOI] [PubMed] [Google Scholar]
- 53.Palacios, G., O. Jabado, N. Renwick, T. Briese, and W. I. Lipkin. 2005. Severe acute respiratory syndrome coronavirus persistence in Vero cells. Chin. Med. J. 118451-459. [PubMed] [Google Scholar]
- 54.Pavio, N., M. H. Buc-Caron, and F. Colbere-Garapin. 1996. Persistent poliovirus infection of human fetal brain cells. J. Virol. 706395-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavio, N., T. Couderc, S. Girard, J. Y. Sgro, B. Blondel, and F. Colbere-Garapin. 2000. Expression of mutated poliovirus receptors in human neuroblastoma cells persistently infected with poliovirus. Virology 274331-342. [DOI] [PubMed] [Google Scholar]
- 56.Pelletier, I., T. Couderc, S. Borzakian, E. Wyckoff, R. Crainic, E. Ehrenfeld, and F. Colbere-Garapin. 1991. Characterization of persistent poliovirus mutants selected in human neuroblastoma cells. Virology 180729-737. [DOI] [PubMed] [Google Scholar]
- 57.Pelletier, I., G. Duncan, and F. Colbere-Garapin. 1998. One amino acid change on the capsid surface of poliovirus sabin 1 allows the establishment of persistent infections in HEp-2c cell cultures. Virology 2411-13. [DOI] [PubMed] [Google Scholar]
- 58.Sawicki, S. G., J. H. Lu, and K. V. Holmes. 1995. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 695535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schafer, D. A., J. B. Miller, and F. E. Stockdale. 1987. Cell diversification within the myogenic lineage: in vitro generation of two types of myoblasts from a single myogenic progenitor cell. Cell 48659-670. [DOI] [PubMed] [Google Scholar]
- 60.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79961-979. [DOI] [PubMed] [Google Scholar]
- 61.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. Raamsman, and A. A. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 736335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stueckemann, J. A., M. Holth, W. J. Swart, K. Kowalchyk, M. S. Smith, A. J. Wolstenholme, W. A. Cafruny, and P. G. Plagemann. 1982. Replication of lactate dehydrogenase-elevating virus in macrophages. 2. Mechanism of persistent infection in mice and cell culture. J. Gen. Virol. 59263-272. [DOI] [PubMed] [Google Scholar]
- 63.Tijms, M. A., D. D. Nedialkova, J. C. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2007. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 8110496-10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timoney, P. J. 1988. Equine viral arteritis: epidemiology and control. J. Equine Vet. Sci. 854-59. [Google Scholar]
- 65.Timoney, P. J., and W. H. McCollum. 1993. Equine viral arteritis. Vet. Clin. N. Am. Equine Pract. 9295-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timoney, P. J., W. H. McCollum, and M. L. Vickers. 1997. The carrier stallion as a reservoir of equine arteritis virus. Equine Dis. Q. 62. [Google Scholar]
- 67.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 873473-3482. [DOI] [PubMed] [Google Scholar]
- 68.Wagner, H. M., U. B. Balasuriya, and N. James MacLachlan. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26251-260. [DOI] [PubMed] [Google Scholar]
- 69.Yamate, M., M. Yamashita, T. Goto, S. Tsuji, Y. G. Li, J. Warachit, M. Yunoki, and K. Ikuta. 2005. Establishment of Vero E6 cell clones persistently infected with severe acute respiratory syndrome coronavirus. Microbes Infect. 71530-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yunis, J. J. 1976. High resolution of human chromosomes. Science 1911268-1270. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, J., Y. Y. Go, N. J. MacLachlan, B. J. Meade, P. J. Timoney, and U. B. R. Balasuriya. Amino acid substitutions in the structural or non-structural proteins of a vaccine strain of equine arteritis virus are associated with its attenuation. Virology, in press. [DOI] [PubMed]
- 72.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81853-879. [DOI] [PubMed] [Google Scholar]