Abstract
Human metapneumovirus (HMPV) is a major causative agent of severe bronchiolitis and pneumonia. Its fusion (F) protein must be cleaved by host proteases to cause membrane fusion, a critical step for virus infection. By generating Vero cells constitutively expressing the transmembrane serine protease TMPRSS2 and green fluorescent protein-expressing recombinant HMPV, we show that TMPRSS2, which is expressed in the human lung epithelium, cleaves the HMPV F protein efficiently and supports HMPV multiplication. The results indicate that TMPRSS2 is a possible candidate protease involved in the development of lower respiratory tract illness in HMPV-infected patients.
Human metapneumovirus (HMPV) was discovered in 2001 as a major causative agent of respiratory tract infectious diseases (25); 5 to 15% of cases of severe bronchiolitis and pneumonia in young children are caused by the virus (27, 28). The clinical features of HMPV infection are similar to those of respiratory syncytial virus (RSV) infection (25, 27). Although both HMPV and RSV are classified in the Pneumovirinae subfamily of the Paramyxoviridae family, they have several differences in their gene organization, as well as in their viral protein functions (6, 24, 25). A striking difference is the cleavability of the fusion (F) protein, which is responsible for membrane fusion between the virus envelope and host cell membrane. The F protein of paramyxoviruses is synthesized as a single polypeptide, the F0 protein, which is then cleaved by cellular proteases at the site just upstream of the hydrophobic fusion peptide (13). This proteolytic cleavage is essential for the function of the paramyxovirus F protein to cause membrane fusion (13, 21). Uniquely, the RSV F protein has an additional cleavage site separated by 27 amino acids from the fusion peptide (10, 32). Both cleavage sites of the RSV F protein have multiple basic amino acid residues (arginine and lysine) and are cleaved by the ubiquitously expressed furin in the Golgi apparatus (6, 13). On the other hand, the cleavage site of the HMPV F protein has only two arginine residues separated by two uncharged residues and thus does not meet the requirements for recognition by furin (24). Under experimental conditions, the HMPV F protein is cleaved by trypsin, like the hemagglutinin (HA) protein of influenza virus (20, 21). Bottcher et al. (5) recently reported that the transmembrane serine proteases TMPRSS2 (17) and HAT (31), which are expressed in the human airway epithelium, can cleave the HA protein. In the present study we show, by generating Vero cells constitutively expressing TMPRSS2 (Vero/TMPRSS2) and green fluorescent protein-expressing HMPV, that TMPRSS2 efficiently supports the cleavage of the HMPV F protein and HMPV multiplication.
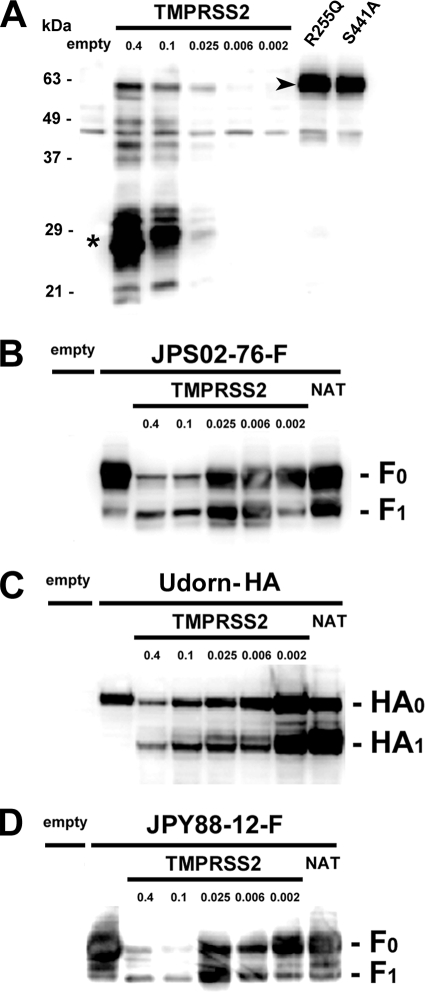
The ability of TMPRSS2 to cleave the HMPV F protein was first analyzed by using a transient-expression system. To generate an expression plasmid for TMPRSS2, total RNA was extracted from a human lung carcinoma cell line, Calu-3, and the cDNA encoding TMPRSS2 was amplified by reverse transcription and PCR using a primer pair, TATGAATTCCACCATGGCTTTGAACTCAGG and TATGCGGCCGCTTAGCCGTCTGCCCTCATT. The amplified fragment was then cloned into pcDNA3 vector (Invitrogen), generating pcDNA-TMPRSS2. The cDNA encoding the F protein was derived from the Japanese HMPV JPS02-76 strain (subtype 2B [11] or B1 [26]) isolated in 2002 (9) and cloned into pCA7 vector (16, 23). For detection by Western blotting, the HA tag sequence was added to the cytoplasmic tail of the F protein. TMPRSS2 was detected with the rabbit polyclonal antibody raised against the amino-terminal (cytoplasmic) region of TMPRSS2 (Abcam). Monolayers of 293T cells were transfected with different amounts of the expression plasmid pcDNA-TMPRSS2 (Fig. 1A). The major band of ∼28 kDa that appeared to be the membrane-anchored cleaved fragment of TMPRSS2 was detected in these cells (Fig. 1A, asterisk), as reported previously (2, 14, 29). To confirm that it is indeed the cleaved form generated by the autocatalytic activity of TMPRSS2, two TMPRSS2 mutants possessing a arginine-to-glutamine substitution at amino acid position 255 (R255Q) or a serine-to-alanine substitution at position 441 (S441A), respectively, were expressed in 293T cells. The former mutation is located in the proteolytic cleavage site of TMPRSS2 and disrupts the motif for recognition by the protease, while the latter is located in the protease domain and causes a loss of the catalytic activity of TMPRSS2 (2). As expected, the uncleaved full-length TMPRSS2 was detected as the major band in cells expressing either of these TMPRSS2 mutants (Fig. 1A, arrowhead).
FIG. 1.
Cleavage activity of TMPRSS2 for HMPV F and influenza virus HA proteins. (A) 293T cells were transfected with empty vector or different amounts of pcDNA-TMPRSS2 (0.4, 0.1, 0. 025, 0.006, and 0.002 μg, respectively). 293T cells were also transfected with pCA7 vectors expressing TMPRSS2 mutants, R255Q and S441A. Asterisk, cleaved form; arrowhead, full-length form. (B) 293T cells were transfected with empty pCA7 vector or pCA7 encoding the JPS02-76F protein (pCA7-JPS02-76-F) (lanes 2 to 8). Several samples were cotransfected with different amounts of pcDNA-TMPRSS2 (0.4, 0.1, 0. 025, 0.006, and 0.002 μg, respectively). A sample was treated with NAT. (C) A similar experiment to that in panel B using the expression plasmid encoding influenza virus HA protein (pCAGGS-HA) instead of pCA7-JPS02-76-F. (D) A similar experiment to that in panel B using pCA7 encoding the JPY88-12F protein instead of pCA7-JPS02-76-F. For all four panels, at 48 h posttransfection, proteins isolated from the cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and TMPRSS2, the F0 and F1 proteins, or the HA0 and HA1 proteins were detected by Western blotting.
Next, various amounts of pcDNA-TMPRSS2 were transfected into 293T cells, together with pCA7 vector encoding the HMPV F protein. Only 0.006 μg of pcDNA-TMPRSS2 was sufficient to cleave ∼50% of the F protein, even though expression of TMPRSS2 was below the detectable level in the Western blotting (Fig. 1A and B). Higher levels of TMPRSS2 expression reduced the F-protein expression levels (Fig. 1B). Similar data were obtained for the influenza virus HA protein (H3 subtype of A/Udorn/72 strain). About 50% of the HA protein was cleaved by transfecting cells with 0.002 μg of pcDNA-TMPRSS2 (Fig. 1C). The F protein of the HMPV JPY88-12 strain (9), classified in another subtype 1B (11) or A2 (26), was also cleaved efficiently by TMPRSS2 (Fig. 1D). These data indicate that TMPRSS2 cleaves the HMPV F protein efficiently as it does the influenza virus HA protein.
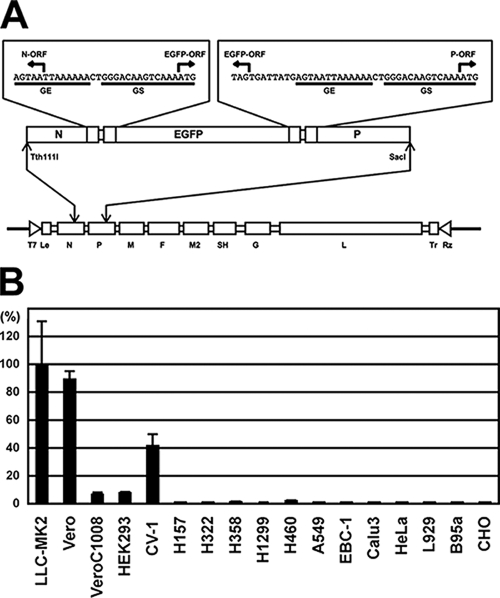
Since the above Western blotting data do not prove that the F protein is properly activated in the context of infectious virus, assays using HMPV were performed. HMPV replicates relatively slowly compared to other paramyxoviruses and often needs a few weeks to exhibit a detectable cytopathic effect (4, 25). Furthermore, no convenient method to titrate HMPV has been developed. The use of HMPV genetically engineered to express a fluorescent protein is therefore advantageous for analysis of HMPV infection (3, 8). In the present study we constructed a plasmid encoding the full-length genome of the HMPV JPS02-76 strain and established a reverse genetics system for it. An additional transcriptional unit encoding enhanced green fluorescent protein (EGFP) was created between the N and P genes (Fig. 2A). The full-length plasmid was transfected into the T7 RNA polymerase-expressing cell line BHK/T7-9 (kindly provided by N. Ito) (12), together with four support plasmids (pCITE-76N, -76P, -76-M2-1, and -76L) encoding the nucleocapsid, phospho-, M2 ORF1, and large proteins of the JPS02-76 strain, respectively. The transfected cells were cocultured with Vero cells at 3 days posttransfection. Infectious HMPV expressing EGFP (rJPS02-76EGFP) was generated efficiently when these cells were cultured in media supplemented with 2.0 μg of N-acetyl-trypsin (NAT; Sigma-Aldrich)/ml. Various cell lines were infected with rJPS02-76EGFP and cultured in appropriate culture media in the absence of trypsin. At 30 h postinfection (p.i.), infected (EGFP-expressing) cell numbers were counted under a fluorescence microscope. rJPS02-76EGFP infected Vero and LLC-MK2 cells most efficiently among the cell lines tested (Fig. 2B), a finding consistent with the results of previous studies (1, 3, 7, 18). When transiently expressed in Vero and LLC-MK2 cells, TMPRSS2 did not support multiple step growth of HMPV very well (data not shown). We speculate that innate immune responses elicited by plasmid transfection may inhibit HMPV growth and that the expression of TMPRSS2 may not be maintained at optimal levels for several days, which are needed for HMPV multiplication. To obtain cells stably expressing TMPRSS2, Vero and LLC-MK2 cells were cotransfected with pCA7 vector encoding TMPRSS2 (pCA7-TMPRSS2) and pCXN2 encoding the neo gene (16) and cultured in the presence of 1.0 mg of Geneticin (G418; Nacalai Tesque)/ml. Several Geneticin-resistant clones were obtained, but most of them grew very slowly and attached to culture plates weakly, even in the absence of Geneticin. One healthy Vero cell clone named Vero/TMPRSS2 was used for the following experiments.
FIG. 2.
Infectivity of HMPV in various cell lines as analyzed by using EGFP-expressing HMPV. (A) The full-length genome cDNA of the HMPV JPS02-76 strain flanked by the T7 promoter (T7) and hepatitis delta virus ribozyme (Rz) sequence was cloned into pBluescript vector (Stratagene). An additional transcription unit for EGFP was created between the N and P genes. Restriction enzyme recognitions sites for Tth111I and SacI were used for cloning. GE, gene end sequence; GS, gene start sequence; Le, leader sequence; Tr, trailer sequence. (B) Infectivity titers of rJPS02-76EGFP were determined in various cell lines. Cell lines were infected with the same stock of rJPS02-76EGFP and incubated in appropriate culture media in the absence of trypsin. At 30 h p.i., EGFP-expressing cell numbers (cell infectious units) were counted. The number determined in LLC-MK2 cells was set to 100%. LLC-MK2, Vero, Vero C1008, and CV-1 cells are monkey kidney-derived cell lines. HEK293 cells are a human kidney-derived cell line. H157, H322, H358, H1299, H460, A549, EBC-1, and Calu-3 cells are human lung carcinoma cell lines. HeLa cells are a human cervical adenocarcinoma cell line. L929 cells are a mouse fibroblast line. B95a cells are a marmoset B-lymphoblastoid cell line. CHO cells are a hamster ovary-derived cell line.
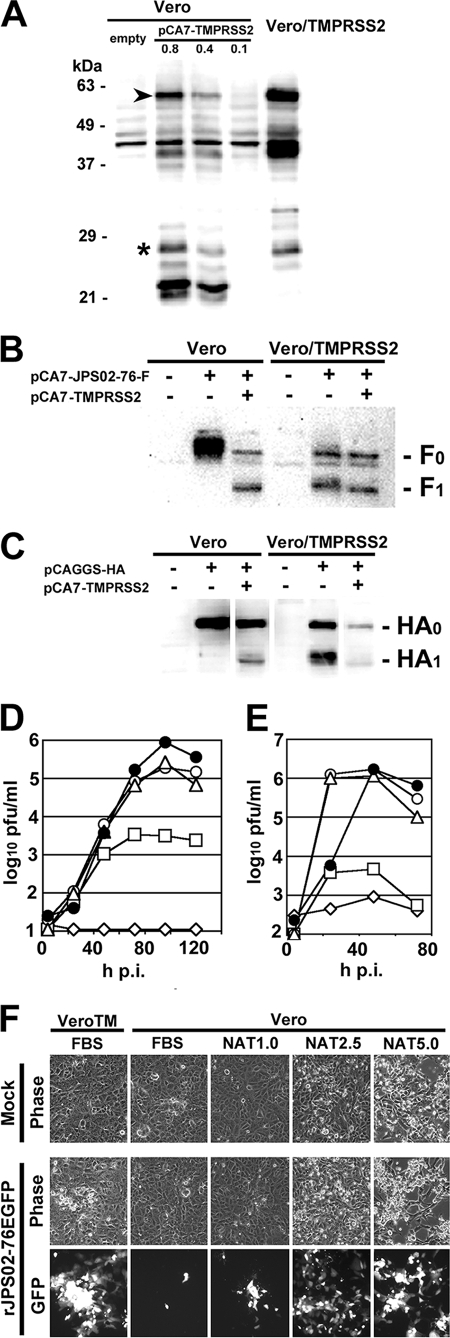
Expression of TMPRSS2 in Vero/TMPRSS2 was verified by Western blotting (Fig. 3A). As controls, Vero cells were transfected with empty pCA7 vector or different amounts of pCA7-TMPRSS2. Western blot analyses detected several TMPRSS2-specific products (Fig. 3A). TMPRSS2 appeared to exhibit a lower autocatalytic activity in Vero cells than in 293T cells, judging from the ratios between the cleaved (asterisk) and uncleaved full-length (arrowhead) forms of TMPRSS2 (Fig. 1A and 3A). Although Western blot analyses also detected other products of ∼40 and ∼22 kDa (Fig. 3A), their exact characters are currently unknown. When the HMPV F protein was expressed alone in Vero cells, only uncleaved F0 protein was detected (Fig. 3B). When the F protein was coexpressed with TMPRSS2 (using 0.1 μg of pCA7-TMPRSS2) in Vero cells, it was cleaved efficiently (Fig. 3B). In contrast, in Vero/TMPRSS2 cells the F protein was cleaved efficiently, even when it was expressed alone (Fig. 3B). Similar results were obtained for the influenza virus HA protein. Unlike in Vero cells, it was cleaved efficiently in Vero/TMPRSS2 cells without pCA7-TMPRSS2 transfection (Fig. 3C). Then, the growth kinetics of HMPV (rJPS02-76EGFP) and influenza virus (A/Udorn/72) was analyzed in Vero and Vero/TMPRSS2 cells (Fig. 3D and E). Without trypsin supplementation, neither rJPS02-76EGFP nor A/Udorn/72 replicated in Vero cells, whereas they grew efficiently in Vero/TMPRSS2 cells, showing maximum titers of ∼1 × 106 PFU/ml at 96 h p.i. and ∼2 × 106 PFU/ml at 48 h p.i., respectively (Fig. 3D and E). Although trypsin supplementation supported HMPV growth in Vero cells, the maximum titer obtained was still lower than that in Vero/TMPRSS2 cells (Fig. 3D). The lowered titers in Vero cells supplemented with trypsin, compared to those in Vero/TMPRSS2 cells, may be partly explained by the fact that the cells were damaged by trypsin (Fig. 3F). These data indicate that TMPRSS2 supports efficient multiple-step growth of HMPV and influenza virus.
FIG. 3.
Multiple-step growth of HMPV and influenza virus in Vero and Vero/TMPRSS2 cells. (A) Vero cells were transfected with empty pCA7 vector, or different amounts of pCA7-TMPRSS2 (0.8, 0.4, and 0.1 μg, respectively). TMPRSS2 in these cells at 48 h posttransfection and that in Vero/TMPRSS2 cells were detected by Western blotting. Asterisk, cleaved form; arrowhead, full-length form. (B) Vero or Vero/TMPRSS2 cells were transfected with pCA7-JPS02-76-F alone or together with pCA7-TMPRSS2. At 48 h posttransfection, F0 and F1 proteins were detected by Western blotting. (C) Vero or Vero/TMPRSS2 cells were transfected with pCAGGS-HA alone or together with pCA7-TMPRSS2. At 48 h posttransfection HA0 and HA1 proteins were detected by Western blotting. (D) Vero and Vero/TMPRSS2 cells were infected with rJPS02-76EGFP at a multiplicity of infection of 0.001. Vero/TMPRSS2 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 7.5% fetal bovine serum (FBS) (•). Vero cells were cultured in DMEM supplemented with 7.5% FBS (⋄) or different concentrations of NAT (1.0, 2.5, and 5.0 μg/ml) (□, ○, and ▵, respectively). Every day after infection, half of the culture media (1 ml) was obtained for plaque titration and replaced with the same amount of respective fresh medium. Plaque assays were performed on Vero/TMPRSS2 cells cultured in DMEM containing 1% methylcellulose and 7.5% FBS. (E) Vero and Vero/TMPRSS2 cells were infected with A/Udorn/72 at an MOI of 0.01. Cells were cultured under conditions as described in panel D, and virus titers in culture medium were determined by plaque assay on MDCK cells. (F) Images of rJPS02-76EGFP-infected Vero/TMPRSS2 (VeroTM) and Vero cells at 2 days p.i. obtained using phase-contrast (phase) and fluorescence (GFP) microscopes. Mock, mock-infected cells; rJPS02-76EGFP, rJPS02-76EGFP-infected cells.
We show here that TMPRSS2 can activate the HMPV F protein, as well as the influenza virus HA protein (5). Thus, the Vero/TMPRSS2 cells generated in the present study might be useful for the isolation of HMPV or influenza virus from clinical specimens. The site of influenza virus multiplication in vivo is in part regulated by the distribution pattern of cellular proteases, and growth of human strains of influenza virus is mostly restricted to the airway epithelium in spite of the ubiquitous expression of its receptor, sialic acid (30). On the other hand, most cell lines support HMPV infection poorly, irrespective of the trypsin supplementation. Therefore, other factors than the cleavage of the F protein should be also involved in the cell tropism of HMPV (Fig. 2B). Inconsistent with the tropism of HMPV in vivo, all three cell lines that supported HMPV entry (Vero, LLC-MK2, and CV1) are derived from monkey kidney, and none of the human lung carcinoma cell lines investigated supported its entry (Fig. 2B). Lack of TMPRSS2 expression in vivo may restrict HMPV growth in the human kidney (17), while proteases, including TMPRSS2, might greatly contribute to its growth in the lung in vivo. Our data do not exclude the possibility that other proteases may also cleave the HMPV F protein, as HAT does the influenza virus HA protein (5). It is also possible that TMPRSS2 indirectly supports the cleavage of the HMPV F protein by activating an enzymatic cascade. Previous studies have indicated that the distribution pattern of cellular proteases and the cleavability of viral proteins are major determinants of virus virulence (15, 19, 22, 30). Elucidation of the factors involved in cleavage of the F protein will thus advance our understanding of the molecular bases for HMPV pathogenicity and provide us with strategies to control HMPV and other related viruses.
Acknowledgments
We thank R. A. Fouchier for providing the reagents required for optimizing our HMPV reverse genetics system. We also thank N. Ito for allowing us to use BHK/T7-9 cells in our system.
This study was supported by the grants from the Naito Foundation and the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Abiko, C., K. Mizuta, T. Itagaki, N. Katsushima, S. Ito, Y. Matsuzaki, M. Okamoto, H. Nishimura, Y. Aoki, T. Murata, H. Hoshina, S. Hongo, and K. Ootani. 2007. Outbreak of human metapneumovirus detected by use of the Vero E6 cell line in isolates collected in Yamagata, Japan, in 2004 and 2005. J. Clin. Microbiol. 451912-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afar, D. E., I. Vivanco, R. S. Hubert, J. Kuo, E. Chen, D. C. Saffran, A. B. Raitano, and A. Jakobovits. 2001. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 611686-1692. [PubMed] [Google Scholar]
- 3.Biacchesi, S., M. H. Skiadopoulos, K. C. Tran, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2004. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology 321247-259. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 1861330-1334. [DOI] [PubMed] [Google Scholar]
- 5.Bottcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 809896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, P. L., and J. E. J. Crowe. 2007. Respiratory syncytial virus and metapneumovirus, p. 1601-1645. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 7.Deffrasnes, C., S. Cote, and G. Boivin. 2005. Analysis of replication kinetics of the human metapneumovirus in different cell lines by real-time PCR. J. Clin. Microbiol. 43488-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Graaf, M., S. Herfst, E. J. Schrauwen, B. G. van den Hoogen, A. D. Osterhaus, and R. A. Fouchier. 2007. An improved plaque reduction virus neutralization assay for human metapneumovirus. J. Virol. Methods 143169-174. [DOI] [PubMed] [Google Scholar]
- 9.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, H. Ishiko, M. Hara, Y. Takahashi, and K. Kobayashi. 2004. Human metapneumovirus infection in Japanese children. J. Clin. Microbiol. 42126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 989859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiguro, N., T. Ebihara, R. Endo, X. Ma, H. Kikuta, H. Ishiko, and K. Kobayashi. 2004. High genetic diversity of the attachment (G) protein of human metapneumovirus. J. Clin. Microbiol. 423406-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, N., M. Takayama-Ito, K. Yamada, J. Hosokawa, M. Sugiyama, and N. Minamoto. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47613-617. [DOI] [PubMed] [Google Scholar]
- 13.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 14.Lucas, J., L. True, S. Hawley, M. Matsumura, C. Morrissey, R. Vessella, and P. Nelson. 2008. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 215118-125. [DOI] [PubMed] [Google Scholar]
- 15.Nagai, Y. 1995. Virus activation by host proteinases. n>A pivotal role in the spread of infection, tissue tropism and pathogenicity. Microbiol. Immunol. 391-9. [DOI] [PubMed] [Google Scholar]
- 16.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 17.Paoloni-Giacobino, A., H. Chen, M. C. Peitsch, C. Rossier, and S. E. Antonarakis. 1997. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics 44309-320. [DOI] [PubMed] [Google Scholar]
- 18.Reina, J., F. Ferres, E. Alcoceba, A. Mena, E. R. de Gopegui, and J. Figuerola. 2007. Comparison of different cell lines and incubation times in the isolation by the shell vial culture of human metapneumovirus from pediatric respiratory samples. J. Clin. Virol. 4046-49. [DOI] [PubMed] [Google Scholar]
- 19.Rott, R., H. D. Klenk, Y. Nagai, and M. Tashiro. 1995. Influenza viruses, cell enzymes, and pathogenicity. Am. J. Respir. Crit. Care Med. 152S16-S19. [DOI] [PubMed] [Google Scholar]
- 20.Schickli, J. H., J. Kaur, N. Ulbrandt, R. R. Spaete, and R. S. Tang. 2005. An S101P substitution in the putative cleavage motif of the human metapneumovirus fusion protein is a major determinant for trypsin-independent growth in Vero cells and does not alter tissue tropism in hamsters. J. Virol. 7910678-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schowalter, R. M., S. E. Smith, and R. E. Dutch. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 8010931-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 2581-20. [DOI] [PubMed] [Google Scholar]
- 23.Takeda, M., S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, and Y. Yanagi. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 7914346-14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295119-132. [DOI] [PubMed] [Google Scholar]
- 25.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. Osterhaus, and R. A. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 413043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, S., B. Greer, J. Hooper, A. Zijlstra, B. Walker, J. Quigley, and S. Hawthorne. 2005. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 388967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright, P. F., G. Neumann, and Y. Kawaoka. 2007. Orthomyxoviruses, p. 1691-1740. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 31.Yamaoka, K., K. Masuda, H. Ogawa, K. Takagi, N. Umemoto, and S. Yasuoka. 1998. Cloning and characterization of the cDNA for human airway trypsin-like protease. J. Biol. Chem. 27311895-11901. [DOI] [PubMed] [Google Scholar]
- 32.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 27631642-31650. [DOI] [PubMed] [Google Scholar]