Abstract
Measles virus, a member of the Morbillivirus family, infects millions of people each year despite the availability of effective vaccines. The V protein of measles virus is an important virulence factor that can interfere with host innate immunity by inactivating alpha/beta interferon (IFN-α/β) and IFN-γ signaling through protein interactions with signal transducer and activator of transcription proteins STAT1 and STAT2. Here we demonstrate that although STAT1 interference results from protein interactions within a V protein N-terminal region encompassed by amino acids 110 to 130, detection of STAT1 interaction and IFN-γ signaling inhibition requires the presence of cellular STAT2. Cell-specific variability in STAT1 interference was observed to correlate with V protein expression level. A more direct target for measles virus V protein-mediated IFN-α/β evasion is STAT2. Results indicate that the widely conserved C-terminal zinc finger domain of measles virus V protein is both necessary and sufficient to bind STAT2 and disrupt IFN-α/β signal transduction. Mutagenesis and molecular modeling define a contact surface for STAT2 association that includes aspartic acid residue 248 as critical for STAT2 interference and IFN antiviral immune suppression. These findings clearly define the molecular determinants for measles virus IFN evasion and validate specific targets as candidates for therapeutic intervention.
Measles virus is a leading cause of death among young children despite the availability of a safe and effective vaccine for the past 40 years (28). Vaccination has greatly limited the spread of measles virus, and yet sufficient vaccine coverage has been difficult to achieve in developing countries. Factors such as immigration and public distrust of vaccine safety have contributed to local measles outbreaks even in developed countries, including the United States (5, 6, 29). A greater understanding of the molecular mechanisms underlying host evasion by this pathogen would facilitate the design of new therapeutic strategies by identifying targets for pharmacological inhibition that could augment or replace vaccinations in some situations.
Measles virus belongs to the Morbillivirus genus of the large Paramyxoviridae family (reviewed in reference 21). Most of these viruses share common genetic features, including a polycistronic gene that encodes two or more viral proteins from overlapping open reading frames (ORFs). In measles virus, a single gene encodes three proteins (C, P, and V) from a series of overlapping ORFs. The P/V/C locus of measles virus, like that of other paramyxoviruses, is associated with host immune evasion, and paramyxoviruses use these gene products for interference with the antiviral cytokines in the interferon (IFN) family. This interference includes inhibition of the critical antiviral IFN signaling (9) as well as the reported prevention of apoptosis (16, 48), cell cycle alterations (24), inhibition of double-stranded RNA signaling (16, 36), and prevention of IFN biosynthesis (16, 36, 48). In most cases, these activities are ascribed to the V protein, but specific cases of P- and C-mediated host evasion have been revealed (8, 10). The ORF encoding the P protein overlaps partially with a second ORF encoding the V protein. Access to the hidden ORF is achieved by cotranscriptional insertion of nontemplated guanine nucleotides at a precise location, or “editing site,” to generate alternate mRNAs that differ only by the presence or absence of one or two additional nucleotides. Due to this unusual coding strategy, the paramyxovirus P and V proteins share an amino terminus but have unique carboxyl termini (4, 45). Paramyxovirus V proteins are identifiable by their C-terminal domain (CTD), which codes for a conserved cysteine-rich region (21, 35, 45). The CTDs among all paramyxovirus V proteins are approximately 50% identical and invariably include one histidine and seven cysteine residues capable of binding two atoms of zinc (25, 35). Aside from this stoichiometry, which is similar to that of some cellular metalloproteins, the spacing of CTD cysteine residues is not consistent with that of known zinc-binding domains and no cellular V protein homologues have been described. Recent X-ray crystallographic studies confirm that the V protein CTD forms a unique zinc finger fold (23).
IFN family cytokines have long been recognized as fundamental mediators of innate antiviral responses (18). Alpha IFN (IFN-α) subtypes and IFN-β, referred to collectively here as IFN-α/β, are the principal antiviral cytokines produced by mammalian cells and act directly on target cells by blocking virus replication and enhancing adaptive immunity. IFNs have diverse effects on a variety of cell types, and both IFN-α/β and IFN-γ (a related but distinct cytokine) can cause diminished virus replication by using a number of mechanisms initiated by changes in gene expression upon IFN receptor stimulation (13). The principal intracellular signaling apparatus downstream of IFN-α/β receptors culminates in the assembly of an active transcription factor complex, ISGF3, which contains two signal transducer and activator of transcription (STAT) proteins, STAT1 and STAT2, and an IFN regulatory factor, IRF9 (12, 20). ISGF3 is responsible for directing the expression of the antiviral effector gene expression program leading to an antiviral state. Similar signaling downstream of the IFN-γ receptor leads to activation of a tyrosine-phosphorylated STAT1 homodimer that regulates a distinct subset of cellular genes that help shape the IFN-γ-mediated antiviral response.
Measles virus has been demonstrated to antagonize IFN-α/β and IFN-γ responses by V protein interference with STAT signal transduction. Prior investigations of measles virus V protein-mediated IFN signaling evasion produced several reports confirming IFN-α/β interference, but IFN-γ signaling interference was not always observed (3, 8, 10, 33, 44). This apparent discrepancy was attributed to differences in the virus strains or passage histories, host cell lines tested, or procedures used. The finding that measles virus can interfere with dendritic cell development and expansion by altering IFN-α/β responses that are STAT2 dependent and STAT1 independent (15) suggested that STAT2 may be an important target for measles virus-induced immune suppression (32).
Like other paramyxovirus V proteins, measles virus V protein can inhibit IFN antiviral responses at least in part through associations with both STAT1 and STAT2 (3, 33). Despite the importance of these protein interactions in determining measles virus virulence, little mechanistic information has been obtained regarding the nature of V protein interactions with STATs. Here, we resolve in molecular detail the biochemical basis for measles virus V protein engagement of STAT1 and STAT2. Detection of STAT1 association was found to be influenced by the V protein expression level and was highly stabilized by the presence of cellular STAT2. The STAT1 contact site was mapped to amino acids 110 to 130, in general agreement with prior reports of P and V protein interference (3, 4, 10, 31). In contrast, STAT2 was found to be a primary target for measles virus V protein IFN-α/β evasion, associating with specific residues of the conserved zinc finger domain, a unique mechanism among the paramyxoviruses.
MATERIALS AND METHODS
Cell culture.
Human HEK293T-, 2fTGH-, and 2fTGH-derived U6A (STAT2-deficient) or U3A (STAT1-deficient) cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% Cosmic calf serum (HyClone) and 1% penicillin-streptomycin (Gibco-BRL) as described previously (34).
For the stable cell lines, 2fTGH cells were transfected with pEF-FLAG vector or measles virus V or D248F plasmid along with a puromycin-resistant plasmid (pBABE-puro) (27). Individual clones were selected and then probed by FLAG immunoblotting for V protein expression. The cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% cosmic calf serum (HyClone), 1% penicillin-streptomycin (Gibco-BRL), and 1 μg/ml of puromycin (Sigma).
Plasmids, transfection, and reporter gene assays.
Measles virus V plasmid DNA (Edmonston ATCC strain) was used for PCR amplification of different fragments to add restriction endonuclease recognition sequences for direct cloning of the PCR products into a mammalian expression plasmid (pEF FLAG) downstream of an in-frame N-terminal FLAG epitope tag. The point mutations were made using Stratagene's QuikChange II XL mutagenesis kit. All plasmid constructs were verified by DNA sequencing.
High-efficiency transient transfection of HEK293T cells was carried out on 100-mm-diameter dishes by use of standard calcium phosphate procedures (2). The 2fTGH cells were transfected using SuperFect (Qiagen) or PEI (Polysciences Inc.) transfection reagents according to the instructions of the manufacturers.
Luciferase reporter gene assays were performed using HEK293T or 2fTGH cells as described previously (47). Briefly, the cells were transfected with a reporter gene, Renilla luciferase, and either empty vector or the cDNA expression plasmids. For IFN-γ response determinations, the reporter gene used contained four copies of m67-SIE (GAS) linked to a TATA box and the firefly luciferase ORF. The IFN-α/β-responsive reporter gene contains five copies of the ISG54 IFN-stimulated response element (ISRE) upstream of the TATA box and the firefly luciferase ORF. After 24 h, transfection medium was replaced with fresh medium or medium supplemented with 1,000 U of IFN-α/ml or 5 ng of IFN-γ/ml. At 14 h after IFN treatment, the cells were harvested and assayed for firefly and Renilla luciferase activity (dual luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those obtained with Renilla luciferase.
For MDA5 signaling assays, cells were transfected with myc-tagged MDA5 construct, MD5, −110 IFN-β luciferase reporter gene, and Renilla luciferase. After 24 h of transfection, the cells were stimulated with 5 μg of poly(I:C)/ml (Amersham Pharmacia) by transfection with Lipofectamine or Lipofectamine 2000 (Invitrogen). The cells were harvested after 6 h and assayed for firefly and Renilla luciferase activity. The data are graphed to represent the average values for triplicate samples normalized to cotransfected Renilla luciferase activity (expressed as a percentage of the value for the control stimulated sample).
Cell extracts, immunoblotting, and immunoprecipitation.
For preparation of cell extracts, samples were washed once with ice-cold phosphate-buffered saline and subsequently lysed with whole-cell-extract buffer (WCEB) as described previously (34, 46). For immunoblotting, proteins were separated, transferred to nitrocellulose, probed with antibodies, and visualized by chemiluminescence (NEN Life Sciences). For immunoprecipitation, lysates were prepared in WCEB and precleared with Sepharose beads. Antibody-protein complexes were purified with FLAG M2 (Sigma) or glutathione beads (GE Healthcare) and washed with WCEB. After elution with sodium dodecyl sulfate, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and processed for immunoblotting. The antibodies used were STAT1 (Santa Cruz C20), STAT2 (Santa Cruz, C24), FLAG (Sigma), and glutathione S-transferase (GST) (Sigma).
Indirect immunofluorescence.
For immunofluorescence, 2fTGH cells were grown on Permanox chamber slides (Nalgene Nunc) and transfected and stained exactly as described previously (40). Images were obtained using a Leica TCSSP confocal microscope, and representative fields with both transfected and untransfected cells in the same field are shown wherever possible for direct comparison.
Antiviral assays.
Antiviral responses measured by green fluorescent protein (GFP) reporter virus assays were conducted as reported previously (11, 17). Briefly, 2fTGH stable cell lines expressing empty vector, wild-type (WT) V protein, and mutant V protein were pretreated with IFN for 8 h. Cells were washed with serum-free media and infected with vesicular stomatitis virus-GFP (VSV-GFP) (9 × 105 PFU/well) for 1 h and cultured for 22 h in the presence of 2% serum-containing media. After 22 h, the cells were photographed and harvested for analysis by flow cytometry.
RESULTS
STAT2 is a primary target for measles virus V protein.
To better understand the diverse observations regarding measles virus V protein host interference, IFN signaling evasion assays were carried out using a variety of mammalian cell lines, including the human cell lines 2fTGH and HEK293T. Uniform IFN-α/β interference was observed for all lines tested, but the degree of IFN-γ signaling interference varied in a fashion that correlated with the intrinsic susceptibility of the cell line to plasmid transfection and the expressed V protein accumulation level. For example, our estimates indicate that transient transfection of equal amounts of plasmid vector into a confluent monolayer of 2fTGH or HEK293T cells results at least fourfold more protein for the HEK293T than for the 2FTGH cell line per microgram of total protein in a 24-h period (data not shown). We routinely observed modest IFN-γ signaling interference in the 2fTGH cells compared to a high degree of interference in the HEK293T cells (Fig. 1A and B).
FIG. 1.
STAT2 is a primary target of measles virus V protein. IFN-α/β and IFN-γ signaling interference by measles virus V (MeV) was tested using HEK293T (A) and 2fTGH (B) cells. Cells were transfected with an ISRE or GAS luciferase (Luc) reporter gene along with empty vector or measles virus V expression plasmid and were stimulated with IFN-α or IFN-γ (+) or left unstimulated (−) for 14 h prior to luciferase assays. (C) Analysis of STAT1 and STAT2 coprecipitation with measles virus V in 2fTGH cells [WT (2f)], U3A cells lacking STAT1 [U3A (−S1)], and U6A cells lacking STAT2 [U6A (−S2)]. STAT1 deficiency does not disrupt STAT2 interactions, but STAT2 deficiency disrupts STAT1 interactions. Complementation of STAT defects [(+S1), (+S2)] restores coprecipitation. C, control vector; V, measles virus V; IP, immunoprecipitation.
The ability of V proteins to interfere with STAT function generally relies on formation of stable protein interactions. To test the specificity of measles virus V protein interactions with STAT1 and STAT2 under moderate expression conditions, a V protein coimmunoprecipitation assay was carried out with transfected 2fTGH- or 2fTGH-derived cell lines harboring single gene defects in STAT1 (U3A) or STAT2 (U6A) (22, 30). Expressed FLAG-tagged measles virus V protein was immunoprecipitated using FLAG-M2 agarose beads and tested for coprecipitation of endogenous STATs. Results indicate that 2fTGH cells can support both STAT1 and STAT2 copurification with the expressed FLAG-tagged V protein (Fig. 1C). In the absence of STAT1 expression (U3A cells), the V protein association with STAT2 was retained. The converse was not true: in the absence of STAT2 (U6A cells), greatly reduced STAT1 association was observed. The defect in STAT1 coprecipitation could be rescued by complementing the cells with stable STAT2 expression. These data indicate that genetic deficiency in STAT2 can influence the association with STAT1. Apparently, STAT2 is a primary target for measles virus V-mediated IFN evasion, but STAT1 interference requires the presence of cellular STAT2 and was observed to correlate with V protein expression levels.
The measles virus V protein CTD is necessary and sufficient for STAT2 binding.
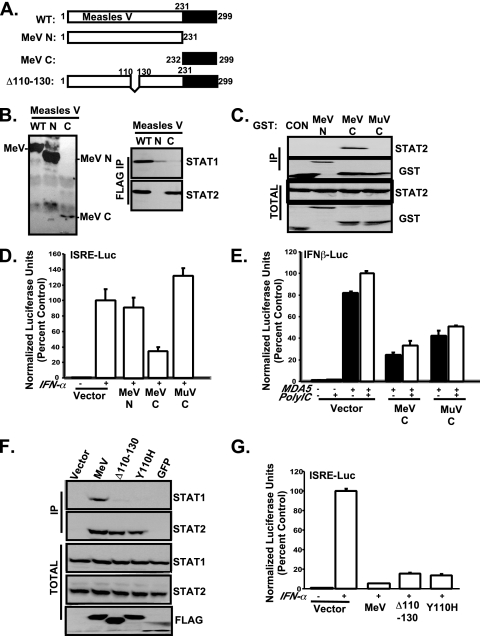
To guide the mapping of the V protein-STAT interaction sites, two FLAG-tagged measles virus V protein fragments were expressed. One of these encompassed the N-terminal 231 amino acids that are shared between measles virus V and P proteins, and the other was the isolated cysteine-rich CTD, consisting of residues 232 to 299 (Fig. 2A). While the N-terminal fragment accumulates to steady-state levels similar to those seen with the WT protein, the much smaller CTD (68 amino acids) was difficult to detect in the transfected cells. FLAG immunoprecipitation revealed that the full-length measles virus V protein coprecipitated both STAT1 and STAT2, but both fragments were greatly incapacitated for STAT1 interaction (Fig. 2B). A small amount of STAT1 coprecipitated with the N-terminal fragment, but STAT2 was readily detected in association with the CTD fragment despite its low level of accumulation. Therefore, the CTD is both necessary and sufficient to mediate STAT2 association. This result stands in contrast with those seen with other paramyxovirus V proteins for which CTD is not sufficient to mediate STAT interaction (see, e.g., references 38 and 46).
FIG. 2.
Measles virus V CTD is necessary and sufficient to bind STAT2. (A) Schematic diagram of measles virus V (MeV) protein and variants. Numbers indicate amino acid positions; the shaded boxes represent the CTD. N, N terminal; C, C terminal. (B) Coimmunoprecipitation of endogenous STATs with V protein fragments. HEK293 T cells were transfected to express V proteins, cell extracts were prepared for immunoprecipitation (IP) with FLAG M2 affinity resin, and the eluates were evaluated by STAT immunoblotting. The left panel illustrates total lysate results; the right panel shows the results obtained after FLAG immunoprecipitation. (C) Details are similar to those described for panel B except that cells were transfected with GST-V protein constructs and lysates were precipitated with glutathione agarose prior to immunoblotting. Control mumps virus CTD (MuV C) did not bind STAT2. (D and E) Effects of GST fusion proteins on IFN-α/β signaling (D) and MDA5 signaling (E). HEK293 T cells were transfected with ISRE-luc or −110-IFN-β-luc reporter genes along with GST-V expression plasmids and MDA5 construct. Cells were left unstimulated (−) or were stimulated (+) with 1,000 U of IFN/ml (8 h) or subjected to transfection with 5 μg of poly:(IC)/ml (6 h) prior to luciferase assays. Bars illustrate average values (n = 3); standard deviations are indicated. (F) STAT1 interference maps to residues 110 to 130. Coimmunoprecipitation of V protein and variants with endogenous STAT1 in HEK293T cells was performed in an experiment similar to that described for panel B. (G) Effect of V protein variants on IFN-α/β signaling inhibition. A luciferase assay for IFN-α/β signaling was executed as described for panel D.
To verify this result, GST fusions were constructed to help stabilize the CTD fragment. Glutathione purification of the expressed fusions confirmed that the measles virus V protein N terminus did not associate with STAT2. STAT2 bound specifically to the measles virus V CTD but not to the control mumps virus V protein CTD (Fig. 2C). The ability of the V protein fragments to interfere with IFN signaling was tested in an IFN-responsive luciferase reporter gene assay. Only the measles virus V CTD disengaged IFN-induced ISRE luciferase reporter gene activity, but neither the measles N-terminal domain nor the mumps virus CTD inhibited reporter gene activity (Fig. 2D). It has been demonstrated that the V protein interacts with MDA5 via the intact zinc finger domain (1). To verify the proper folding of both CTD fusions, their ability to disrupt signaling by the RNA signaling-helicase protein MDA5 was tested. Both measles virus and mumps virus CTD fusions were capable of disrupting MDA5-dependent signaling to the IFN-β promoter reporter gene (Fig. 2E). Based on these experiments, we conclude that the CTD interaction with STAT2 is a specific and unique adaptation of measles virus for overcoming IFN antiviral signal transduction.
Measles virus V IFN-α/β evasion is independent of the STAT1 binding site.
STAT1 can readily coprecipitate with full-length measles virus V protein in a STAT2-dependent manner but does not associate well with either the N terminus or the C terminus in isolation. In order to determine the protein interaction site for STAT1, a series of engineered overlapping deletions were screened for STAT1 coimmunoprecipitation and IFN-γ signaling interference (data not shown). The results implicated amino acids 110 to 130 as a potential STAT1 contact site (Fig. 2A). Deletion of this segment resulted in a failure to coprecipitate STAT1 but not STAT2 (Fig. 2F). This region coincides with a subset of residues common to the P and V proteins previously noted for extensive sequence conservation among members of the Morbillivirus family (8). Specifically, tyrosine residue 110 (Y110) of the P protein was implicated as important in regulating STAT1 activity in cells infected with V-deficient measles virus (8) and mutation of Y110 to histidine decreased V protein binding to overexpressed STAT1 (3). This mutant, Y110H, was defective for interaction with endogenous STAT1 in our assay, but STAT2 interaction remained intact (Fig. 2F). Importantly, these mutant V proteins remained potent antagonists of IFN-α-induced transcription (Fig. 2G). While residues 110 to 130 and, specifically, Y110 are required for STAT1 association, they are not required for STAT2 binding and IFN-α/β signal interference.
CTD structure and amino acid sequence specify STAT2 interaction.
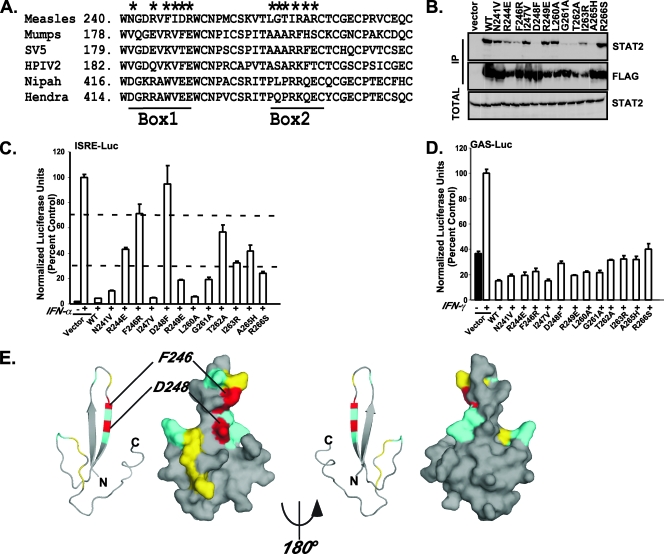
The paramyxovirus V protein CTD is highly conserved in amino acid sequence and has been characterized biochemically and structurally as a cysteine-rich zinc binding domain (25, 35). The crystal structure of SV5 V protein revealed that the conserved CTD forms a unique zinc finger fold as the result of the presence of the invariant histidine and seven cysteine residues coordinating two zinc atoms. The zinc binding produces a first finger that is 20 amino acids long and consists of two antiparallel β strands and a second finger of 12 residues (23). Amino acid sequence comparison of measles virus and mumps virus V CTDs (Fig. 3A) revealed three nonconserved regions, referred to as Box1, Box2, and Box3, that we hypothesized may contribute to the specific STAT2 association. Box1 and Box2 are embedded in the first and second zinc fingers, respectively, and Box3 is a unique C-terminal extension found only in the measles virus V protein.
FIG. 3.
CTD structure and amino acid sequence specify STAT2 interaction. (A) Comparison of measles virus and mumps virus V protein CTD sequences. Box1, Box2, and Box3 and zinc fingers 1 and 2 are indicated; five-pointed stars represent individual residues targeted for alanine substitution. (B) Coimmunoprecipitation (Co-IP) of box swap mutants Box1 (B1), Box2 (B2), and Box3 (B3), histidine substitution (H), or histidine/cysteine triple substitution (HC) with endogenous STAT2 in HEK293T cells. Vec, vector. (C) Effects of the V proteins on IFN-α/β signaling in HEK293T cells were tested as described for Fig. 1. (D) Effects of the box swap mutants on MDA5 signaling to the IFN-β reporter gene were tested as described for Fig. 2. (E) Effects of the V proteins on IFN-γ signaling in HEK293T cells were tested as described for Fig. 1.
To test the importance of these three regions in STAT2 antagonism, three measles virus V variants were constructed to remove Box3 or to replace Box1 and Box2 with the analogous residues of mumps virus V protein. This swapping strategy was used to preserve the overall zinc finger structure of the CTD but to replace the variable sequences from the STAT2-binding measles virus V protein CTD with those of the non-STAT2-binding mumps virus V protein CTD. In order to test the importance of the finger structure itself, alanine substitution mutations were engineered for the histidine (H232A) or the histidine plus the first two cysteines (H232A/C251A/C255A) in order to disrupt the zinc coordination (Fig. 3A).
The WT and variant V proteins were expressed in HEK293T cells, and FLAG immunoprecipitation was carried out to test for STAT2 binding (Fig. 3B). The WT protein bound well to STAT2, and deletion of Box3 had little effect. In contrast, both substitutions for Box1 and Box2 and the point mutations resulted in proteins that failed to precipitate STAT2. However, all the mutants retained their ability to bind STAT1.
The ability of these V proteins to disrupt IFN-α/β signal transduction was tested with an ISRE luciferase reporter gene assay. All of the proteins that bound STAT2 also retained the ability to inhibit IFN-α/β signaling (Fig. 3C). Importantly, despite the loss of STAT2 binding, V proteins with Box1 and Box2 substitutions retained the ability to inhibit MDA5 signaling, indicating that the overall CTD structure remained intact irrespective of the substitutions (Fig. 3D). In addition, all the mutants also retained the ability to inhibit IFN-γ/STAT1 signaling in HEK293T cells (Fig. 3E). Together, these data indicate that both the zinc finger structure and the specific amino acids within the fingers are required for measles virus V to engage STAT2.
Point mutations disengage V-STAT2 association.
Alignment of V protein CTDs revealed several measles-specific residues within Box1 and Box2 that could account for the ability to associate with STAT2. To further examine the amino acids important for STAT2 association, point mutations were designed to replace measles virus amino acids within Box1 and Box2 with the respective mumps virus residues (Fig. 4A). The mutant V proteins were tested for STAT2 coimmunoprecipitation (Fig. 4B) and IFN-α/β (Fig. 4C) or IFN-γ (Fig. 4D) signaling inhibition. These parameters differed greatly among the six Box1 and six Box2 mutations tested. Based on the ISRE luciferase reporter gene assay results, mutations N241V, I247V, and L260A retained more than 80% of the WT level of activity whereas others exhibited from 30% to 70% of WT activity (Fig. 4C). One mutation, D248F, produced the strongest defect in STAT2 binding and failed to disrupt IFN signaling, and a second, F246R, was severely compromised. Despite the differential effects on STAT2, all these mutant V proteins retained the ability to disengage IFN-γ/STAT1 signaling in HEK293T cells (Fig. 4D).
FIG. 4.
Point mutations disengage V-STAT2 association. (A) Sequence alignment of measles virus V protein with other paramyxovirus V proteins in the zinc finger region. Positions of Box1 and Box2 are shown; the residues indicated by five-pointed stars were replaced by the corresponding mumps virus V protein residues as described in the text. SV5, simian virus 5; HPIV2, type 2 human parainfluenza virus. (B) Coimmunoprecipitation (IP) of the point mutants with endogenous STAT2. (C) Effect of the mutants on IFN-α/β signaling. Dashed lines indicate 30% and 70% signaling interference thresholds. (D) Effect of the mutants in a IFN-γ signaling assay. (E) The measles virus V protein sequence was modeled on the SV5 structure (PDB accession no. 2B5L) by use of the Swiss PDB viewer available at http://www.expasy.org/spdbv/ (14). Each pair of images illustrates a ribbon view on the left and a surface view on the right, with front and back faces indicated by 180° rotation. Coloration corresponds to the activity level of mutations for each position. Gray represents nonmutated residues, cyan indicates neutral substitutions, yellow represents mutations that retain 30 to 80% activity, and red represents mutations that are more than 70% defective.
D248F disrupts V-mediated STAT2 cytoplasmic retention and antiviral signaling evasion.
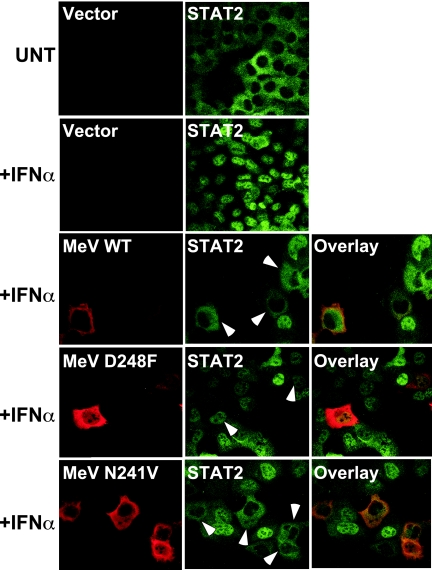
The disruption of measles virus V protein function by D248F mutation was characterized further in sensitive biological assays for IFN antiviral signaling. Measles virus V protein is known to prevent IFN-induced STAT2 nuclear translocation (33) and thereby to interfere with formation of the antiviral state. Indirect immunofluorescence was used to visualize the subcellular localization of STAT2 in 2fTGH cells in the presence of measles virus V proteins (Fig. 5). STAT2 is found in the cytoplasm of unstimulated cells, but IFN stimulation induces rapid translocation and nuclear accumulation (41). In the presence of measles virus V protein, IFN fails to induce STAT2 nuclear accumulation (33). The D248F mutant did not retain STAT2 in the cytoplasm, but a control mutant, N241V (which binds to STAT2), functioned like the WT V protein.
FIG. 5.
D248F mutation prevents V-mediated STAT2 cytoplasmic sequestration. 2fTGH cells were transiently transfected with empty vector or expression plasmids for the WT or mutant measles virus V (MeV) proteins indicated. Cells were left unstimulated (UNT) or were treated with 1,000 U of IFN-α/ml for 30 min prior to fixation, permeabilization, and sequential staining for a FLAG epitope tag to detect V protein and antiserum to detect endogenous STAT2. Arrows point to V-expressing cells.
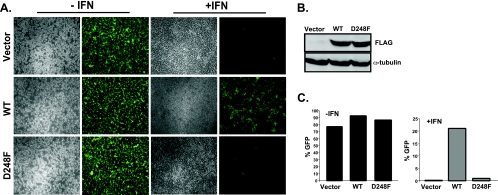
To verify the defective IFN evasion activity of the D248F mutant V protein in a biologically relevant context, a standard cytopathic effect assay was used to evaluate IFN antiviral responses. Stable cell lines expressing measles virus V protein or a D248F mutant were generated, and cell lines with similar levels of V protein abundance (Fig. 6B) were treated with IFN-α for 8 h followed by infection with a reporter virus containing a GFP transgene (VSV-GFP). Control cells with no expressed V protein were highly susceptible to infection, but IFN treatment dramatically reduced the number of fluorescent cells. Expression of WT measles virus V protein disrupted protection by IFN, resulting in uniformly higher GFP expression. In contrast, cells expressing the D248F V protein were unhindered in establishing an IFN-induced antiviral state, protecting the cells from VSV-GFP replication (Fig. 6A and C). Together, these findings confirm that aspartic acid residue 248 of measles virus V protein CTD is a critical determinant of host immune suppression.
FIG. 6.
D248F mutation is required for antiviral evasion. (A) Reporter virus assay for antiviral effects. 2fTGH stable cell lines expressing the empty vector or the V proteins indicated were left untreated (−IFN) or treated with IFN-α (+IFN) for 8 h and then infected with 9 × 105 PFU of VSV-GFP/well for 22 h prior to fluorescence microscopy. (B) Demonstration of V protein expression in stable cell lines with FLAG antibody and anti-α-tubulin control. (C) Quantification of VSV-GFP. The cells described for panel A were harvested, and 10,000 cells were analyzed by flow cytometry for GFP fluorescence.
DISCUSSION
Host immune responses provide a strong selective pressure for the evolution of viral evasion tactics. The ability to overcome host defenses enables the pathogen to succeed, allowing further diversification of the viral armamentarium. The results presented here demonstrate that measles virus has developed distinct mechanisms for disengaging the IFN-γ and IFN-α/β signal transduction machinery through separate associations with STAT1 and STAT2.
STAT2 appears to be a primary target for V protein-mediated IFN-α/β evasion and is engaged in the absence of STAT1 or the V protein's STAT1 binding site. Mapping the protein interaction domain for STAT2 revealed that the V protein CTD is necessary and sufficient for this interaction. This finding is unique to measles virus V protein and differs from the results obtained with other paramyxoviruses described to date. For example, members of the Rubulavirus family require both the N- and C-terminal domains for their STAT-directed evasion properties (34), and the V protein CTD is totally dispensable for STAT binding and IFN evasion for members of the Henipavirus family (38). Results demonstrate that despite the high degree of conservation in the paramyxovirus V protein CTD, sufficient differences exist in measles virus V protein for specific targeting of STAT2. Both amino acid sequence and zinc finger integrity were found to contribute to STAT2 association. The high conservation in the paramyxovirus V protein CTD enabled computer modeling of the measles virus CTD on the SV5 structure (Fig. 4E). This model illustrates that the Box1 and the Box2 regions lie on the same surface of the zinc fingers. Plotting the positions of incapacitating mutations revealed the close proximity of D248 and F246 within a three-dimensional pocket formed by both fingers that apparently serves to coordinate STAT2 associations along a single V protein interface. As this fold requires intact zinc coordination, such a model accounts for the observation that both structural integrity and amino acid sequence are required for STAT2 interaction. The reverse side of the CTD is free of STAT2-binding residues. It is tempting to speculate that this more conserved face of the CTD is responsible for multimeric interactions (46) or for association with other viral or cellular proteins such as MDA5, the RNA signaling helicase (1).
Results indicate that STAT1 can be targeted by V protein under optimal experimental conditions. This STAT1 interaction is mediated by an amino terminal peptide requiring residues 110 through 130, a region noted for high sequence conservation among members of the Morbillivirus family (8), that is present in both the P and V proteins. It is interesting that Nipah virus and Hendra virus, distantly related paramyxoviruses of the Henipavirus genus, also bind to STAT1 with residues in the P/V common region, via amino acids 100 to 160 for the Nipah virus P, V, and W proteins (26, 39, 42). For measles virus, this region originates with tyrosine 110, a residue that has been implicated in control of IFN responses. The original measles virus infectious clone, Ed-TAG, was shown to be defective in IFN evasion (7, 31, 37), and sequence comparisons revealed the presence of two amino acid substitutions. One of these changed cysteine 272 to arginine, destroying the CTD zinc finger, and a second was a substitution of histidine for tyrosine 110 (Y110H) (31). Subsequent reports of studies using the Moraten, Schwartz, or CAM70 strains confirmed the importance of Y110 in STAT1 interference (3, 8, 10). Our data indicate that this is also an important site of STAT1 physical interaction for the Edmonston ATCC strain, as deletion of amino acids 110 to 130 or Y110H mutation results in V proteins defective for endogenous STAT1 association and IFN-γ signaling evasion.
The common P/V N terminus is considered to be natively unstructured but able to adopt essential conformations upon engagement by a relevant cellular or virus-encoded apparatus (19). Interference with IFN-γ/STAT1-mediated signaling in transfected cell lines was differentially observed (Fig. 1), and the N-terminal STAT1 interaction site was apparently of relatively low affinity when presented without the CTD attached (Fig. 2). Both P and V have the intrinsic capacity to engage STAT1 via residues 110 to 130, but this outcome is influenced greatly by extrinsic factors such as protein abundance, expression context or host cell type, stability, and physical associations with other cellular and viral components. At high expression levels, mass action may drive STAT1 association with the V protein in the absence of the V protein CTD or cellular STAT2, whereas at moderate expression levels of V protein, efficient STAT1 binding to the V protein requires cellular STAT2. In the absence of V protein expression, the P protein's impact on STAT1 would be revealed. As is consistent with our findings, infectious measles virus lacking V protein expression retains the ability to antagonize STAT1 signaling via the P protein in a Y110-dependent reaction (8). This partial redundancy in STAT1 antagonism may have been acquired as a result of the unique P/V coding strategy but could be retained due to an advantage obtained by targeting IFN-γ versus IFN-α/β signals during the virus life cycle in vivo. These differential effects on signaling may become more apparent in the infection of animal models or in the context of natural measles virus infections with WT strains. Further, these actions may be additionally supported by the C protein, which was demonstrated to have additive effects on IFN signaling interference (8, 10).
It is clear from the present experiments and prior investigations (44) that IFN-α/β signal interference is more effectively achieved by measles virus V protein than IFN-γ interference. While P and V proteins share the STAT1 binding site and have the capacity to block IFN-γ signaling, results demonstrate that STAT2 is a primary target of measles virus V protein and that that targeting results in uniform IFN-α/β signaling interference. The specificity for STAT2 is likely related to the important role of STAT2 in mediating IFN-α/β responses in dendritic cells, an important target for measles virus and lymphocytic choriomeningitis virus immune suppression (15, 32).
We demonstrated here the importance of the measles virus V protein CTD in IFN-α/β signal interference and identified specific residues important for STAT2 recognition. Despite the availability of vaccines, measles virus infection is still a leading cause of illness and death among children. The results presented here give insight into the development of small molecules that can be used for the improvement of existing measles virus vaccines and as adjuvants for the use of measles virus as an oncolytic vector for human therapeutics (43), especially for patients with intact IFN systems.
Acknowledgments
We are grateful to Patricia Devaux and Roberto Cattaneo for insightful discussions, members of the Horvath Laboratory for critical evaluation and comments on the manuscript, and Louise E. Ludlow for generating the Y110H mutant.
This work was supported by NIH grant R01AI050707.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 3.Caignard, G., M. Guerbois, J. L. Labernardiere, Y. Jacob, L. M. Jones, F. Wild, F. Tangy, and P. O. Vidalain. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368351-362. [DOI] [PubMed] [Google Scholar]
- 4.Cattaneo, R., K. Kaelin, K. Baczko, and M. A. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56759-764. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2008. Multistate measles outbreak associated with an international youth sporting event—Pennsylvania, Michigan, and Texas, August-September 2007. MMWR Morb. Mortal. Wkly. Rep. 57169-173. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2008. Outbreak of measles—San Diego, California, January-February 2008. MMWR Morb. Mortal. Wkly. Rep. 57203-206. [PubMed] [Google Scholar]
- 7.Combredet, C., V. Labrousse, L. Mollet, C. Lorin, F. Delebecque, B. Hurtrel, H. McClure, M. B. Feinberg, M. Brahic, and F. Tangy. 2003. A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice. J. Virol. 7711546-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devaux, P., V. von Messling, W. Songsungthong, C. Springfeld, and R. Cattaneo. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 36072-83. [DOI] [PubMed] [Google Scholar]
- 9.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontana, J. M., B. Bankamp, W. J. Bellini, and P. A. Rota. 2008. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 37471-81. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, R. M. 1981. Interferons: a primer. Academic Press, New York, NY.
- 12.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 878555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312879-882. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 15.Hahm, B., M. J. Trifilo, E. I. Zuniga, and M. B. Oldstone. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity 22247-257. [DOI] [PubMed] [Google Scholar]
- 16.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 30315-32. [DOI] [PubMed] [Google Scholar]
- 17.Horvath, C. M., and J. E. Darnell, Jr. 1996. The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 protein. J. Virol. 70647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaacs, A., and J. Lindemann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B. 147258-267.13465720 [Google Scholar]
- 19.Karlin, D., S. Longhi, V. Receveur, and B. Canard. 2002. The N-terminal domain of the phosphoprotein of Morbilliviruses belongs to the natively unfolded class of proteins. Virology 296251-262. [DOI] [PubMed] [Google Scholar]
- 20.Kessler, D. S., S. A. Veals, X. Y. Fu, and D. E. Levy. 1990. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 41753-1765. [DOI] [PubMed] [Google Scholar]
- 21.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 22.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 151312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, T., X. Chen, K. C. Garbutt, P. Zhou, and N. Zheng. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124105-117. [DOI] [PubMed] [Google Scholar]
- 24.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 749152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198399-404. [DOI] [PubMed] [Google Scholar]
- 26.Ludlow, L. E., M. K. Lo, J. J. Rodriguez, P. A. Rota, and C. M. Horvath. 2008. Henipavirus V protein association with polo-like kinase reveals functional overlap with STAT1 binding and interferon evasion. J. Virol. 826259-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss, W. J., and D. E. Griffin. 2006. Global measles elimination. Nat. Rev. Microbiol. 4900-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulholland, E. K. 2006. Measles in the United States, 2006. N. Engl. J. Med. 355440-443. [DOI] [PubMed] [Google Scholar]
- 30.Müller, M., C. Laxton, J. Briscoe, C. Schindler, T. Improta, J. E. Darnell, Jr., G. R. Stark, and I. M. Kerr. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 124221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno, S., N. Ono, M. Takeda, K. Takeuchi, and Y. Yanagi. 2004. Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 852991-2999. [DOI] [PubMed] [Google Scholar]
- 32.Oldstone, M. B. 2006. Viral persistence: parameters, mechanisms and future predictions. Virology 344111-118. [DOI] [PubMed] [Google Scholar]
- 33.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 777635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 764190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208121-131. [DOI] [PubMed] [Google Scholar]
- 36.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 30333-46. [DOI] [PubMed] [Google Scholar]
- 37.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 145773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 785358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez, J. J., and C. M. Horvath. 2004. Host evasion by emerging paramyxoviruses: Hendra virus and Nipah virus v proteins inhibit interferon signaling. Viral Immunol. 17210-219. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 7611476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257809-813. [DOI] [PubMed] [Google Scholar]
- 42.Shaw, M. L., A. García-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 785633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Springfeld, C., V. von Messling, M. Frenzke, G. Ungerechts, C. J. Buchholz, and R. Cattaneo. 2006. Oncolytic efficacy and enhanced safety of measles virus activated by tumor-secreted matrix metalloproteinases. Cancer Res. 667694-7700. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, K., S. I. Kadota, M. Takeda, N. Miyajima, and K. Nagata. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545177-182. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304160-166. [DOI] [PubMed] [Google Scholar]
- 47.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 776385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 7610109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]