Abstract
The innate immune response is the first line of defense against foreign pathogens. The recognition of virus-associated molecular patterns, including double- and single-stranded RNA, by pattern recognition receptors initiates a cascade of signaling reactions. These result in the transcriptional upregulation and secretion of proinflammatory cytokines that induce an antiviral state. Many viruses have evolved mechanisms to antagonize these responses in order to help them establish a productive infection. We have previously shown that West Nile virus (WNV) is able to inhibit Toll-like receptor 3 (TLR3)-mediated activation of interferon (IFN) regulatory factor 3 (IRF3) (F. Scholle and P. W. Mason, Virology 342:77-87, 2005). In the present study, the WNV nonstructural (NS) proteins were analyzed individually for their ability to antagonize signal transduction mediated by TLR3. We report that expression of WNV NS1 inhibits TLR3-induced transcriptional activation of the IFN-β promoter and of an NF-κB-responsive promoter. This inhibition was due to a failure of the TLR3 ligand poly(I:C) to induce nuclear translocation of IRF3 and NF-κB. Furthermore, NS1 expression also inhibited TLR3-dependent production of interleukin-6 and the establishment of an antiviral state. The function of NS1 in flavivirus infection is not well understood. NS1 is required for viral RNA replication and is also secreted from mammalian cells but not from insect cells. Here, we identify a previously unrecognized role for NS1 in the modulation of signaling pathways of the innate immune response to WNV infection.
Since its introduction into the United States in 1999, West Nile virus (WNV) has spread across North America and into Canada, South America, and the Caribbean. In 2006, a total of 4,261 human cases of WNV disease were reported to the CDC, making WNV the leading cause of arboviral encephalitis in the United States (6). Although most human infections are subclinical, some develop into a neuroinvasive disease such as West Nile meningitis, West Nile encephalitis, and West Nile poliomyelitis (55). Those populations most susceptible to neuroinvasive disease include the elderly, children, and the immunocompromised. Currently, there is no effective treatment or vaccine available for WNV.
WNV is a member of the Flaviviridae family and the genus Flavivirus. It is a mosquito-borne single-stranded RNA (ssRNA) virus with a genome of positive polarity that exists in an enzootic transmission cycle between mosquitoes and birds. Humans and horses are dead-end hosts that can suffer significant morbidity and mortality after infection. The WNV genome is approximately 11 kilobases in length, 5′ capped, and contains an open reading frame that is translated as a single polyprotein (35). Co- and posttranslational processing by viral and cellular proteases results in three structural proteins, C, M, and E, and seven nonstructural (NS) proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. NS2A has been reported to inhibit JAK-STAT signaling from the alpha/beta interferon (IFN-α/β) receptor (39-41), and mutations in NS2A acquired during selection of replicon harboring cells lead to reduced genome replication, attenuation, and enhanced ability to establish a persistent infection (50, 51). In addition NS2A has been shown to play a role in virus assembly (28, 38). NS2B is a cofactor to NS3, the viral serine protease, and NS5 is the RNA-dependent RNA polymerase and methyl transferase (35). The function(s) of NS4A and NS4B is largely not understood although NS4B has been implicated in inhibition of IFN signaling (44, 45). NS1 is required for viral RNA replication and can be trans-complemented (26, 36, 37, 58). Within infected cells NS1 is known to be translocated into the lumen of the endoplasmic reticulum, can be secreted from mammalian but not mosquito cells, and is also found cell surface associated (5, 41, 42). NS1 is secreted to high levels during flavivirus infection, resulting in production of NS1-specific antibodies (24, 27, 31, 42).
To induce a productive infection, a virus or invading pathogen must avoid detection and eradication by the immune system, including the innate immune response. The innate immune system provides a rapid response aimed at inhibiting the progression of infection. Components of the innate immune system include activation of natural killer cells, macrophages, the complement system, and production of proinflammatory cytokines through recognition of pathogen-associated molecular patterns (PAMPs), including double-stranded RNA (dsRNA) or ssRNA, by pattern recognition receptors (PRRs) (59). PRR stimulation results in a cascade of signaling reactions that lead to transcriptional upregulation and secretion of type I IFN and proinflammatory cytokines, resulting in establishment of an antiviral state.
The PRRs that recognize virally derived RNA PAMPs include Toll-like receptor 7/8 (TLR7/8) (ssRNA [10]), TLR3 (dsRNA [2]), and the RNA helicases, retinoic-acid-inducible gene I (RIG-I) (5′ triphosphate containing RNA [23]) and melanoma differentiation-associated gene 5 (mda-5) (dsRNA [17]). TLR3 is a transmembrane receptor located on the cell surface in fibroblasts and within endocytic vesicles in myeloid dendritic cells and other cell types (16, 43, 46, 47) Its ectodomain consists of a leucine-rich repeat motif that is responsible for ligand binding and an intracellular Toll/interleukin 1 (IL-1) receptor domain responsible for adaptor molecule binding (3, 49). TLR3, through its interactions with the adaptor molecule TRIF (Toll/IL-1 receptor domain-containing adaptor inducing IFN), induces activation of the noncanonical IκB kinases (IKKs) TBK-1 and IKK-ɛ leading to phosphorylation (at Ser 396), dimerization, and nuclear translocation of IFN regulatory factor 3 (IRF3) (13, 62). TLR3 signaling also activates the transcription factors NF-κB and AP-1. Detection of their respective PAMPs by RIG-I and mda-5 also leads to activation of IRF3, NF-κB, and AP-1 through the use the adaptor molecule IPS-1 (25). Once activated by either pathway, these transcription factors can cooperate in the transcriptional activation of the IFN-β promoter (19, 33, 48, 63). Following translation, IFN-β is secreted and binds to the IFN-α/β receptor (IFNR) in an autocrine and paracrine fashion. Activation of the IFNR induces a signaling cascade that leads to the establishment of an antiviral state through the transcriptional upregulation of IFN-stimulated genes (19, 22). In addition to type 1 IFN, TLR3 also stimulates expression of proinflammatory cytokines such as RANTES, IL-6, and IL-8 (2).
Many viruses have evolved mechanisms to antagonize these signaling cascades in order to establish a productive infection. Several studies have demonstrated interference of flavivirus NS proteins with signal transduction from the IFNR. These proteins include NS2A, NS4B, and NS5, depending on the virus species (4, 20, 34, 41, 45, 53). In addition to interfering with IFN signaling functions, expression of the WNV NS proteins has also been shown to interfere with TLR3 signaling (53).
In this study we show that the WNV NS1 protein is responsible for the interference with TLR3 activation of transcription factors, TLR3-induced gene expression, and TLR3-dependent establishment of an antiviral state.
MATERIALS AND METHODS
Viruses and cell lines.
WNV replicon particles (VRPs) were produced as previously described (12, 52) and quantitated by titration on HeLa cells, followed by immunohistochemical detection. Vesicular stomatitis virus (VSV) strain Hazelhurst (Charles River Laboratories) was propagated in Vero cells, and titers were determined on HeLa cells by plaque assay (see below).
A list of cell lines used in this study and their characteristics are provided in Table 1. HeLa 1.1.1 and HeLa 2.1 cells, harboring a WNV replicon that expresses the WNV NS genes 1 to 5 and a neomycin resistance cassette (WNR NS1-5ET2AN) (53) were grown in Dulbecco's modification of minimal essential medium (DMEM; Cellgro) supplemented with 10% HyClone fetal bovine serum (Cellgro), 1% antibiotics, 20 μg/ml gentamicin, and 400 μg/ml G418 at 37°C. HeLa C cells are a “cured” derivative of HeLa 1.1.1, established by eliminating the WNV replicon through several passages in culture medium containing 200 U/ml IFN-α. HeLa C cells were grown in the same medium as above without G418. Constitutive TLR3-expressing HEK293 cells (293/TLR3) (InvivoGen) were grown in the above-described medium with the addition of 10 μg/ml blasticidin (InvivoGen), and Vero cells were grown in DMEM with 5% fetal bovine serum, 1% antibiotics, and 20 μg/ml gentamicin. 293TLR3/rep cells were established by infection of 293/TLR3 cells with WNR NS1-5ET2AN VRPs (51), which encode neomycin acetyltransferase as a selectable marker. Replicon-harboring cells were selected and maintained in medium containing 600 μg/ml of G418.
TABLE 1.
Cell lines used in this study
| Cell line | Source | Characteristic(s) |
|---|---|---|
| HeLa C | Cervical carcinoma | IFN-cured derivative of HeLa 1.1.1 |
| HeLa 1.1.1 | Cervical carcinoma | Harbors WNV replicon |
| HeLa 2.1 | Cervical carcinoma | Harbors WNV replicon; different clone from HeLa 1.1.1 |
| HeLa pc6 | Cervical carcinoma | pc6-transfected vector control line |
| HeLa G | Cervical carcinoma | Stably expresses NS1; polyclonal pool |
| HeLa D-24 | Cervical carcinoma | Stably expresses NS1; clone |
| 293/TLR3 | Embryonic kidney | Stably expresses TLR3 |
| 293/TLR3/rep | Embryonic kidney | 293/TLR3 harboring WNV replicon |
Plasmids and generation of cell lines stably expressing NS1.
The mammalian expression vector pcDNA6 (pc6) (Invitrogen) was used to express cloned cDNAs of each individual WNV NS protein (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) as well as an NS2B-3 fusion protein expressing the active protease. Briefly, each coding region was PCR amplified with primers incorporating a 5′ BamHI and a 3′ XbaI restriction site and cloned into the respective restriction sites of pc6. A C-terminal hemagglutinin (HA) tag was added to facilitate detection, and all constructs were sequenced to confirm fidelity of amplification.
HeLa C cell lines that stably express NS1 and a pc6 vector control cell line were created by TransIT-LT1 transfection (Mirus) with each construct, followed by selection for plasmid maintenance with blasticidin (10 μg/ml). Clonal and polyclonal populations of NS1-expressing cells were isolated following blasticidin selection.
Reporter assays.
Reporter assays using the luciferase (Luc) reporter gene under control of the IFN-β promoter (IFN-β pGL3; a gift from J. Hiscott) (32) or an NF-κB response element (NF-κB-Luc) (Stratagene) have been described previously (53). Briefly, 100 ng each of either IFN-β pGL3 or NF-κB-Luc and pCMV (where CMV is cytomegalovirus) β-galactosidase (β-Gal) (Invitrogen) was cotransfected into HeLa C or 293/TLR3 cells, respectively, along with 500 ng of NS expression construct using the TransIT-LT1 transfection reagent (Mirus). In experiments where various amounts of NS1 plasmid were transfected, pc6 was used to keep the overall DNA concentration constant. At 24 h posttransfection, cells were either left untreated or were treated for 4 h with 20 μg/ml poly(I:C) (pIC; Calbiochem). Following treatment, cells were lysed in reporter lysis buffer (Promega) containing 0.1% Triton X-100 and assayed for Luc and β-Gal activities using a Promega Luc assay system and an ONPG (o-nitrophenyl-β-d-galactopyranoside)-based β-Gal assay. β-Gal activity was used to normalize the Luc data for all experiments. All data are expressed as relative light units/mU of β-Gal activity.
Immunofluorescence.
HeLa C cells were seeded onto LabTek chamber slides and transfected with each NS protein expression construct or the pc6 vector control. At 24 h posttransfection cells were treated with 20 μg/ml pIC for 2.5 h or left untreated and prepared for indirect immunofluorescence analysis (IFA) for NF-κB subcellular localization and identification of NS protein expression. Briefly, cells were washed with phosphate-buffered saline, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked (2% bovine serum albumin, 5% normal horse serum, and 10 mM glycine in phosphate-buffered saline). The cells were then incubated with a rabbit anti-NF-κB p65 antibody (Santa Cruz) and a mouse anti-HA antibody (Sigma), followed by incubation with appropriate secondary antibodies (AlexaFluor 568-conjugated anti-rabbit antibody and AlexaFluor 488-conjugated anti-mouse antibody [Invitrogen]). The cells were then analyzed for NF-κB subcellular localization and HA expression by immunofluorescence using a Zeiss Axioskop 2 microscope. IFA for NF-κB translocation in cell lines constitutively expressing NS1 was performed similarly using a fluorescein isothiocyanate-conjugated anti-rabbit antibody (Kirkegaard and Perry Laboratories) as a secondary antibody.
Western blot analysis.
Subconfluent stable NS1-expressing cells (HeLa G and D24 cells), pc6-expressing cells, and HeLa 1.1.1 and HeLa C cells were either treated with 20 μg/ml pIC for 4 h or left untreated. Following treatment, protein extracts were prepared in cell lysis buffer (300 mM NaCl, 50 mM Tris-HCl, 0.1% Triton X-100, pH 7.6). Following a 10-min incubation on ice, lysates were clarified by centrifugation, and protein concentrations were determined using a detergent-compatible protein assay kit (Bio-Rad). Equal amounts of protein were electrophoretically separated on 4 to 12% Nu-PAGE gels (Invitrogen) and electroblotted onto polyvinylidene difluoride membranes (Immobilon-P transfer membrane; Millipore). Following a blocking step with Tris-buffered saline-0.1% Tween-20 with 5% dry milk, a phospho-specific IRF3 (Ser 396) antibody (Cell Signaling) or anti-IRF3 antibody was diluted in Tris-buffered saline-0.1% Tween-20 with 5% bovine serum albumin and used to blot the membrane, followed by horseradish peroxidase-conjugated secondary antibodies (KPL). Bound horseradish peroxidase was visualized with an ECL Plus kit (Amersham Biosciences). Membranes were stripped and reprobed with a mouse anti-β-actin antibody (Sigma) as a loading control. NS1 was detected using a WNV-specific mouse hyperimmune serum (murine hyperimmune ascitic fluid [MHIAF]) as the primary antibody.
Real-time RT-PCR assays.
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's specifications. Extracts were DNase I (Ambion)-treated to remove contaminating genomic DNA, precipitated, and resuspended in nuclease-free water. Reverse transcription (RT) was carried out with the ImProm II RT kit (Promega) using random hexamers as primers. Real-time PCR analysis was carried out using the iQ Sybr Green Supermix kit (Bio-Rad) and the following primers: for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GGATTTGGTCGTATTGGGCG-3′ and 5′-TGGAAGATGGTGATGGGATTTC-3′; for IL-6, 5′-GAGAAGATTCCAAAGATGTAGCCG-3′ and 5′-ACATTTGCCGAAGAGCCCTC-3′.
Reactions were set up in 96-well PCR plates (Eppendorf). Amplifications were carried out for 50 cycles, followed by a melt curve analysis of resulting products to confirm the specificity of the reactions. To construct standard curves, total RNA was isolated from the cells, and 300- to 600-bp fragments of the gene(s) of interest were amplified by RT-PCR using the appropriate primer sets. PCR fragments were gel purified and quantitated, and the copy number was calculated. Serial 10-fold dilutions were prepared for use as templates to create standard curves in real-time PCRs. All samples were normalized to GAPDH. To control for plate-to-plate variation, GAPDH reactions were run for all samples on the same plate as the respective real-time PCR for the gene of interest. All data are expressed as the ratio of copy numbers of IL-6 per 106 copies of GAPDH for samples run in triplicate.
ELISA.
IL-6 levels in the supernatant of cells treated with 20 μg/ml pIC for 6 h or left untreated were determined using an enzyme-linked immunosorbent assay (ELISA) kit (E-bioscience) according to the manufacturer's specifications. Each experimental set was performed in triplicate.
siRNA knockdown.
The TLR3-specific small interfering RNA (siRNA) 1 and 2 and controls (siCon, a scrambled siRNA; siGLO, a fluorescently labeled nontargeting siRNA) were purchased from Dharmacon. A total of 50 nM of siRNA was transfected using Dharmafect DF-1 reagent according to the manufacturer's recommendation. At 48 h posttransfection cells were treated with 10 μg/ml pIC for 1 h or left untreated. Following treatment, cells were challenged with WNV VRPs expressing Luc at a multiplicity of infection (MOI) of 0.2. At 24 h postinfection cell lysates were made and assayed for Luc expression. The siRNA sense sequences were as follows: siRNA 1, 5′-GAACUAAAGAUCAUCGAUUUU-3′; siRNA 2, 5′-GCUAUGUUUGGAAUUAGCAAA-3′; and siCON, 5′-UGGUUUACAUGUCGACUAA-3′.
Successful reduction of TLR3 mRNA levels was confirmed by semiquantitative RT-PCR using GAPDH primers described above and the following TLR3 primers: Forward, 5′-TCACTTGCTCATTCTCCCTT-3′; Reverse, 5′-GACCTCTCCATTCCTGGC-3′.
Plaque assays.
HeLa cells that stably express NS1 (D24 G cell lines), WNV replicon (1.1.1), or pc6 vector control were seeded to confluence in 24-well plates and left untreated or treated with 20 μg/ml pIC for 4 h, followed by infection with 10-fold serial dilutions of virus. VSV infections were performed at 37°C with agitation every 15 min for 1 h, followed by overlay with 1× DMEM-0.6% tragacanth gum. At 24 to 36 h postinfection the cells were stained with 5% crystal violet, and viral titers were determined.
Virus growth curve analysis.
For analysis of virus growth kinetics, HeLa G, HeLa D24, pc6, and 1.1.1 cells were seeded onto 24-well plates to 80% confluence, left untreated or treated with 20 μg/ml pIC for 3 h, and infected with VSV at an MOI of 0.01 (HeLa-specific MOI). At the indicated times, cell supernatants were collected for each condition. VSV titers were determined by plaque assay on Vero cell monolayer.
RESULTS
Inhibition of TLR3-induced transcription in WNV replicon-bearing cells.
We have previously demonstrated that pIC stimulation of the IFN-β promoter in HeLa cells is mediated through TLR3 (53). TLR3-dependent nuclear translocation of IRF3 and transcriptional activation of the IFN-β promoter and IRF3 response elements in HeLa cells are inhibited in WNV-infected and WNV replicon-bearing HeLa cells (53) (Fig. 1A). TLR3 stimulation also results in activation of NF-κB and subsequent transcriptional upregulation of NF-κB-responsive cytokine promoters. Thus, we were interested in determining whether WNV RNA replication and protein expression would also negatively affect NF-κB activation.
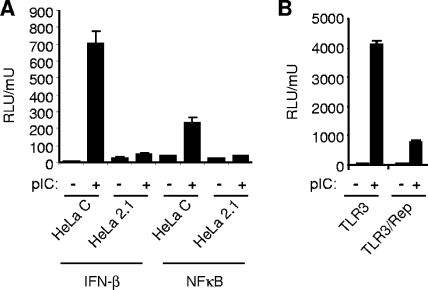
FIG. 1.
Inhibition of IFN-β-promoter and NF-κB activation by TLR3 in WNV replicon cells and pIC-induced activation of the IFN-β promoter and NF-κB reporter in HeLa WNV replicon cells (HeLa 2.1) and IFN-cured replicon cells (HeLa C). (A) Cells were transiently transfected with IFN-β pGL3 or NF-κB-Luc and pCMV-β-Gal and then either treated with 20 μg/ml of pIC for 4 h or left untreated. Cell lysates were analyzed for Luc and β-Gal activities. (B) 293/TLR3 cells (TLR3) and WNV replicon-bearing 293/TLR3 cells (TLR3/rep) were assayed for pIC-induced transcriptional activation of an NF-κΒ response element. Cells were transfected with NF-κB Luc and pCMV-β-Gal, and 24 h posttransfection the cells were treated with 20 μg/ml pIC for 4 h or left untreated. Cell lysates were analyzed for Luc and β-Gal activities. Data are expressed as Luc activity normalized to β-Gal activity from duplicate samples. Error bars represent the standard deviations. RLU, relative light units.
Reporter assays were performed to monitor NF-κB-dependent transcriptional stimulation following TLR3 engagement. HeLa C cells and HeLa WNV replicon-bearing cells (HeLa 2.1) were transfected with NF-κB-Luc and pCMV-β-Gal for normalization of transfection efficiency. In the absence of pIC stimulation, each cell line displayed low levels of Luc activity. pIC treatment induced a strong NF-κB response in HeLa C cells but not in replicon-bearing HeLa 2.1 cells (Fig. 1A). The same results were obtained with 293/TLR3 and 293/TLR3 cells containing a constitutively replicating WNV replicon (293TLR3/rep) (Fig. 1B).
WNV NS1 inhibits TLR3-induced transcription.
The data above, obtained with WNV replicons with deletions of the structural protein coding region, argue that either WNV RNA replication or expression of the WNV NS proteins is responsible for TLR3 inhibition. Flavivirus NS proteins have been shown to inhibit signal transduction from the IFN-α/β receptor (4, 20, 34, 41, 44, 53). Thus, we hypothesized that one or more NS proteins could be responsible for inhibition of TLR3 signal transduction as well. Expression constructs for each of the individual WNV NS proteins, as well as for the fusion protein encoding the active protease, NS2B-3, were established. Expression of individual NS proteins was assayed after transient transfection into HeLa cells. NS2B and NS4A were detected in IFAs in individual cells without problems (see Fig. 3) but were expressed at levels too low to be detected by Western blotting. In contrast, all other NS proteins were readily detected by Western analysis (data not shown).
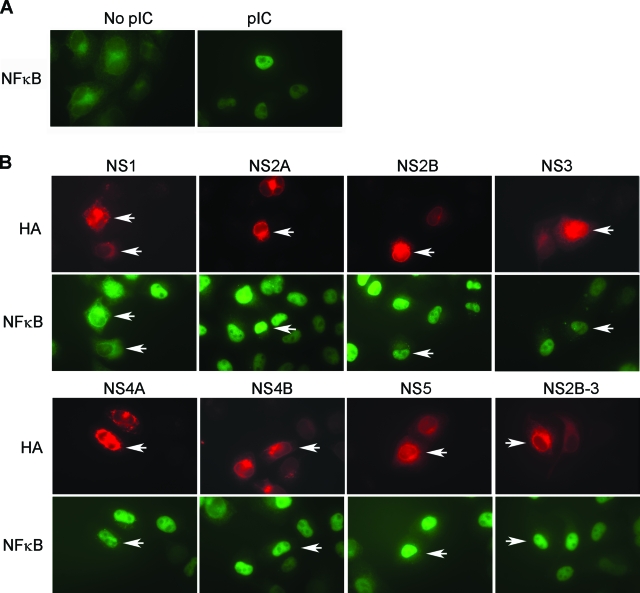
FIG. 3.
WNV NS1 inhibits the TLR3-induced nuclear translocation of NF-κB. (A) HeLa C cells were transfected with the empty vector pc6 and either left untreated or treated with 20 μg/ml pIC for 2.5 h. NF-κB was detected by immunofluorescence. (B) HeLa C cells were transfected with NS protein expression constructs. At 24 h posttransfection cells were challenged with 20 μg/ml pIC for 2.5 h. Cells were dually stained for NF-κB subcellular localization using an NF-κB-specific antibody and NS protein expression using an HA epitope-specific antibody. Red indicates NF-κB, and green indicates NS expression. All NF-κB pictures were taken under identical conditions using the same exposure settings. For NS proteins, exposure times had to be adjusted slightly to account for variation of levels of expression between the different constructs.
To identify the individual NS protein(s) that might interfere with TLR3 signaling, reporter assays using the IFN-β promoter in HeLa C cells cotransfected with individual NS expression constructs were performed. TLR3 engagement induced strong transcriptional activation of the IFN-β promoter in all cells except those cells transfected with the NS1 expression construct (Fig. 2A). NS1 expression strongly inhibited IFN-β promoter-driven Luc expression in response to TLR3 engagement compared to cells transfected with the empty vector control. Similar results were obtained when NF-κB-dependent reporter activity was assayed in 293/TLR3 cells transfected with the individual NS protein expression constructs (Fig. 2B). Due to the low level of expression of NS2B and NS4A, reporter assays with those two constructs are not shown.
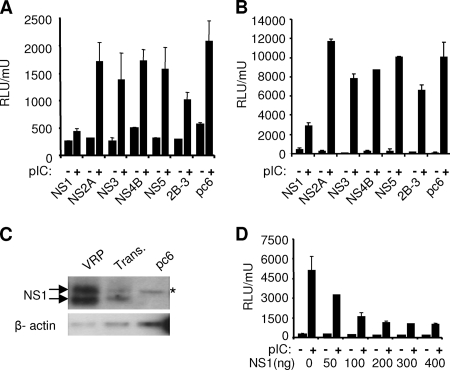
FIG. 2.
WNV NS1 inhibits the pIC-induced transcriptional activation of the IFN-β promoter and NF-κB response element. (A) HeLa C cells were transfected with individual expression constructs for NS proteins together with the IFN-β pGL3 reporter construct and pCMV-β-Gal. At 24 h posttransfection cells were treated with 20 μg/ml pIC for 4 h or left untreated. Cell lysates were assayed for Luc and β-Gal activities. (B) NS expression constructs were transfected into 293/TLR3 cells along with NF-κB-Luc and pCMV β-Gal. Cells were treated with pIC as above, and Luc and β-Gal activities were determined. (C) HeLa C cells were transfected (Trans) with 500 ng of NS1 expression plasmid or the same amount of empty vector (pc6) or infected at an MOI of 1 with VRPs. Cell lysates were prepared 24 h after transfection or infection, and levels of NS1 expression were analyzed by Western blotting using a WNV-specific MHIAF. A nonspecific band close to the molecular weight of NS1 (denoted by an asterisk) is seen in pc6-transfected vector control cells. As a loading control the blot was stripped and reprobed for β-actin. (D) Various amounts of NS1 expression plasmid were transfected into HeLa C cells along with the IFN-β pGL3 reporter and β-Gal expression plasmids. Following mock or pIC treatment, cells were harvested and assayed for Luc and β-Gal activity. Data are expressed as Luc activity relative to β-Gal activity from duplicate samples. Error bars represent the standard deviations. RLU, relative light units.
To rule out the possibility that the effects on TLR3 signaling observed in NS1-expressing cells are not due to protein overexpression, NS1 expression levels in transiently transfected cells were compared to cells infected with VRPs at an MOI of 1 (Fig. 2C). These results illustrate that NS1 is expressed to significantly lower levels in transient transfection than in VRP infection. Our transfection efficiency in HeLa cells was routinely between 60 to 70%.
To determine whether TLR3 inhibition was dependent on the level of NS1 expression, a dose response experiment was performed. As little as 50 ng of transfected NS1 construct was able to inhibit IFN-β promoter activation after pIC treatment, with a maximum inhibition reached at 200 ng of NS1 expression plasmid under our assay conditions (Fig. 2D).
The reporter assays do not address a role for NS2B and NS4A in TLR3 inhibition. However, reporter activation was not significantly inhibited in cells transfected with the NS2B-3 fusion construct from which NS2B is expressed as a cofactor for NS3, and other experiments, discussed below, indicate that NS4A expressed by itself is not involved in inhibition of TLR3 signaling (see Fig. 3).
All NS proteins, with exception of the NS2B-3 fusion protein, were expressed individually and contain epitope tags at the C terminus. Interestingly, even at the highest amounts of NS1 plasmid transfected, we did not observe TLR3 inhibition to the degree found in replicon-bearing cells. We do not have a definite explanation for this observation at this time, but we cannot rule out the possibility that NS proteins other than NS1 could contribute to TLR3 inhibition when expressed in their natural context as part of the viral polyprotein. Alternatively, the presence of the epitope tag might interfere with some of their functions. Despite these caveats, NS1 has been reported to function in viral replication when expressed in trans and can also function in this context when tagged with a C-terminal epitope (26, 58). Our experiments demonstrate that NS1 expressed by itself can inhibit TLR3 signal transduction. Thus, the subsequent experiments focus on NS1.
WNV NS1 blocks the TLR3-induced nuclear translocation of NF-κB.
Based on the observed inhibition of NF-κB-dependent transcription in WNV replicon-bearing cells and NS1-expressing cells (Fig. 1 and 2), we investigated whether TLR3-induced nuclear translocation of NF-κB was affected in NS protein-expressing cells. In untreated vector control transfected cells, NF-κB localized predominantly to the cytoplasm. Stimulation with pIC induced nuclear translocation of NF-κB in approximately 90% of the cells (Fig. 3A). Next, HeLa C cells, transfected with each individual NS expression construct, were treated with pIC (Fig. 3B) or left untreated (data not shown). NS protein expression and NF-κB subcellular localization were analyzed by immunofluorescence. As indicated above, all NS proteins could be detected by IFA including NS2B and NS4A. None of the NS proteins induced NF-κB translocation in the absence of pIC stimulation (data not shown). Upon treatment with pIC, nuclear translocation of NF-κB was evident in almost all NS protein-expressing cells except for those cells transfected with the NS1 expression plasmid. NS1-expressing cells showed a cytoplasmic NF-κB staining pattern regardless of pIC stimulation. The immunofluorescence data also demonstrate that NS2B and NS4A expression does not interfere with TLR3-induced NF-κB translocation.
HeLa cells that express NS1 constitutively are deficient in TLR3 signaling.
For subsequent investigations into the biological significance of NS1-mediated TLR3 inhibition, HeLa cell lines that constitutively express NS1 were established. Several NS1-expressing polyclonal and clonal populations were isolated. NS1 expression levels in stable cell lines were significantly lower than observed in replicon-bearing cells (Fig. 4A). To verify TLR3 inhibition in these cell lines, we monitored activation of NF-κB and IRF3. Inhibition of NF-κΒ translocation was assayed by immunofluorescence (Fig. 4B). Vector control and NS1-expressing cells (pc6 and D24, respectively) were either treated with pIC or left untreated. In all untreated cells, NF-κΒ was localized to the cytoplasm. In vector control cells, pIC readily induced the nuclear translocation of NF-κΒ. This was in sharp contrast to the cytoplasmic staining pattern observed in D-24 cells following pIC treatment (Fig. 4B).
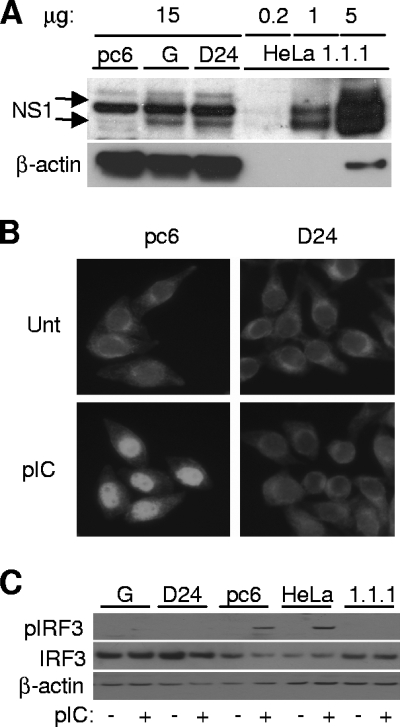
FIG. 4.
Inhibition of transcription factor activation in cell lines constitutively expressing NS1. (A) NS1 expression levels in NS1-expressing cell lines. Cell lysates from NS1-expressing cell lines were compared to 1.1.1 replicon cells in terms of NS1 expression levels. HeLa G is a polyclonal NS1-expressing population, and D24 is an individual NS1-expressing clone. HeLa 1.1.1 cells are WNV replicon-bearing cells, and pc6 is a vector control cell line. Fifteen micrograms of total protein per lane was loaded for pc6, G, and D24 cells, and 0.2, 1, and 5 μg was loaded for 1.1.1 lysates. The WNV MHIAF used as primary antibody cross-reacts with a nonspecific cellular protein with a molecular weight very similar to the glycosylated higher-molecular-weight form of NS1 and partially obscures its detection. (B) NF-κB nuclear translocation in NS1-expressing cell lines. NS1-expressing D-24 and vector control cells were treated for 2 h with 20 μg/ml pIC or left untreated (Unt). Cells were then stained to detect NF-κΒ subcellular localization. (C) Phosphorylation of IRF3 in response to TLR3 stimulation in NS1-expressing cell lines. Following 4 h of pIC treatment, cell lysates were analyzed for phospho-IRF3 (Ser 396) by immunoblotting (top panel). The middle panel shows the levels of total IRF3, and the bottom panel shows levels of β-actin as a loading control.
IRF3 activation was investigated by analyzing the phosphorylation of IRF3 in response to TLR3 engagement in NS1-expressing cell lines, WNV replicon-bearing cells, and vector control cells. Following a mock or pIC treatment, cell lysates derived from NS1-expressing HeLa cell lines (polyclonal pool, G; clonal isolate, D24), vector control cells (pc6), and WNV replicon cells (1.1.1) were analyzed by Western blotting using an antibody specific for phospho-IRF3 (Ser 396). Phospho-IRF3 was not detected in lysates from unstimulated cells across the panel of cell lines tested. In pIC-stimulated cells, phospho-IRF3 was detected in lysates from HeLa C and HeLa pc6 cells (Fig. 4C). In contrast, pIC did not induce phosphorylation of IRF3 in replicon-bearing HeLa cells and the NS1-expressing clonal cell line D24. The NS1-expressing HeLa pool G showed only very minimal IRF3 phosphorylation. In agreement with these results, inhibition of IRF3 nuclear translocation in response to pIC was also observed in D24, G, and 1.1.1 cells (data not shown). In summary, NS1-expressing HeLa cell lines showed the same phenotypes of TLR3 inhibition with regard to transcription factor activation previously detected with transiently NS1 transfected cells and replicon-bearing or WNV-infected cells (53) (Fig. 1).
NS1 inhibits TLR3-mediated production of IL-6.
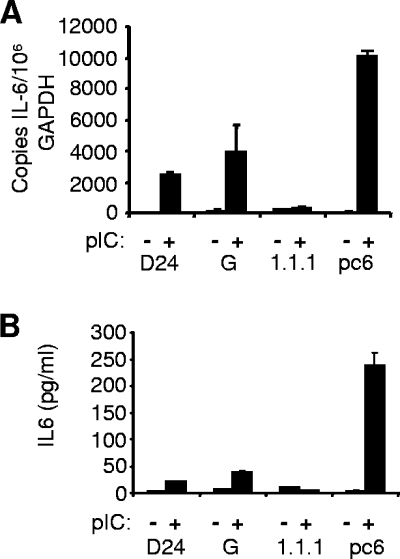
To investigate whether NS1 had an inhibitory effect on endogenous gene induction by TLR3, we examined cytokine production in HeLa cells in response to pIC treatment. In contrast to the reporter assay data, when IFN-β production in response to pIC was investigated by bioassay and real-time RT-PCR, IFN-β transcript levels were only slightly elevated, and levels of secreted IFN-β were below the limit of detection (results not shown) (see Discussion). We therefore monitored upregulation of IL-6 as an example of a cytokine that has been shown to be induced by several different viruses in a TLR3-dependent manner (18, 29, 61). Induction of IL-6 following pIC treatment was readily observed at both the mRNA and the protein level (Fig. 5). Compared to vector control cells, induction of IL-6 mRNA and protein levels after pIC stimulation were greatly reduced in D24 and HeLa 1.1.1 replicon cells. The polyclonal pool HeLa G showed intermediate reduction in IL-6 expression. These results confirm that expression of NS1 is able to interfere with TLR3-mediated induction of endogenous genes as well as reporter genes.
FIG. 5.
WNV NS1 inhibits TLR3-induced transcriptional activation and secretion of IL-6. (A) IL-6 mRNA levels were determined by real-time RT-PCR from isolated RNA in the indicated cell lines following a mock or 6-h treatment with 20 μg/ml pIC. Results are displayed as the copy number of IL-6 transcripts per 106 copies of GAPDH. (B) IL-6 protein levels were determined by ELISA from the culture supernatants recovered from cells treated as in panel A. RT-PCR data are from a representative experiment run in duplicate, and ELISA data are a representative experiment run in triplicate. Error bars represent standard deviations.
WNV NS1 inhibits establishment of a TLR3-induced antiviral state.
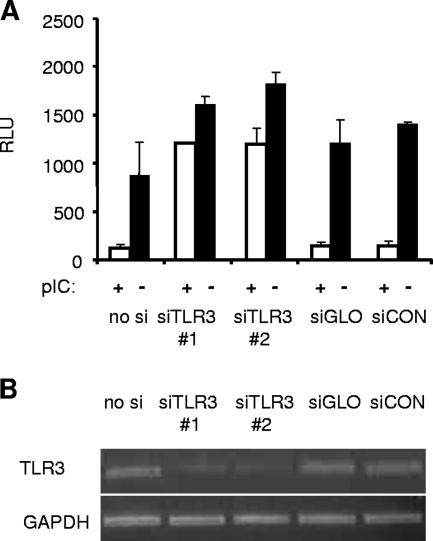
Preliminary studies on HeLa cells demonstrated that pIC treatment efficiently induces an antiviral state in these cells. To confirm that this pIC-induced antiviral effect was indeed mediated by TLR3, TLR3 expression was suppressed using two different TLR3-specific siRNAs targeting distinct regions of the TLR3 mRNA. As a control, a scrambled nonspecific siRNA was utilized (siCON). pIC-treated or nontreated siRNA-transfected cells were subsequently infected with Luc-expressing WNV VRPs, and Luc activity was determined as a measure of replication. In control siRNA-transfected cells (siGLO, siCON, and no siRNA), pIC treatment almost completely eliminated VRP infection. However, cells transfected with either TLR3-specific siRNA (1 or 2), pIC only slightly reduced the infectivity of VRPs (Fig. 6A). Semiquantitative RT-PCR was used to confirm reduction of TLR3 mRNA levels in cells transfected with TLR3 targeting siRNAs (Fig. 6B). These results confirm that signaling through TLR3 is required for a pIC-induced antiviral state in this cell type.
FIG. 6.
pIC induces a TLR3-dependent antiviral effect in HeLa cells. (A) HeLa C cells were transfected with two different TLR3-specific siRNAs (1 and 2) and three different controls (siGLO, siCON, or no siRNA [no si]) (see Materials and Methods). Following a 1-h treatment with 10 μg/ml pIC or mock treatment, cells were infected with WNV VRPs expressing Luc at an MOI of 0.2. At 24 h postinfection cell lysates were assayed for Luc expression. The representative experiment shown was conducted in duplicate. Error bars represent standard deviations. (B) TLR3 and GAPDH levels in siRNA-transfected cells were analyzed by semiquantitative RT-PCR using specific primers for both TLR3 and GAPDH. RLU, relative light units.
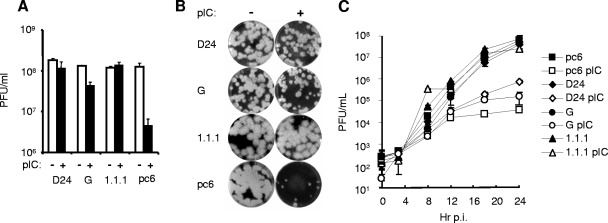
To investigate the ability of NS1 to inhibit this antiviral effect, titrations of VSV were performed on pIC-treated or untreated monolayers of HeLa pc6, G, D24, and 1.1.1 cells. Plaque assays demonstrated that the VSV infectious titer on pIC-treated HeLa pc6 cells was reduced by approximately 1.5 logs, while HeLa 1.1.1 cells and D24 were resistant to the antiviral action of pIC. The polyclonal population HeLa G showed intermediate sensitivity to pIC, resulting in a slight decrease of VSV infectivity after pIC treatment (Fig. 7A). VSV plaque size on pIC-treated monolayers of the various cell lines corresponded with the infectivity data. HeLa pc6 cells demonstrated both a reduction in plaque number as well as a significant decrease in plaque size while plaque sizes were only slightly reduced in pIC-treated D24 and 1.1.1 cells. HeLa G showed an intermediate phenotype (Fig. 7B).
FIG. 7.
Expression of WNV NS1 inhibits the TLR3-induced antiviral effect. (A) HeLa pc6, D24, G, and 1.1.1 cells were pretreated with 20 μg/ml pIC for 4 h or left untreated. Cells were subsequently infected with serial 10-fold dilutions of VSV. At 24 h postinfection plaques were visualized by crystal violet staining, and VSV infectious titers were determined. (B) VSV plaque size and morphology in an experiment conducted as described above. Cells were untreated (−) or treated with pIC (+) as described above. Pictures shown are from wells infected with a 106 dilution of VSV. (C) Multistep growth curve of VSV on the indicated cell lines. Cells were left untreated or pretreated with pIC for 4 h followed by infection with VSV at an MOI of 0.01. Supernatants were harvested every 4 h, and virus production was determined by plaque assay on Vero cells. Values indicate VSV PFU/ml of supernatant. Error bars indicate the standard deviations. Closed symbols denote virus recovered from untreated cells. Open symbols denote virus recovered from pIC-treated cells. Squares, HeLa pc6; triangles, HeLa 1.1.1; diamonds, HeLa D24; circles, HeLa G.
To more carefully investigate the effect of TLR3 signaling on viral growth, multistep growth curves of VSV on D24, G, HeLa 1.1.1, and pc6 control cells were determined over a time course of 24 h (Fig. 7C). In the absence of pIC treatment, VSV growth kinetics and virus production were nearly identical on all four cell lines. pIC pretreatment severely inhibited VSV production in pc6 cells (ca. 4 logs) compared to only a 2-log decrease in D24 cells (Fig. 7C). As in the previous experiments, HeLa G cells showed an intermediate phenotype, and the HeLa 1.1.1 line was nonresponsive to pIC treatment.
DISCUSSION
The ability of many viruses to successfully induce a productive infection involves evasion or modulation of host antiviral responses. Our previous work reported that both WNV-infected and WNV replicon-bearing cells are deficient in TLR3-induced IFN-β promoter activation and activation of IRF3 in HeLa cells (53). In the present study we identified the WNV NS1 protein as responsible for TLR3 inhibition. Using our approach of separately expressing individual NS proteins, we did not observe TLR3 inhibition with any of the other NS protein expression constructs tested. As alluded to above, this approach does not completely rule out possible contributions of other NS proteins when they are expressed in their natural context as part of the viral polyprotein, and we did observe a low level of residual stimulation in cells transfected with NS1 alone (Fig. 2D).
Compared to all of our constitutively NS1-expressing cell lines, HeLa 1.1.1 replicon cells consistently exhibited stronger TLR3 inhibition. Western blot analysis demonstrated that HeLa 1.1.1 cells express approximately 20 times as much NS1 as either HeLa D-24 or HeLa G. These findings are in line with our observations that TLR3 inhibition is dependent on the level of NS1 expression (Fig. 2D).
NS1 inhibited TLR3-stimulated IFN-β promoter and NF-κB-dependent promoter activation in reporter assays, nuclear translocation of NF-κB and IRF3, expression of IL-6, and establishment of an antiviral state. Interestingly, we noted a marked discrepancy between activation of the IFN-β promoter in reporter assays and activation of the endogenous IFN-β promoter in HeLa cells. We were not able to observe significant secretion of type I IFN by bioassay in culture supernatants of pIC-treated HeLa cells (data not shown). However, in coculture experiments HeLa cells were able to protect non-TLR3-expressing cells (Huh7) from VRP infection after pIC treatment (results not shown). It is therefore possible that low levels of type I IFN are produced by HeLa cells in response to TLR3 engagement and are able to act in a para-and/or autocrine fashion to establish an antiviral state. Alternatively, other TLR3-induced genes might be responsible for conferring an antiviral state independent of type I IFN. The identification of the exact mechanism of establishment of an antiviral state in HeLa cells by TLR3 is beyond the scope of this investigation and will be pursued separately.
Interference with TLR3 signaling by NS1 and WNV replicons was not only observed in HeLa cells but also in TLR3-expressing HEK293 cells and in WNV replicon-bearing mouse embryo fibroblasts (F. Gilfoy and P. Mason, personal communication). Our findings are in direct contrast to a previous report describing no interference of WNV infection with TLR3 signal transduction in several different cell lines (14). Some of these discrepancies might be attributed to the use of different cell lines in that study. However, TLR3-expressing HEK293 cells were used both in the former study and in our experiments. It is possible that amounts of NS1 insufficient to inhibit TLR3 were expressed in the former study since the pIC challenges of infected cells were conducted anywhere from 3 to 6 h postinfection (14). We previously described TLR3 inhibition in WNV-infected cells at 10 to 12 h postinfection (53) and demonstrate in the present study that the degree of TLR3 inhibition is dependent on the expression level of NS1.
The role of TLR3 in the control of virus infections is somewhat controversial. In vitro studies have demonstrated that flaviviruses can be recognized by several different PRRs, including RIG-I, mda-5, and protein kinase R (7, 14, 15, 16a). Flaviviruses have also been reported to activate the ssRNA PRR TLR7 in plasmacytoid dendritic cells (56, 60). Interestingly, flaviviruses do not seem to engage TLR3 during infection in cell culture systems, as described by Fredericksen et al. (14) and according to our own unpublished observations. Most of these studies were performed using common cell lines that respond to TLR3 ligands. It is reasonable to speculate that TLR3 engagement might be different in vivo in the context of specific cell-cell interactions. As an example, a role for TLR3 in cross-priming by TLR3-expressing CD8+ dendritic cells in the activation of cytotoxic T lymphocytes has been described (54), and several studies clearly demonstrate an involvement of TLR3 in the innate response to virus infection in vivo.
While Edelmann et al. described no differences in the pathogenesis of several viruses in TLR3-deficient mice compared to wild-type animals (11), several other studies, conducted with a variety of viruses, demonstrate that TLR3 can have either protective or detrimental influences on viral pathogenesis. In most cases, TLR3-deficient mice displayed reduced production of proinflammatory cytokines in response to virus infection including tumor necrosis factor alpha, IL-6, or monocyte chemotactic protein 1 (18, 21, 29, 61). However, the outcome in terms of pathogenesis can be quite distinct, depending on the virus. Influenza A virus infection in TLR3-deficient mice yields higher virus titers and reduced cytokine responses, yet these mice have a survival advantage (29). Similarly, TLR3 signaling in WNV-infected mice leads to increased permeability of the blood-brain barrier and increased WNV invasion into the central nervous system, whereas TLR3-deficient mice showed decreased central nervous system invasion (61). In both cases these results were attributed to TLR3-dependent production of proinflammatory cytokines, which cause detrimental effects when they are produced at high levels. In both influenza A virus and WNV infection, higher levels of virus production could be detected in the absence of functional TLR3. In contrast, encephalomyocarditis virus infection in TLR3-deficient mice led to decreased cytokine production accompanied by higher viral loads and increased mortality. Interestingly, levels of IFN-β were not decreased in encephalomyocarditis virus-infected TLR3-deficient animals (21). It is evident from these examples that the role of TLR3 in viral infection can vary with the virus species and the cell type(s) infected.
NS1 is a glycoprotein that is required for flavivirus replication (37) and is also secreted to high levels during flavivirus infection in vivo (24, 27, 31, 42). Two recent studies have addressed a potential role of NS1 in immunomodulation. NS1 was shown to interfere with activation of complement factor H in vitro (8), and cell surface-associated NS1 was found to mediate phagocytosis of WNV-infected cells and thus might play a role in clearance of virally infected cells (9). The mechanism of NS1-mediated inhibition of TLR3 is currently under investigation. Both TLR3 and NS1 associate with membranes of the endoplasmic reticulum for maturation and targeting to their respective destination(s). It is possible that direct interactions of NS1 and TLR3 are responsible for TLR3 inhibition, or, alternatively, NS1 could interfere with downstream signaling such as recruitment of the adaptor protein TRIF or activation of TBK-1. Dengue NS1 protein can be endocytosed by several different cell types (1), and an intriguing alternative is that secreted NS1 might inhibit TLR3 signaling after being endocytosed by target cells, possibly by interfering with TLR3 signaling in the endosome. This hypothesis is currently under investigation.
Several viruses have evolved independent mechanisms to interfere with TLR3. The hepatitis C virus NS3/4A protease interferes with TLR3-dependent IRF3 activation by cleaving the TLR3 adaptor molecule TRIF (30), and vaccinia virus interferes with TLR3 through expression of its AR46 protein (57). Our results add another example to this list and delineate a potentially different mechanism of targeting TLR3. The fact that several different viruses and, in particular, two small RNA viruses containing genomes with limited coding capacity have evolved ways to interfere with this innate immune signaling pathway strongly argues for an interplay between TLR3 and the virus. These interactions are bound to have important implications for the biology of these viruses while their significance might not be fully understood at present.
Acknowledgments
We thank Kristen Belanger for critical reading of the manuscript and Peter Mason and Felicia Gilfoy for helpful discussions.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases to F.S. (grant number K22AI067925) and the North Carolina Agricultural Research Service (project 06849).
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Alcon-LePoder, S., M. T. Drouet, P. Roux, M. P. Frenkiel, M. Arborio, A. M. Durand-Schneider, M. Maurice, I. Le Blanc, J. Gruenberg, and M. Flamand. 2005. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J. Virol. 7911403-11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413732-738. [DOI] [PubMed] [Google Scholar]
- 3.Bell, J. K., G. E. Mullen, C. A. Leifer, A. Mazzoni, D. R. Davies, and D. M. Segal. 2003. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24528-533. [DOI] [PubMed] [Google Scholar]
- 4.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 7912828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinton, M. A. 2002. The molecular biology of West Nile virus: a new invader of the Western hemisphere. Annu. Rev. Microbiol. 56371-402. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2007. West Nile virus activity—United States, 2006. MMWR Morb. Mortal. Wkly. Rep. 56556-559. [PubMed] [Google Scholar]
- 7.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8157-171. [DOI] [PubMed] [Google Scholar]
- 8.Chung, K. M., M. K. Liszewski, G. Nybakken, A. E. Davis, R. R. Townsend, D. H. Fremont, J. P. Atkinson, and M. S. Diamond. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. USA 10319111-19116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, K. M., B. S. Thompson, D. H. Fremont, and M. S. Diamond. 2007. Antibody recognition of cell surface-associated NS1 triggers Fc-γ receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J. Virol. 819551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diebold, S. S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 3031529-1531. [DOI] [PubMed] [Google Scholar]
- 11.Edelmann, K. H., S. Richardson-Burns, L. Alexopoulou, K. L. Tyler, R. A. Flavell, and M. B. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology 322231-238. [DOI] [PubMed] [Google Scholar]
- 12.Fayzulin, R., F. Scholle, O. Petrakova, I. Frolov, and P. W. Mason. 2006. Evaluation of replicative capacity and genetic stability of West Nile virus replicons using highly efficient packaging cell lines. Virology 351196-209. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 14.Fredericksen, B. L., and M. Gale, Jr. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 802913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredericksen, B. L., B. C. Keller, J. Fornek, M. G. Katze, and M. Gale, Jr. 2008. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funami, K., M. Matsumoto, H. Oshiumi, T. Akazawa, A. Yamamoto, and T. Seya. 2004. The cytoplasmic “linker region” in Toll-like receptor 3 controls receptor localization and signaling. Int. Immunol. 161143-1154. [DOI] [PubMed] [Google Scholar]
- 16a.Gilfoy, F. D., and P. W. Mason. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 8111148-11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowen, B. B., J. D. Hoopes, M. H. Wong, K. H. Jung, K. C. Isakson, L. Alexopoulou, R. A. Flavell, and R. W. Sidwell. 2006. TLR3 deletion limits mortality and disease severity due to phlebovirus infection. J. Immunol. 1776301-6307. [DOI] [PubMed] [Google Scholar]
- 19.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 765532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 791343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardarson, H. S., J. S. Baker, Z. Yang, E. Purevjav, C. H. Huang, L. Alexopoulou, N. Li, R. A. Flavell, N. E. Bowles, and J. G. Vallejo. 2007. Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. Am. J. Physiol. Heart Circ. Physiol. 292H251-H258. [DOI] [PubMed] [Google Scholar]
- 22.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-κB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010237-248. [DOI] [PubMed] [Google Scholar]
- 23.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 24.Huang, J., C. Teng, and R. Shyn. 2001. Enzyme-linked immunosorbent assay using recombinant NS1 antigens for serodiagnosis of dengue virus infection, p.267. Abstr. 101st Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 25.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 26.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 7310272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konishi, E., S. Pincus, B. A. Fonseca, R. E. Shope, E. Paoletti, and P. W. Mason. 1991. Comparison of protective immunity elicited by recombinant vaccinia viruses that synthesize E or NS1 of Japanese encephalitis virus. Virology 185401-410. [DOI] [PubMed] [Google Scholar]
- 28.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 764773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Goffic, R., V. Balloy, M. Lagranderie, L. Alexopoulou, N. Escriou, R. Flavell, M. Chignard, and M. Si-Tahar. 2006. Detrimental contribution of the Toll-like receptor (TLR) 3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 1022992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libraty, D. H., P. R. Young, D. Pickering, T. P. Endy, S. Kalayanarooj, S. Green, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 1861165-1168. [DOI] [PubMed] [Google Scholar]
- 32.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 206342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 182986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 805908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. G. Griffin, R. A. Lamb, M. A. Martin, and B. Roizman (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 734611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 719608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 777804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, W. J., H. B. Chen, X. J. Wang, H. Huang, and A. A. Khromykh. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 7812225-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 802396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 791934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macdonald, J., J. Tonry, R. A. Hall, B. Williams, G. Palacios, M. S. Ashok, O. Jabado, D. Clark, R. B. Tesh, T. Briese, and W. I. Lipkin. 2005. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 7913924-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto, M., K. Funami, M. Tanabe, H. Oshiumi, M. Shingai, Y. Seto, A. Yamamoto, and T. Seya. 2003. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 1713154-3162. [DOI] [PubMed] [Google Scholar]
- 44.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 798004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 10014333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishiya, T., and A. L. DeFranco. 2004. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 27919008-19017. [DOI] [PubMed] [Google Scholar]
- 47.Nishiya, T., E. Kajita, S. Miwa, and A. L. Defranco. 2005. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J. Biol. Chem. 28037107-37117. [DOI] [PubMed] [Google Scholar]
- 48.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NF-κB- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 996322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossi, S. L., R. Fayzulin, N. Dewsbury, N. Bourne, and P. W. Mason. 2007. Mutations in West Nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 364184-195. [DOI] [PubMed] [Google Scholar]
- 51.Rossi, S. L., Q. Zhao, V. K. O'Donnell, and P. W. Mason. 2005. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology 331457-470. [DOI] [PubMed] [Google Scholar]
- 52.Scholle, F., Y. A. Girard, Q. Zhao, S. Higgs, and P. W. Mason. 2004. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J. Virol. 7811605-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholle, F., and P. W. Mason. 2005. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology 34277-87. [DOI] [PubMed] [Google Scholar]
- 54.Schulz, O., S. S. Diebold, M. Chen, T. I. Naslund, M. A. Nolte, L. Alexopoulou, Y. T. Azuma, R. A. Flavell, P. Liljestrom, and C. Reis e Sousa. 2005. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433887-892. [DOI] [PubMed] [Google Scholar]
- 55.Sejvar, J. J. 2007. The long-term outcomes of human West Nile virus infection. Clin. Infect. Dis. 441617-1624. [DOI] [PubMed] [Google Scholar]
- 56.Silva, M. C., A. Guerrero-Plata, F. D. Gilfoy, R. P. Garofalo, and P. W. Mason. 2007. Differential activation of human monocyte-derived and plasmacytoid dendritic cells by West Nile virus generated in different host cells. J. Virol. 8113640-13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stack, J., I. R. Haga, M. Schroder, N. W. Bartlett, G. Maloney, P. C. Reading, K. A. Fitzgerald, G. L. Smith, and A. G. Bowie. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 2011007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tajima, S., T. Takasaki, and I. Kurane. 2008. Characterization of Asn130-to-Ala mutant of dengue type 1 virus NS1 protein. Virus Genes 36323-329. [DOI] [PubMed] [Google Scholar]
- 59.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21335-376. [DOI] [PubMed] [Google Scholar]
- 60.Wang, J. P., P. Liu, E. Latz, D. T. Golenbock, R. W. Finberg, and D. H. Libraty. 2006. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J. Immunol. 1777114-7121. [DOI] [PubMed] [Google Scholar]
- 61.Wang, T., T. Town, L. Alexopoulou, J. F. Anderson, E. Fikrig, and R. A. Flavell. 2004. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 101366-1373. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 1696668-6672. [DOI] [PubMed] [Google Scholar]
- 63.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 171087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]