Abstract
While a diversity of immunogens that elicit qualitatively different cellular immune responses are being assessed in clinical human immunodeficiency virus vaccine trials, the consequences of those varied responses for viral control remain poorly understood. In the present study, we evaluated the induction of virus-specific T-cell responses in rhesus monkeys using a series of diverse vaccine vectors. We assessed both the magnitude and the functional profile of the virus-specific CD8+ T cells by measuring gamma interferon, interleukin-2, and tumor necrosis factor alpha production. We found that the different vectors generated virus-specific T-cell responses of different magnitudes and with different functional profiles. Heterologous prime-boost vaccine regimens induced particularly high-frequency virus-specific T-cell responses with polyfunctional repertoires. Yet, immediately after a pathogenic simian-human immunodeficiency virus (SHIV) challenge, no significant differences were observed between these cohorts of vaccinated monkeys in the magnitudes or the functional profiles of their virus-specific CD8+ T cells. This finding suggests that the high viral load shapes the functional repertoire of the cellular immune response during primary infection. Nevertheless, in all vaccination regimens, higher frequency and more polyfunctional vaccine-elicited virus-specific CD8+ T-cell responses were associated with better viral control after SHIV challenge. These observations highlight the contributions of both the quality and the magnitude of vaccine-elicited cellular immune responses in the control of immunodeficiency virus replication.
Evidence for the critical importance of cellular immunity in the control of human immunodeficiency virus (HIV) replication has served as an impetus for creating an AIDS vaccine that elicits potent CD8+ cytotoxic T-lymphocyte (CTL) responses (2, 20, 22). Studies of humans showing an association between CTL responses and clinical status, as well as studies of nonhuman primate models demonstrating a critical requirement for CD8+ lymphocytes in the clearance of a primate lentivirus, highlighted the importance of cellular immunity in HIV control. These findings provided an impetus for exploring a variety of vaccine strategies for eliciting HIV-specific cellular immune responses. In fact, HIV vaccine modalities that elicit high-frequency cellular immune responses are now being assessed in advanced human clinical efficacy trials (8, 9, 14).
While the evaluation of HIV vaccine candidates has, for the most part, depended on quantifying the magnitude of vaccine-elicited T-lymphocyte gamma interferon (IFN-γ) responses, emerging data are indicating that different vaccine modalities can induce cellular immune responses that are qualitatively very different (3-7, 12, 13, 16, 18, 23). Some vaccine modalities bias T-lymphocyte responses to CD8+ and others to CD4+ cells. Different vaccine platforms also induce T-lymphocyte responses with very different functional profiles. The consequences of these qualitative differences for viral control and clinical disease progression following infection remain poorly understood.
The present study was initiated to systematically explore the qualitative differences in the cellular immune responses generated in rhesus monkeys using different vaccine modalities. We had the opportunity to do this evaluation by comparing the immune responses elicited with different vaccine vectors that express the same gene inserts. We were also able to evaluate the ramifications of these qualitative differences in vaccine-induced T cells on lentiviral control and clinical disease progression in the vaccinated monkeys following primate lentivirus challenge.
MATERIALS AND METHODS
Selection of rhesus monkeys.
Heparinized blood samples were obtained from Mamu-A*01− rhesus monkeys (Macaca mulatta). All animals were maintained in accordance with the Guide for the Care and Use of Laboratory Animals (15) and with the approval of the Institutional Animal Care and Use Committee of Harvard Medical School and the National Institutes of Health.
Immunization and challenge of rhesus monkeys.
Archived peripheral blood lymphocytes (PBL) from 45 rhesus monkeys from previously reported studies (10, 11, 17, 19, 21) were assigned to five experimental groups that received different homologous or heterologous prime-boost immunizations. Plasmid DNA, rAd5, and rPox vaccine vectors were constructed as previously described and administered by intramuscular injection using a needle-free Biojector system. The immunization and challenge schedules for each experimental group of monkeys are summarized in Table 1. All experimentally vaccinated and 14 Mamu-A*01− control monkeys were challenged intravenously with 50 50% monkey infectious doses of pathogenic simian-human immunodeficiency virus SHIV-89.6P from the same original virus stock.
TABLE 1.
Immunization and challenge schedule
| Group | Priming immunization | Boosting immunization | SHIV-89.6P challenge at wk | Reference |
|---|---|---|---|---|
| rAd5-rAd5 (n = 6) | rAd5 (1012 particles; wk 0 and 8) | rAd5 (1012 particles; wk 26) | 48 | |
| rPox-rPox (n = 6) | rPox (109 PFU; wk 0 and 8) | rPox (109 PFU; wk 26 and 43) | 64 | 19 |
| DNA-DNA (n = 4) | DNA (5 mg; wk 0, 4, and 8); IL-2/Ig plasmid (5 mg; wk 0 and 4) | DNA (5 mg; wk 42) | 60 | 17 |
| DNA-rAd5 (n = 17) | DNA (4 mg; wk 0, 4, and 8) | rAd5 (1012 particles; wk 26) | 38 | 10 |
| DNA-rPox (n = 12) | DNA (5 mg; wk 0, 4, and 8); IL-2/Ig plasmid (5 mg; wk 0 and 4) | rPox (109 PFU; wk 42) | 60 | 17 |
CD4+ T-lymphocyte counts and plasma viral RNA levels.
Peripheral blood CD4+ T-lymphocyte counts were calculated by multiplying the total lymphocyte count by the percentage of CD3+ CD4+ T cells determined by monoclonal antibody (MAb) staining and flow cytometric analysis. Plasma viral RNA levels were measured by an ultrasensitive branched DNA amplification assay with a detection limit of 125 copies per ml (Siemens Diagnostics, Berkeley, CA).
Antibodies.
The antibodies used in this study were directly coupled with fluorescein isothiocyanate, phycoerythrin (PE), PE-Texas Red, peridinium chlorophyll protein-Cy5.5, PE-Cy7, AmCyan, Pacific Blue, allophycocyanin, Alexa Fluor 700, and Quantum-Dot 605. All reagents were validated and titrated using rhesus monkey PBL. The following MAbs were used: anti-tumor necrosis factor alpha (TNF-α)-fluorescein isothiocyanate (MAb11; BD Biosciences, San Jose, CA), anti-CD95-PE-Texas Red (DX2; BD Biosciences), anti-CD28-peridinium chlorophyll protein-Cy5.5 (L293; BD Biosciences), anti-IFN-γ-PE-Cy7 (B27; BD Biosciences), anti-CD3-Pacific Blue (SP34-2; BD Biosciences), anti-interleukin-2 (IL-2)-allophycocyanin (MQ1-17H12; BD Biosciences), anti-CD8α-Alexa Fluor 700 (RPA-T8; BD Biosciences), and anti-CD4-Quantum-Dot 605 (unconjugated CD4 antibody was obtained from BD Biosciences; Quantum-Dot 605 was obtained from Invitrogen, Carlsbad, CA). A violet fluorescent reactive dye (ViViD; Invitrogen) was also used as a viability marker to exclude dead cells in the analysis.
PBL stimulation and intracellular cytokine staining.
Purified PBL were isolated from EDTA-anticoagulated blood and frozen in the vapor phase of liquid nitrogen. The cells were later thawed and allowed to rest for 6 h at 37°C in a 5% CO2 environment. The viability of these cells was >90%. Peripheral blood mononuclear cells (PBMCs) were then incubated at 37°C in a 5% CO2 environment for 6 h in the presence of RPMI medium-10% fetal calf serum alone (unstimulated), a pool of 15-mer Gag or Env peptides (2 μg/ml of each peptide; AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, Germantown, MD), or staphylococcal enterotoxin B (5 μg/ml; Sigma-Aldrich, St. Louis, MO) as a positive control. All cultures contained monensin (GolgiStop; BD Biosciences) as well as 1 μg/ml anti-CD49d (BD Biosciences). Anti-CD28 and anti-CD49d MAbs are usually used to costimulate T-cell activation in intracellular cytokine-staining assays. Since we included anti-CD28 antibody in our staining panel of MAbs, we excluded the anti-CD28 MAb in the stimulation phase of the assay to avoid the downregulation of this molecule. The cultured cells were stained with MAbs specific for cell surface molecules, including CD3, CD4, CD8, CD28, and CD95. After fixing the cells with Cytofix/Cytoperm solution (BD Biosciences), the cells were permeabilized and stained with antibodies specific for IFN-γ, TNF-α, and IL-2. The labeled cells were fixed in 1% formaldehyde-phosphate-buffered saline.
The vaccine-induced cellular immune responses in the monkeys that received DNA-DNA, rAd5-rAd5, DNA-rAd5, and DNA-rPox vaccine regimens were assessed using a pool of 15-mer simian immunodeficiency virus SIVmac239 Gag peptides. Because the monkeys that received rPox-rPox immunizations developed very low frequency Gag-specific T-cell responses (19), Env-specific T-cell responses were evaluated using a pool of 20-mer HIV type 1 (HIV-1) 89.6P (KB9) Env peptides in these animals.
Flow cytometric analysis.
Samples were collected on a BD LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR). Approximately 500,000 to 1,000,000 events were collected per sample. Doublets were excluded by forward scatter (FSC) area versus FSC height. Dead cells were excluded by staining with amine reactive dye. CD4+ and CD8+ T cells were determined by their expression of CD3, CD4, or CD8. A functional analysis was done by plotting the expression of each cytokine molecule against another, and a Boolean combination of single functional gates was generated using FlowJo software. The frequency of cells producing IFN-γ, TNF-α, and IL-2, either individually or in any combination, was determined by FlowJo, formatted in PESTLE, and analyzed using SPICE software (both the PESTLE and SPICE software were provided by M. Roederer, NIH, Bethesda, MD). All values used for the analysis are background subtracted. Responses were considered positive when the percentage of total cytokine-producing cells was at least twice that of the background, and the cutoff for a positive response was 0.05%.
Statistical analyses.
Statistical analyses and graphical presentations were computed with GraphPad Prism. The Kruskal-Wallis test for multiple groups (or its equivalent Mann-Whitney test for two groups) was used to compare the naïve CD4+ T lymphocytes, peak and plateau viral RNA levels, and cellular immune responses of the different groups of experimental animals. Comparison of the cytokine distributions between groups was performed using a one-sided permutation test in SPICE.
RESULTS
Study design.
Archived viably frozen PBL from previously reported preclinical rhesus monkey vaccine studies of homologous and heterologous prime-boost immunization regimens were studied (10, 11, 17, 19, 21). The immunization and challenge schedules for each experimental group of monkeys are summarized in Table 1. In the homologous prime-boost experimental groups, six monkeys received 1012 particles of rAd5 as priming immunizations at weeks 0 and 8, followed by homologous rAd5 boosting at week 26. Six monkeys received 109 PFU of recombinant modified vaccinia virus Ankara (MVA) at weeks 0 and 8, followed by a homologous boosting at weeks 26 and 43. Another four monkeys received 5 mg plasmid DNA priming immunizations at weeks 0, 4, and 8, followed by a boosting immunization with homologous DNA plasmid at week 42. This group of experimental monkeys also received 5 mg inoculations of IL-2/immunoglobulin (Ig) plasmid on day 2 after both the week 0 and week 4 vaccinations. In the heterologous DNA-rAd5 experimental group, 17 monkeys received 4-mg inoculations of plasmid DNA as a priming immunization at weeks 0, 4, and 8, followed by a boosting immunization with 1012 particles of rAd5 at week 26. Twelve monkeys in the heterologous DNA-rPox experimental group received 5-mg inoculations of plasmid DNA as a priming immunization at weeks 0, 4, and 8 followed by a boosting immunization with 109 PFU of recombinant MVA, Vac, or fowlpox virus at week 42. Five milligrams of IL-2/Ig plasmid were also inoculated on day 2 after the week 0 and 4 vaccinations. At 12 weeks (DNA-rAd5), 18 weeks (DNA-rPox, DNA-DNA), 21 weeks (rPox-rPox), or 22 weeks (rAd5-rAd5) after the final immunizations, all experimental and 14 control monkeys were challenged intravenously with 50 50% monkey infectious doses of pathogenic SHIV-89.6P from the same original virus stock.
Vaccine-elicited cellular immune responses.
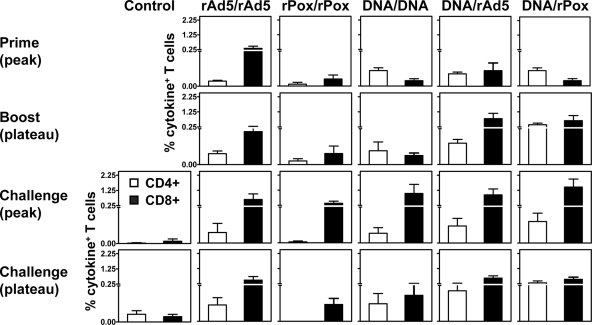
Since cellular immune responses contribute to primate lentivirus containment, we sought to determine whether these different vaccine strategies induce qualitatively or quantitatively different virus-specific T-lymphocyte responses. PBL from the monkeys receiving homologous or heterologous vaccination regimens were exposed to pools of overlapping peptides spanning the SIV Gag or HIV-1 Env protein, and the fractions of CD4+ or CD8+ T cells producing IFN-γ, TNF-α, or IL-2 were determined by intracellular cytokine staining. The sum total of the production of these three cytokines in response to these pooled-peptide stimulations is shown in the top two rows of Fig. 1.
FIG. 1.
Virus-specific cellular immune responses following vaccination and following SHIV-89.6P challenge. Six groups of Mamu-A*01− rhesus monkeys were evaluated: (i) control, comprising the monkeys immunized with a sham vaccine; (ii) rAd5-rAd5, comprising the monkeys immunized with a homologous rAd-rAd5 regimen; (iii) rPox-rPox, comprising the monkeys immunized with a homologous rPox-rPox regimen; (iv) DNA-DNA, comprising the monkeys immunized with a homologous DNA-DNA immunization regimen; (v) DNA-rAd5, comprising the monkeys immunized with a heterologous DNA-rAd5 regimen; and (vi) DNA-rPox, comprising the monkeys immunized with a heterologous DNA-rPox regimen. All experimental and 14 Mamu-A*01− control monkeys were then challenged with SHIV-89.6P. PBL isolated at the indicated times following vaccination or challenge were exposed to pools of overlapping peptides spanning the Gag or Env proteins, and the fractions of CD4+ or CD8+ T cells producing IFN-γ, TNF-α, or IL-2 were determined by intracellular cytokine staining. Data are presented as the mean frequencies of the cytokine-producing CD4+ or CD8+ T cells from each group of monkeys ± the standard errors of the means.
Monkeys primed with rAd5 developed high-frequency, sustained virus-specific CD8+ T-cell responses. In contrast, monkeys primed with rPox developed low-frequency, transient virus-specific CD8+ T-cell responses. Monkeys primed with plasmid DNA developed low-frequency virus-specific T-cell responses that included both CD4+ and CD8+ T cells, whereas animals that received a DNA immunization augmented with IL-2/Ig administration developed low-frequency virus-specific T-cell responses that predominantly included CD4+ T cells.
Homologous rAd5, rPox, or DNA boosting elicited no further increase in the magnitude of the antigen-specific T-cell responses. However, the monkeys that received DNA and IL-2/Ig immunizations developed a dramatic expansion of their virus-specific CD4+ and CD8+ T-cell responses following heterologous rPox boosting. A dramatic expansion of virus-specific T-cell responses, biased to CD8+ T cells, was seen after heterologous rAd5 boosting. Thus, a heterologous DNA prime, rAd5, or rPox boost immunization elicited the highest-frequency virus-specific T-cell responses.
Cellular immune responses following SHIV-89.6P challenge.
Since early control of primate lentivirus replication is mediated by the cellular immune response in the infected individual (22), we were interested in determining whether these different vaccination regimens affected the magnitude or quality of the cellular immune responses that developed in these monkeys following virus challenge (Fig. 1, bottom two rows). Interestingly, the magnitudes of the virus-specific CD8+ T-cell responses of these groups of vaccinated monkeys were indistinguishable 2 weeks after the challenge. The magnitudes of the mean cytokine responses were 0.64% ± 0.37% (rAd5-rAd5), 0.70% ± 0.16% (rPox-rPox), 1.04% ± 0.59% (DNA-DNA), 0.95% ± 0.39% (DNA-rAd5), and 1.46% ± 0.53% (DNA-rPox). Further, these virus-specific CD8+ T cells were preserved 12 weeks following the SHIV-89.6P challenge. As expected, the control monkeys generated only low-frequency virus-specific CD8+ T-cell responses following challenge, and highly significant differences were found between the magnitudes of the responses in the control and vaccinated groups of monkeys both 2 weeks and 12 weeks following SHIV-89.6P challenge (Kruskal-Wallis tests, P = 0.0001).
Functional profiles of virus-specific CD8+ T cells elicited by homologous or heterologous prime-boost vaccination.
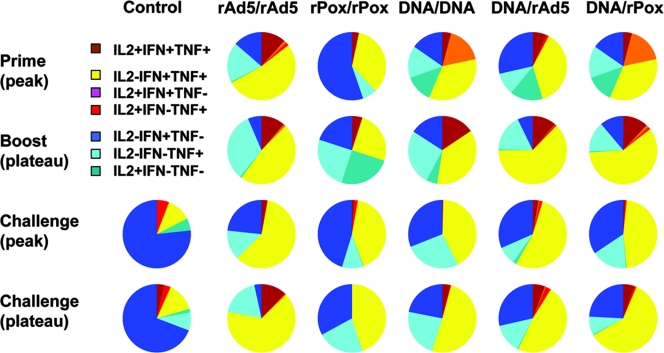
Recent studies have suggested that the heterogeneity of the functional capacities of virus-specific CD8+ T cells may be important in the control of disease progression in infected individuals (1). We therefore used MAb staining and polychromatic flow cytometry to assess the pooled-peptide-stimulated production of three cytokines to characterize the quality of the virus-specific T-cell responses elicited by the different vaccine strategies. Virus-specific cytokine-producing CD8+ T cells were divided into seven distinct populations based on their production of IFN-γ, TNF-α, and IL-2, either individually or in any combination (Fig. 2). The profiles of the functional capacities of the cells are shown by expressing each type of cytokine response as a proportion of the total response. The mean values for the animals in each experimentally vaccinated group are shown in a series of pie charts (Fig. 3, top two rows).
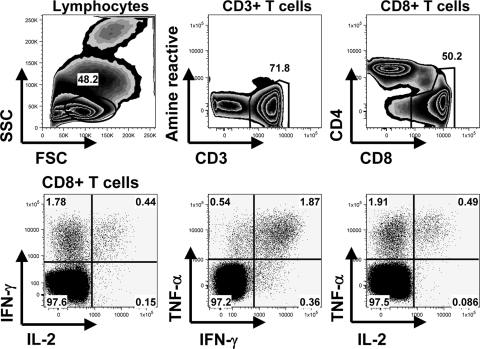
FIG. 2.
Gating strategy used to identify CD8+ T lymphocytes and cytokine expression. The top row shows the gating strategy used to define lymphocytes, based on FSC and side scatter (SSC), and CD8+ T lymphocytes, based on the expression of CD3 and CD8. Dead cells were excluded by amine reactive dye staining. The bottom row shows IFN-γ, TNF-α, or IL-2 expression in CD8+ T lymphocytes.
FIG. 3.
Cytokine profiles of antigen-specific CD8+ T cells following vaccination and following SHIV-89.6P challenge. PBL isolated from the different cohorts of rhesus monkeys at the indicated times following vaccination and following virus challenge were exposed to pools of overlapping peptides spanning the Gag or Env proteins, and cytokine production was measured by staining with MAbs and flow cytometric analysis. The antigen-specific CD8+ T cells were divided into seven distinct populations based on their production of IFN-γ, TNF-α, and IL-2. The cytokine profiles of these cells were determined by expressing each cytokine response as a proportion of the total antigen-specific cytokine-producing CD8+ T-cell response. Data were analyzed using SPICE software and are presented as the mean values from each experimental group in pie charts.
The predominant populations of polyfunctional cells in the vaccinated monkeys were those that produced both IFN-γ and TNF-α. IL-2 production was also detected in association with these other cytokines, but cell populations with these functions contributed less to the overall response. Monkeys primed with rAd5 or DNA developed polyfunctional virus-specific CD8+ T-cell responses that were predominantly IFN-γ+ TNF-α+ or IL-2+ TNF-α+. Animals primed with rPox developed polyfunctional responses as well, but these responses were biased toward the production of IFN-γ alone (blue).
Homologous boosting following rAd5, rPox, or DNA priming did not alter the quality of the vaccine-elicited CD8+ T-cell responses. However, the monkeys that received DNA immunizations demonstrated a dramatic expansion of their polyfunctional virus-specific CD8+ T-cell responses following heterologous rAd5 or rPox boosting, with three-fourths of the responses made up of either IFN-γ+ TNF-α+ or IFN-γ+ TNF-α+ IL-2+ cells.
Cytokine profiles of virus-specific CD8+ T cells following SHIV-89.6P challenge.
Interestingly, the cytokine profiles of the virus-specific CD8+ T cells of these groups of vaccinated monkeys were indistinguishable 2 weeks after challenge, with most cells being IL-2− IFN-γ+ TNF-α+ (Fig. 3, bottom two rows). Further, the cytokine profiles of these cells were for the most part unchanged 12 weeks following SHIV-89.6P challenge. In contrast, IFN-γ single-positive T cells dominated the virus-specific CD8+ T-cell responses of the control monkeys, and highly significant differences were found between the control and vaccinated groups in the proportion of polyfunctional virus-specific CD8+ T cells that were observed following SHIV-89.6P challenge (Kruskal-Wallis tests, P = 0.0001).
Plasma viral RNA levels and naïve CD4+ T lymphocytes following SHIV-89.6P challenge.
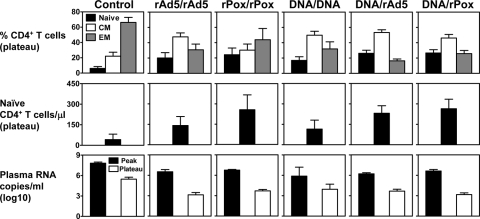
We then sought to determine whether viral replication and the rate of disease progression following SHIV-89.6P challenge differed in monkeys immunized with these different vaccine regimens (Fig. 4). Plasma viral RNA levels were maximal at day 14 after challenge in all control and experimentally vaccinated monkeys. The experimentally vaccinated monkeys had significantly lower peak plasma viral RNA levels than did the control animals (Kruskal-Wallis tests, P = 0.0001). However, the five groups of experimentally vaccinated monkeys did not demonstrate significant differences in their peak plasma viral RNA levels. The set-point viral RNA levels were evaluated in all of the monkeys on day 84 after challenge. The control monkeys had over 2-log-higher plasma viral RNA levels on day 84 than the vaccinated animals, and the difference in these levels between the control and vaccinated monkeys was highly significant (Kruskal-Wallis tests, P = 0.0001). However, no statistically significant differences were observed between the five experimental groups in their set-point plasma viral RNA levels.
FIG. 4.
Peripheral blood naïve CD4+ T lymphocytes and plasma viral RNA levels of different cohorts of control and vaccinated rhesus monkeys following SHIV-89.6P challenge. The monkeys were immunized by a homologous rAd5-rAd5, rPox-rPox, or DNA-DNA or heterologous DNA-rAd5 or DNA-rPox regimen and then challenged with SHIV-89.6P. PBL were evaluated for CD4+ T-lymphocyte subsets on day 84 after SHIV-89.6P challenge. Peripheral blood CD4+ T lymphocytes were divided into naïve, central memory (CM), and effector memory (EM) subpopulations based on their expression of CD28 and CD95. The relative distribution of these CD4+ T-cell subsets for each cohort of monkeys is shown in the top row, and the mean absolute naïve CD4+ T-cell counts for each cohort of monkeys is shown in the middle row. A comparison of the peak plasma virus RNA levels on day 14 and the plateau plasma virus RNA levels on day 84 after SHIV-89.6P challenge is shown in the bottom row. Data are presented as the mean values for each group of monkeys ± the standard errors of the means.
Because naïve CD4+ T lymphocytes are selectively infected and eliminated by the CXCR4-tropic virus SHIV-89.6P, we also evaluated the peripheral blood naïve CD4+ T cells in the challenged monkeys. The peripheral blood CD4+ T lymphocytes were divided into naïve, central memory, and effector memory subpopulations based on their expression of CD28 and CD95. The relative representation of each CD4+ T-cell subset on day 84 after the challenge was assessed (Fig. 4, top two rows). Naïve CD4+ T cells were preserved in the vaccinated animals. In contrast, this population of cells was selectively depleted in the control animals. In fact, a highly significant difference was apparent in the percentages of circulating naïve CD4+ T cells between the control and vaccinated monkeys (Kruskal-Wallis tests, P = 0.0007). As with the plasma viral RNA levels, no significant differences were found in the relative representation of peripheral blood naïve CD4+ T cells between the five experimental groups of monkeys. Consistent with the relative proportion of peripheral blood naïve CD4+ T cells in these animals, absolute naïve CD4+ T cells were also preserved in the vaccinated but not the control monkeys, and no significant differences in the absolute naïve CD4+ T-cell counts were observed between the groups of experimentally vaccinated monkeys.
Magnitude and quality of vaccine-induced virus-specific CD8+ T-cell responses were associated with control of viral replication following SHIV-89.6P challenge.
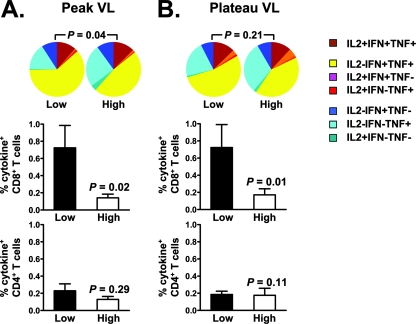
While the extent of protection against viral replication was indistinguishable between the cohorts of animals receiving different experimental vaccine regimens, the variability in the quantity and quality of the virus-specific CD8+ T cells within each group of vaccinated monkeys may have obscured the contributions of these aspects of the immune response to the control of virus. To evaluate this possibility, we divided all 33 animals that received experimental vaccines into two groups, low and high, on the basis of the magnitudes of their peak and plateau plasma viral RNA levels, and both the quantity and quality of their virus-specific CD4+ and CD8+ T-cell responses after the boost immunizations were assessed (Fig. 5). Interestingly, high-magnitude (P = 0.02) and more polyfunctional (P = 0.04) vaccine-elicited virus-specific CD8+ T cells were detected in animals that demonstrated low peak plasma viral RNA levels following SHIV-89.6P challenge (Fig. 5A, top two panels). Furthermore, high-magnitude (P = 0.01) vaccine-elicited virus-specific CD8+ T cells were also seen in the monkeys with low set-point plasma viral RNA levels (Fig. 5B, top two panels). However, no significant differences were observed between these two groups of vaccinated monkeys in the magnitudes and functional profiles of their vaccine-elicited virus-specific CD4+ T cells (Fig. 5, bottom panels). Thus, both the magnitudes and functional profiles of the virus-specific CD8+ T cells generated by vaccination were associated with control of viral replication following SHIV-89.6P challenge.
FIG. 5.
Higher magnitude and more polyfunctional vaccine-elicited antigen-specific CD8+ T-cell responses were associated with reduced plasma viral RNA levels following SHIV-89.6P challenge. The 33 animals that received different vaccine regimens were divided into two groups, low and high, based on the magnitude of their peak (A) or plateau (B) plasma viral RNA levels (VL). PBL isolated following the boost immunization were exposed to pools of overlapping peptides spanning the Gag or Env proteins, and the fraction of CD8+ or CD4+ T cells producing IFN-γ, TNF-α, or IL-2 was determined by intracellular cytokine staining. In the bottom two panels, data are presented as the mean values for each group of monkeys ± the standard errors of the means. Differences in the magnitudes of the cytokine-producing CD8+ or CD4+ T cells between the two experimental groups were analyzed using the Mann-Whitney test. In the top panels, functional profiles of the vaccine-elicited antigen-specific CD8+ T cells were analyzed using SPICE software.
DISCUSSION
These studies demonstrate striking qualitative differences in the lineages of the cellular responses elicited by each of the evaluated vaccine modalities. The rAd5-induced responses predominantly involved CD8+ T lymphocytes, while the plasmid DNA-elicited cells were predominantly CD4+ T lymphocytes. Although these biases did not change following homologous boosting, they changed substantially following heterologous boosting, with the development of high-frequency, balanced responses.
We also evaluated the functional profiles of the virus-specific CD8+ T cells by measuring their IFN-γ, IL-2, and TNF-α production. The expression of several other molecules has been used by other investigators to assess the function of T cells, including β-chemokines (macrophage inflammatory protein 1β [MIP-1β]) and molecules that are associated with cytolytic activity (CD107). Previous studies from our laboratory and the laboratories of others have shown that as HIV/SIV-induced disease progresses, HIV/SIV-specific CD8+ T cells first lose their ability to produce IL-2 and later TNF-α, while their ability to produce IFN-γ and MIP-1β and their expression of CD107 can be preserved at the very late stages of disease. We therefore decided not to evaluate CD107 and MIP-1β expression in the present study, and we used a relatively limited set of anti-cytokine MAbs to evaluate the functional profile of the virus-specific T cells.
Monkeys receiving rAd5, rPox, and plasmid DNA developed virus-specific polyfunctional CD8+ T-lymphocyte responses following the initial administration of a single vaccine immunogen, although there was some bias toward IFN-γ-only production in the rPox-immunized monkeys. This finding of an IFN-γ bias differs from recent reports of polyfunctionality in rPox-induced cellular immune responses in humans (16). However, importantly, when rAd5 or rPox vectors were employed as boosting immunogens following plasmid DNA priming, the profile of the CD8+ T-lymphocyte response was mostly polyfunctional.
Although the different vaccination regimens generated qualitatively different virus-specific T-cell populations, those differences were lost following the virus challenge. While the T lymphocytes of the control vaccinees made only low-frequency virus-specific responses, both the CD8+ and CD4+ T-lymphocyte responses in all groups of experimentally vaccinated monkeys were robust. Further, the profile of cytokine production by the virus-specific T lymphocytes in the control monkeys was heavily biased toward cells that produce only IFN-γ, while the virus-specific T lymphocytes of all of the experimentally vaccinated monkeys following challenge were uniformly polyfunctional. Most importantly, we observed no significant differences between any of the cohorts of vaccinated monkeys following virus challenge in the magnitudes of their virus-specific cellular immune responses, the biases of those responses to CD8+ T lymphocytes, and the functional profiles of those cells. This uniformity of virus-specific T lymphocytes in the vaccinated monkeys likely reflects the overwhelming influence on lymphocyte differentiation of the high levels of viral antigen present in these animals.
Consistent with these findings, we observed lower plasma viral RNA levels and better preservation of naïve CD4+ T lymphocytes in the vaccinated than in the control monkeys. However, there was no significant difference in these clinical parameters between the various groups of experimentally vaccinated monkeys. Thus, while the immunologic profiles of the vaccine-elicited T lymphocytes differed considerably between these groups of vaccinees, the critical cellular immune responses that were mounted following virus infection and the clinical consequences of those infections were indistinguishable between the groups.
Interestingly, when all animals that received different vaccine immunizations were grouped together and divided into two groups based on the magnitudes of their peak plasma viral RNA levels, highly significant differences were observed in both the quantity and quality of the vaccine-elicited virus-specific CD8+ T cells between these two groups. This finding was not apparent when analyzing each cohort of monkeys separately. These observations suggest that both the quantity and quality of the vaccine-induced immunodeficiency virus-specific CD8+ T-cell responses were associated with control of viral replication.
CD8+ T cells mediate multiple effector functions during acute HIV infection but become exhausted and lose their ability to produce some cytokines with the persistence of viral antigenemia during chronic HIV infection. Although many studies provide insights into how the functional capacity of the HIV-specific CD8+ T cells correlates with the clinical course of disease progression, it remains unclear whether the maintenance of CTLs with a “polyfunctional” profile in long-term nonprogressors is an epiphenomenon associated with good viral control or is the mechanism responsible for the good clinical status. Although we found that both the magnitude and polyfunctionality of the vaccine-elicited CD8+ T-cell responses were predictive of the viral set point after infection, it is still possible that polyfunctionality is a consequence of vaccine take in these monkeys.
Virus replication is contained by cellular immune responses during the first days following infection. The magnitude of that cellular response is modified by prechallenge vaccine-elicited cellular immune responses. However, the findings in the present study suggest that the comparable viral antigen load in all cohorts of vaccinated monkeys during the primary infection was associated with comparable functional repertoires of the cellular immune responses generated in response to the infection. These findings likely explain why no significant differences were observed between the different cohorts of vaccinated monkeys in the magnitudes and the functional profiles of their virus-specific CD8+ T cells following viral infection.
There are certainly important caveats that must be acknowledged when interpreting these findings. The available macaque challenge systems do not perfectly model HIV-1 infection in humans. More specifically, the CXCR4-tropic SHIV-89.6P used in these challenge studies causes a disease that is very different than that caused by CCR5-tropic strains of SIV and HIV-1. SHIV-89.6P preferentially infects CXCR4+ naïve CD4+ T cells, whereas SIVmac251 and HIV-1 mainly target activated memory CD4+ T cells. In macaque models, T-cell-based vaccines provide long-term protection against SHIV-89.6P. However, the same live recombinant vaccines only provide short-term control of viral replication after a SIVmac251 challenge. Moreover, the recent STEP human clinical trial suggests that the dramatic protection seen in the SHIV-89.6P macaque model may not correlate with clinical protection against HIV infection in humans. Nevertheless, the findings in the present study raise the possibility that differences in both the magnitude and quality of vaccine-elicited CD8+ T cells generated by different vaccine modalities may be of importance for controlling virus replication.
Acknowledgments
We are grateful to Mark Cayabyab, Avi-Hai Hovav, Robert Seder, John Mascola, and Gary Nabel for helpful conversations and Michelle Lifton for technical assistance.
This work was supported in part by funds from the intramural research program of the Vaccine Research Center, NIAID, NIH; the Harvard Medical School CFAR grant AI060354; and the NIH grant N01-AI30033.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 3.Casimiro, D. R., L. Chen, T. M. Fu, R. K. Evans, M. J. Caulfield, M. E. Davies, A. Tang, M. Chen, L. Huang, V. Harris, D. C. Freed, K. A. Wilson, S. Dubey, D. M. Zhu, D. Nawrocki, H. Mach, R. Troutman, L. Isopi, D. Williams, W. Hurni, Z. Xu, J. G. Smith, S. Wang, X. Liu, L. Guan, R. Long, W. Trigona, G. J. Heidecker, H. C. Perry, N. Persaud, T. J. Toner, Q. Su, X. Liang, R. Youil, M. Chastain, A. J. Bett, D. B. Volkin, E. A. Emini, and J. W. Shiver. 2003. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J. Virol. 776305-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catanzaro, A. T., M. Roederer, R. A. Koup, R. T. Bailer, M. E. Enama, M. C. Nason, J. E. Martin, S. Rucker, C. A. Andrews, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 254085-4092. [DOI] [PubMed] [Google Scholar]
- 5.Edupuganti, S., D. Weber, and C. Poole. 2004. Cytotoxic T-lymphocyte responses to canarypox vector-based HIV vaccines in HIV-seronegative individuals: a meta-analysis of published studies. HIV Clin. Trials 5259-268. [DOI] [PubMed] [Google Scholar]
- 6.Hovav, A.-H., M. W. Panas, C. Osuna, M. J. Cayabyab, P. Autissier, and N. L. Letvin. 2007. The impact of a boosting immunogen on the differentiation of secondary memory CD8+ T cells. J. Virol. 8112793-12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabbari, A., and J. T. Harty. 2006. Secondary memory CD8+ T cells are more protective but slower to acquire a central-memory phenotype. J. Exp. Med. 203919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letvin, N. L. 2006. Progress and obstacles in the development of an AIDS vaccine. Nat. Rev. Immunol. 6930-939. [DOI] [PubMed] [Google Scholar]
- 9.Letvin, N. L. 2005. Progress toward an HIV vaccine. Annu. Rev. Med. 56213-223. [DOI] [PubMed] [Google Scholar]
- 10.Letvin, N. L., Y. Huang, B. K. Chakrabarti, L. Xu, M. S. Seaman, K. Beaudry, B. Korioth-Schmitz, F. Yu, D. Rohne, K. L. Martin, A. Miura, W. P. Kong, Z. Y. Yang, R. S. Gelman, O. G. Golubeva, D. C. Montefiori, J. R. Mascola, and G. J. Nabel. 2004. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J. Virol. 787490-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letvin, N. L., J. R. Mascola, Y. Sun, D. A. Gorgone, A. P. Buzby, L. Xu, Z. Y. Yang, B. Chakrabarti, S. S. Rao, J. E. Schmitz, D. C. Montefiori, B. R. Barker, F. L. Bookstein, and G. J. Nabel. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 3121530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacGregor, R. R., R. Ginsberg, K. E. Ugen, Y. Baine, C. U. Kang, X. M. Tu, T. Higgins, D. B. Weiner, and J. D. Boyer. 2002. T-cell responses induced in normal volunteers immunized with a DNA-based vaccine containing HIV-1 env and rev. AIDS 162137-2143. [DOI] [PubMed] [Google Scholar]
- 13.Masopust, D., S. J. Ha, V. Vezys, and R. Ahmed. 2006. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J. Immunol. 177831-839. [DOI] [PubMed] [Google Scholar]
- 14.McMichael, A. J. 2006. HIV vaccines. Annu. Rev. Immunol. 24227-255. [DOI] [PubMed] [Google Scholar]
- 15.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academic Press, Washington, DC.
- 16.Precopio, M. L., M. R. Betts, J. Parrino, D. A. Price, E. Gostick, D. R. Ambrozak, T. E. Asher, D. C. Douek, A. Harari, G. Pantaleo, R. Bailer, B. S. Graham, M. Roederer, and R. A. Koup. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 2041405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santra, S., D. H. Barouch, B. Korioth-Schmitz, C. I. Lord, G. R. Krivulka, F. Yu, M. H. Beddall, D. A. Gorgone, M. A. Lifton, A. Miura, V. Philippon, K. Manson, P. D. Markham, J. Parrish, M. J. Kuroda, J. E. Schmitz, R. S. Gelman, J. W. Shiver, D. C. Montefiori, D. Panicali, and N. L. Letvin. 2004. Recombinant poxvirus boosting of DNA-primed rhesus monkeys augments peak but not memory T lymphocyte responses. Proc. Natl. Acad. Sci. USA 10111088-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santra, S., M. S. Seaman, L. Xu, D. H. Barouch, C. I. Lord, M. A. Lifton, D. A. Gorgone, K. R. Beaudry, K. Svehla, B. Welcher, B. K. Chakrabarti, Y. Huang, Z. Y. Yang, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Replication-defective adenovirus serotype 5 vectors elicit durable cellular and humoral immune responses in nonhuman primates. J. Virol. 796516-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santra, S., Y. Sun, J. G. Parvani, V. Philippon, M. S. Wyand, K. Manson, A. Gomez-Yafal, G. Mazzara, D. Panicali, P. D. Markham, D. C. Montefiori, and N. L. Letvin. 2007. Heterologous prime/boost immunization of rhesus monkeys by using diverse poxvirus vectors. J. Virol. 818563-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 21.Seaman, M. S., L. Xu, K. Beaudry, K. L. Martin, M. H. Beddall, A. Miura, A. Sambor, B. K. Chakrabarti, Y. Huang, R. Bailer, R. A. Koup, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J. Virol. 792956-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, Y., J. E. Schmitz, A. P. Buzby, B. R. Barker, S. S. Rao, L. Xu, Z. Y. Yang, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2006. Virus-specific cellular immune correlates of survival in vaccinated monkeys after simian immunodeficiency virus challenge. J. Virol. 8010950-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuola, J. M., S. Keating, D. P. Webster, T. Berthoud, S. Dunachie, S. C. Gilbert, and A. V. Hill. 2005. Differential immunogenicity of various heterologous prime-boost vaccine regimens using DNA and viral vectors in healthy volunteers. J. Immunol. 174449-455. [DOI] [PubMed] [Google Scholar]